Abstract
Sugarcane (Saccharum spp.) is an important perennial grass crop for both sugar and biofuel industries. The Louisiana sugarcane breeding program is focused on improving sugar yield by incrementally increasing genetic gain. With the advancement in genotyping and (highthroughput) phenotyping techniques, genomic selection is a promising marker-assisted breeding tool. In this study, we assessed ridge regression best linear unbiased prediction (rrBLUP) and various Bayesian models to evaluate genomic prediction accuracy using a 10-fold cross validation on 95 commercial and elite parental clones from the Louisiana sugarcane breeding program. Datasets (individual and pooled in various combinations) were constructed based on soil type (light—Commerce silty loam, heavy—Sharkey clay) and crop (plant cane, ratoon). A total of 3906 SNPs were used to predict the genomic estimated breeding values (GEBVs) of the clones for sucrose content and cane and sugar yield. Prediction accuracy was estimated by both Spearman’s rank correlation and Pearson’s correlation between phenotypic breeding values and GEBVs. All traits showed significant variation with moderate (42% for sucrose content) to high (85% for cane and sugar yield) heritability. Prediction accuracy based on rank correlation was high (0.47–0.80 for sucrose content; 0.61–0.69 for cane yield, and 0.56–0.72 for sugar yield) in all cross-effect prediction models where soil and crop types were considered as fixed effects. In general, Bayesian models demonstrated a higher correlation than rrBLUP. The Pearson’s correlation without soil and crop type as fixed effects was lower with no clear pattern among the models. The results demonstrate the potential implementation of genomic prediction in the Louisiana sugarcane variety development program.
1. Introduction
Sugarcane (Saccharum spp.) is an economically important perennial grass crop grown primarily for sugar in tropical and subtropical regions of the world. Sugarcane bagasse, the fibrous remains after sugar extraction, is burnt by the sugar mills for electricity to operate the mill [1]. Sugarcane contributes 70% and 45% to the total sucrose production globally and in the United States, respectively [2]. Of the three (Louisiana, Florida, and Texas) major sugarcane producing states of the USA, Louisiana produces about 20% of the total sugar in the U.S. Sugarcane contributes approximately $3 billion to Louisiana’s economy with an annual value of over $800 million [3]. In 2021, the total cane production in Louisiana was 15.2 Mt. grown over 196,000 ha of land, which amounts to 52% of the total production in the U.S., grown on 379,200 ha [2]. In recent years, sugarcane has gained attention as an energy crop. Sugarcane produces the highest amount of biomass per unit area and is considered as a sustainable and renewable source of bioenergy that has the potential for fulfilling the growing demand for energy while simultaneously reducing greenhouse gas emissions [4]. Therefore, it is very important for the breeding programs to improve the sugar and biomass yield traits of this crop. Success in a breeding program depends upon strategies such as the appropriate selection of parents, the experimental design used, and the genetic variation and heritability of the trait(s) to be improved [5].
In sugarcane, most breeding programs focus on the improvement in the sucrose yield, ratooning ability, biotic and abiotic stress resistance, and total biomass yield for bioenergy production [6,7,8]. Phenotypic recurrent selection among clones is carried out in sugarcane breeding, but the time required from initial crossing of the parents to the release of an improved, desired commercial clone using a conventional phenotypic selection approach requires up to 12 years [9,10]. Reducing the length of the breeding cycle without significant loss in the accuracy of the breeding value of selected clones in each generation is an important goal of sugarcane breeding. Additionally, traditional phenotypic recurrent selection can be difficult and expensive, especially for difficult-to-select traits, and inaccurate for traits with low heritability [11].
Marker-assisted selection (MAS) can overcome the limitations of conventional breeding efforts, where indirect selection of the trait(s) of interest is accomplished in experimental breeding clones by using molecular markers linked to desirable traits [12]. While MAS has been successfully implemented in other crops, application in sugarcane has lagged, primarily due to its genetic complexity. Sugarcane is highly heterozygous and a complex polyploid plant. Modern sugarcane cultivars (2n = 100−140) resulted from the interspecific hybridization of S. officinarum as female (2n = 8x = 80; x = 10) and S. spontaneum as male (2n = 5x−16x = 40−128; x = 8), contributing ~80% and ~10–20% to the hybrid genome, respectively [13]. Nevertheless, quantitative trait loci (QTLs) have been identified for several traits in sugarcane [14].
The first successful application of MAS was achieved in sugarcane with the identification of markers linked to a major QTL, Bru1, for brown rust resistance [15], which have been widely used to screen sugarcane clones in various sugar industries worldwide [16,17,18,19,20,21]. However, most of the important traits in sugarcane, especially cane yield and sugar yield, are quantitatively inherited [22]. MAS fails when the trait expression is controlled by a large number of QTLs with small effects [23]. Genomic selection (GS), on the other hand, can overcome such limitations and has been efficiently used in selecting individuals for desirable traits in the early stages by estimating their breeding value using genome-wide marker information with all major and minor QTLs controlling the trait of interest [24,25,26].
The prediction models in GS allowed us to identify the best performing individuals to be used as parents in a breeding program or for next-generation advancement by using their genomic-estimated breeding values (GEBVs) based on SNPs generated by high-throughput genotyping by sequencing [27,28]. A prediction model is developed by using phenotypic and genome-wide marker data of a training population (TP) to determine the GEBVs by best linear unbiased prediction (BLUP) [29]. However, traditional statistical assumptions are violated in GS: (1) multi-collinearity where several independent variables in a model are correlated and (2) model overfitting where phenotypes of a few genotypes in the training population are regressed on thousands of markers, generated by high throughput genotyping by next-generation sequencing. These two issues can cause both the underestimation and overestimation of the marker effect (response) in prediction. Pavlou et al. [30] developed a more accurate risk prediction model by using penalized regression where the number of genotypes (n) in a training population were fewer than markers (predictors, p). Studies showed that the quantitative traits may not be affected by the genome-wide variants [31,32]. Therefore, selecting a subset of SNPs associated with the trait can reduce computational complexity and decrease prediction inaccuracy caused by overestimation. Although an identified subset of genome-wide variants can be efficiently used for genomic selection, it is difficult to overcome the p >> n (large-p small-n) problem while fitting a GS model.
Genome-wide SNP makers have been utilized for the prediction of GEBVs of parents and genotypes through GS in several crops including sugarcane [33]. Since the introduction of GS, many statistical approaches have been developed to counter the p >> n problem and improve prediction accuracy. Though a large number of statistical methods are being developed, a few of them are widely used [34]. In this study, ridge regression best linear unbiased prediction (rrBLUP) and different Bayesian approaches (Bayes A, Bayes B, Bayes C, Bayesian Lasso, Bayesian ridge regression) were used for the comparative evaluation of the prediction accuracies.
2. Material and Methods
2.1. Sugarcane Clones and Phenotypic Data
The present study comprised of 95 sugarcane clones that included cultivars, elite, and historic (foreign) clones from the Louisiana sugarcane breeding program [35,36]. The clones were planted in 3 m plots with a row spacing of 1.8 m in 2015 and 2016 in both light (Commerce silt loam) and heavy soil (Sharkey clay) at the Sugar Research Station, St. Gabriel, Louisiana. Data on traits such as sucrose content, cane and sugar yield from plant cane (2016 and 2017), and ratoon crops (1st ratoon 2017 and 2018; 2nd ratoon 2018 and 2019; and 3rd ratoon 2019) were recorded for each soil type [36]. Briefly, millable stalk counts per plot were made in August each year. These counts were used to determine the mill stalks ha−1. The trials were harvested in December of each crop year. A 6-stalk sample was hand cut and the leaves stripped off and weight per stalk (kg) was recorded. The sample was then shredded and analyzed in a Bruker NIR unit to determine the theoretical recoverable sugar (sucrose content, kg Mg−1). Cane yield (Mg ha−1) was estimated as the product of stalk population and stalk weight. Sugar yield (Mg ha−1) per hectare was calculated as the product of cane yield and sucrose content.
Phenotypic Data Analysis
Based on soil type and/or crop, the dataset was further subdivided into four subsets (light soil and heavy soil) and crop (plant cane and ratoon). Furthermore, all the subsets of data were processed by averaging the phenotypes of each clone to make unique sets. Another set of data was constructed from the original dataset by taking the average of the phenotypes of each clone irrespective of any condition. The resulting five subsets were named as plant cane, ratoon cane, light soil, heavy soil, and all combined. To develop a genomic selection model, the performance of six different statistical models was examined using the five datasets. Phenotypic data were analyzed for the correlation between years, soil type, and crop, and broad-sense heritability using JMP Pro version 14.0.0 as described by Fickett et al. [36]. Narrow-sense heritability was calculated as described by de Los Campos et al. [37].
2.2. Genotypic Data
Genotyping of the clones used for the present study has been described earlier [36]. The 6534 SNPs and InDels distributed over 10 homologous groups of sugarcane corresponding to 10 sorghum chromosomes [36] were further filtered by removing 1020 SNPs with a minor allele frequency of less than 0.1, which were considered as missing values. Missing SNPs were imputed using JMP Genomics, as described earlier [36]. Furthermore, 1608 SNPs with the same genetic information were removed. Ultimately, 3906 SNPs (Supplementary Figure S1) were used for the genomic selection models with 95 clones that were represented under most conditions.
2.3. Models Used for Genomic Prediction
To counter the regression problem associated with large p and small n, various approaches that perform shrinkage of estimates, selection of variables, or both combined are commonly used. As a result, the problem mainly focuses on the type of penalization and shrinkage procedure. In this study, the SNP effects were predicted using six different statistical models: ridge regression best linear unbiased prediction (rrBLUP) [38], Bayesian ridge regression (BRR), Bayesian least absolute shrinkage and selection operator (Bayesian Lasso) [39], Bayes A and B [24], and Bayes C [40]. Model implementation was conducted using the R programming language with different packages and functions (Supplementary Table S1).
For all approaches, a linear mixed model was used for fitting the genotypic information, in other words,
where y is the vector of different phenotypic traits; X is the full rank design matrix associated with fixed effects that includes the general mean; Z is the design matrix associated with random effects u due to the genome-wide variants (or SNPs); is the vector of residuals; and is the intercept or the general mean.
2.4. Model Efficiency
The efficiency of the prediction model was measured by calculating the Pearson’s correlation coefficient between the predicted (GEBV) and actual phenotypic values (BLUE) using the formula,
where is the Pearson’s correlation coefficient between the predicted and actual ; is the covariance between the predicted and actual phenotypes; and and are the standard deviation of the predicted and actual phenotype, respectively.
Sugarcane clones were ranked based on the phenotypes (BLUEs) and GEBVs calculated by the six models. The correlation between the ranks was calculated by the Spearman’s correlation coefficient to check the correlation between the ranks without and with the use of marker data by using the formula,
where is the Spearman’s rank correlation coefficient; is the difference between the ranks estimated by phenotype only and GEBVs of ith clone; and n is the total number of clones used for ranking.
2.5. Prediction Accuracy (without Fixed Effects)
Model predictability was also examined without incorporating any fixed component into the model. For this, total five datasets were retrieved, named, light soil, heavy soil, plant cane, ratoon, and all combined. Each dataset containing 95 genotypes and 3906 SNPs (markers) was partitioned randomly into 10 parts, of which nine parts were used for the training data for every model and the remaining one part for validation. Each random part consisted of nine or ten random genotypes. This process was iterated until all 10 parts were exhausted for the testing set for the traits to perform 10-fold cross-validation. Furthermore, the correlation between the BLUEs and GEBVs of the clones was calculated for every trait by using all datasets.
2.6. Prediction Accuracy: All vs. Individual Conditions
Another approach evaluated different datasets as the training population and the individual conditions as the testing population. For instance, in the models developed for light soil plant cane, the training population was averaged over two years of plant cane data of light soil and the testing populations were the plant cane data of the corresponding two different years of light soil. Likewise, in the model developed for heavy soil plant cane, the training population was averaged over two years of plant cane data of heavy soil and the testing populations were the plant cane data of the corresponding two different years of heavy soil. However, in the models developed for light soil ratoon and heavy soil ratoon, the training datasets were the data averaged over different years as well as different crops (i.e., first ratoon, second ratoon, and third ratoon) and the testing datasets were the data of different crop types of their respective soil type.
Similarly, in the models developed for plant cane, the training datasets were the data averaged over soil types (i.e., light soil and heavy soil) as well as different years and the testing datasets were the data of each year of plant cane in each soil type and the plant cane data of each soil type averaged over the years. In the models developed for ratoon, the training dataset was the data averaged over the soil types, plant types as well as years and the testing datasets were the data of different plant types (i.e., first ratoon, second ratoon, and third ratoon) in different soil types averaged over different years. In the models developed for light soil and heavy soil, the training datasets were the data averaged over different plant types as well as different years and the testing datasets were the data of different plant types averaged over different years in light soil and heavy soil, respectively. Finally, models were developed for all combined data where the training dataset comprised of the data averaged over the soil types, plant types as well as years, and the testing datasets were the data of different plant types in each soil type averaged over the years.
2.7. Cross-Validation: Crop Type/Soil Types as Fixed Effect
In this study, a fixed effect component due to the environment was also introduced into the models. Two different environmental conditions were soil type as light or heavy and crop type as plant cane and first ratoon. For this, three different datasets were retrieved and named as L.Pc-vs.-H.Pc, L.Pc-vs.-L.Fr, and H.Pc-vs.-H.Fr. In the L.Pc-vs.-H.Pc dataset, the average data of plant cane under light soil and average data of plant cane under heavy soil were stacked. Similarly, in L.Pc-vs.-L.Fr, the average data of plant cane under light soil and average data of the first ratoon under light soil were stacked and the same thing was undertaken for heavy soil to obtain the H.Pc-vs.-H.Fr dataset. Furthermore, all the datasets were used to develop prediction models. In the L.Pc-vs.-H.Pc dataset, soil type (i.e., light and heavy soil) was taken as a fixed effect component in the prediction model. However, in L.Pc-vs.-L.Fr and H.Pc-vs.-H.Fr, crop type (i.e., plant cane and only first ratoon) was taken as a fixed effect component in the model. As mentioned earlier, all the datasets were randomly partitioned into ten parts and in each fold of the cross-validation, nine parts were taken for training the model and the remaining one part was used for prediction.
3. Results
3.1. Phenotypic Variation
Data on sucrose content showed that N27 had the lowest value in both crop types, plant cane and ratoon cane, and for both soil types, light as well as heavy soil (Supplementary Table S2). Conversely, Ho 09-803 had the highest sucrose content in plant cane for heavy soil. The ratoon crop of L 06-001 recorded the highest value for sucrose content in light soil. For cane yield and sugar yield, L 09-107 had the highest value for plant cane in heavy soil, whereas L 81-010 was the best in ratoon cane and light soil. L 06-038 recorded the lowest values for cane yield and sugar yield in plant cane, while L CP 86-454 and HoCP 09-841 (for cane yield only) were the clones with the lowest values in ratoon cane for both soil types.
When the data were pooled and averaged over crop type and soil type, N27 had the minimum value for sucrose, whereas L 06-001 recorded the highest value for % sucrose. For cane yield and sugar yield, LCP 86-454 and L 81-010 had the lowest and highest values, respectively (Supplementary Table S2).
The correlation of traits between years were consistently moderate to high for all traits except for cane yield, where the correlation between 2016 and 2017 was exceptionally low in plant cane for heavy soil (Supplementary Table S3). In general, the correlation values were higher for all traits but cane yield in the second ratoon in light soil. On the other hand, the correlation of traits in different soil type and crop datasets were mostly low for all traits except between the first and second ratoon crops for light soil, which was high (0.67–0.68) (Supplementary Table S3).
All the traits had moderate to high broad-sense heritability (H2), ranging from 42% for sucrose in ratoon in heavy soil to 85% for cane yield and sugar yield in combined data, closely followed by 84% for cane yield and sugar yield in light soil (Supplementary Table S2). In general, H2 of the traits was higher in light soil and ratoon cane compared to heavy soil and plant cane. Heritability values of the cane yield and the sugar yield over the crop and soil type were comparable and higher. When the data were pooled over both the crop and soil types, the H2 values were generally higher with 77% for sucrose content and 85% for both the cane yield and sugar yield. Narrow-sense (h) heritability values were expectedly lower than H2 (Supplementary Table S2). For sucrose, h ranged from 24% in heavy soil to 35% in heavy soil plant cane. For both cane yield and sugar yield, h was the lowest (0.18) in heavy soil ratoon crop. Cane yield and sugar yield had the highest h values of 0.32 and 0.35, respectively, in light soil. The Shapiro–Wilk test showed that the cane yield and sugar yield data fit to a normal distribution in most datasets whereas the sucrose content displayed a skewed distribution (Supplementary Figure S2, Supplementary Table S4).
3.2. Prediction Accuracy (without Fixed Effects)
Model predictability was examined with five datasets such as light soil, heavy soil, plant cane, ratoon, and all combined without incorporating any fixed effect component into the model. Table 1 shows the mean of the correlation between the actual and predicted phenotypic values by different models calculated over 10-fold validation. Though the means of the correlation values showed negative results, the fold-wise information (Supplementary Table S5) revealed correlation values, ranging from highly positive to highly negative. In heavy soil, the performance of rrBLUP was better than the other models with correlations of 0.06, 0.16, and 0.19 for sucrose content, cane yield, and sugar yield, respectively (Table 1). However, for sugar yield and cane yield, low to moderate positive correlations were produced by all the models under study. For cane yield, BRR showed the highest correlation at 0.19 and BL showed the lowest at 0.08, while for sugar yield, Bayes-C resulted in the highest correlation (0.23) and BL showed the lowest correlation (0.16).

Table 1.
The Pearson correlation between the actual (BLUEs) and predicted values (GEBVs) using various models on different datasets.
In plant cane, BRR performed better than the rest of the models for sugar yield with a correlation of 0.11 followed by rrBLUP and Bayes-B with a correlation of 0.11 and 0.10, respectively. In the ratoon crop, all models demonstrated a positive correlation for sucrose content and sugar yield with the highest correlation by rrBLUP and BL (closely followed by Bayes-B) for cane yield, respectively (Table 1). However, for cane yield, negative correlations were obtained for all models except rrBLUP, which showed a low positive correlation of 0.04.
With the combined dataset, all models produced a negative prediction accuracy for sucrose content. However, all models had a positive correlation for sugar yield with BRR resulting in the highest correlation (0.23), followed by Bayes B with a correlation of 0.19, while rrBLUP showed the lowest correlation (0.09). On the other hand, only rrBLUP showed a low positive correlation for cane yield at 0.04 with other models, resulting in negative correlations (Table 1).
Overall, the prediction accuracy values were low or most times negative. However, fold-wise information showed that the correlation between the BLUEs and GEBVs was drastically changed based on genotypes in the testing set (Supplementary Table S5).
3.3. Prediction Accuracy with Putative Causal SNPs as Fixed Effects
The SNPs, significantly associated with sucrose content, cane yield, and sugar yield through genome-wide association mapping [36], were used as fixed effect covariates in the rrBLUP model to determine their effect on genomic prediction accuracy of the traits. The results (Table 2) showed no definitive pattern in the increase/decrease in the correlation between the actual and predicted phenotypic values. Except under heavy soil condition, there was an increase in the prediction accuracy for cane yield and sugar yield, with the highest increase from 0.04 (Table 1) to 0.27 for cane yield in ratoon cane and from 0.11 (Table 1) to 0.28 for sugar yield in plant cane. On the other hand, for sucrose content, it decreased in all conditions, except a slight increase from −0.12 (Table 2) to −0.08 for the combined data.

Table 2.
The Pearson correlation between the actual (BLUEs) and predicted values (GEBVs) using rrBLUP with sucrose content, cane yield, and sugar yield associated SNPs as the fixed effect.
3.4. Prediction Accuracy: All vs. Individual Conditions
The graphical representation of the results of the correlation between the actual and predicted phenotypic value for individual conditions as the test data by taking all combined dataset as the training data (Supplementary Table S6) is given in Figure 1.
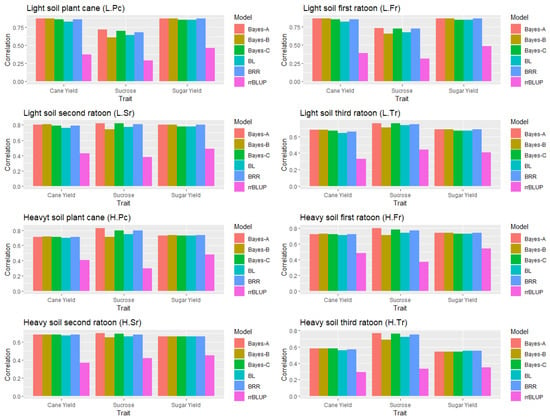
Figure 1.
Rank correlation between the actual (BLUEs) and predicted phenotypic values (GEBVs) by taking all combined dataset as the training data (no fixed effect). Light soil (averaged over crop type); plant cane (averaged over soil type); ratoon cane (averaged over soil type). L—light soil, H—heavy soil, Pc—plant cane, R—ratoon, and Fr—first ratoon.
The cross-validation results showed that the correlation between the actual and predicted phenotype was comparably very high in Bayes-A, Bayes-B, Bayes-C, and in Bayesian ridge regression irrespective of the traits studied. For cane and sugar yield, all models except for rrBLUP had comparable prediction accuracy. In contrast, rrBLUP showed the least correlation between the actual and predicted phenotypic values, ranging from 0.29 for sucrose in plant cane under light soil to 0.54 for sugar yield in the first ratoon under heavy soil. All the additional numeric results are provided in Supplementary Table S7.
3.5. Prediction Accuracy: Soil Type and/or Crop Type as Fixed Effects
With crop type as the fixed effect in the models, the rank correlation values between phenotypic BLUEs and GEBVs increased significantly in both the light and heavy soils (Table 3). Nevertheless, the correlation values were comparable under the light and heavy soil types, except for cane and sugar yield, where the values in heavy soil were significantly higher than the light soil. Sucrose had generally less prediction accuracy than cane and sugar yield for all models, whereas rrBLUP resulted in the lowest values compared to other models for all traits. For sucrose, cane yield, and sugar yield in light soil, the highest correlation values were observed in Bayes-A (0.92), Bayes-A (0.98), followed by Bayes-A (0.97), and BRR (0.95), respectively. On the other hand, rrBLUP recorded the highest correlation values for cane yield (0.98), similar to BL, and sugar yield (0.98) compared to other models.

Table 3.
The rank correlation between the phenotypic (BLUEs) and predicted values (GEBVs) using various models in different datasets.
Rank correlations between phenotypic BLUEs and GEBVs obtained from different models were higher than that without fixed effect when the data were combined over soil types (used as fixed effects) for both the plant cane and ratoon crops (Table 3). Except for the sugar yield in plant cane where a high yet comparable correlation was observed for all models, rrBLUP generated low correlation values in comparison to other models in both the plant cane and ratoon cane. In the plant cane, the highest correlation for sucrose was observed in Bayes-A (0.96); for cane yield, BL, Bayes-A, and Bayes-B had similar values at 0.97; and for sugar yield, the highest correlation was observed in Bayes-A (0.98). In the ratoon cane crop, the Bayes-A model predicted the performance for all three traits with the highest accuracy values.
When the datasets were combined across both the soil and crop types, the lowest rank correlation between the phenotypic BLUEs and GEBVs was observed for all traits in rrBLUP with values of 0.52, 0.63, and 0.71 for the sucrose, cane yield, and sugar yield, respectively (Table 3). For sucrose content, the highest correlation was observed in Bayes-B (0.93); for cane yield, Bayes-C (0.96), followed by BRR (0.95); and for sugar yield, three models (BRR, Bayes-A, and Bayes-C) produced the highest correlation at 0.95.
The results of the 10-fold cross-validation where a fixed effect component due to two different environment conditions (soil and crop type) was introduced into the models are summarized in Supplementary Table S8. Interestingly, all the models gave similar correlation values when averaged over 10-fold for all three traits in L.Pc vs. H.Pc and H.Pc vs. H.Fr. However, in L.Pc vs. L.Fr, rrBLUP showed slightly less correlation (0.47) compared to other models for sucrose only, while for other traits, all models resulted in similar prediction accuracy (Figure 2, Supplementary Table S8).
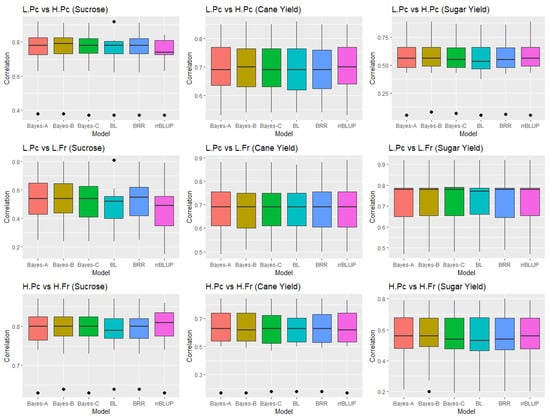
Figure 2.
Boxplots showing rank correlations between the actual and predicted phenotype in 10-fold cross-validation. L—Light soil, H—Heavy soil, Pc—Plant cane, R—Ratoon, and Fr—First ratoon.
4. Discussion
Genomic selection as a promising marker-assisted selection (MAS) tool in sugarcane is now being experimentally tested in a few sugarcane breeding programs [41,42,43,44,45]. In this study, although the population size was low relative to other published reports, our objective was to assess the prediction accuracy of six genomic selection models by cross-validating the GEBvs with actual phenotypic values for three traits, sucrose content, cane yield, and sugar yield. In addition to the overall dataset, individual datasets were constructed based on the soil type (light and heavy) and crop type (plant cane and first, second, and third ratoon). Overall, Bayesian models performed better with higher prediction accuracy than rrBLUP. Many previous studies have also demonstrated a better performance of Bayesian methods compared to rrBLUP, which assigns constant and equal variance to all markers [24,46]. For a trait controlled by a few QTLs with a larger effect, Bayesian methods outperformed the rrBLUP method [34]. Using additive genomic prediction models, Deomano et al. [41] obtained an encouraging prediction accuracy of 0.25–0.45 for sugar yield and cane yield. For the same traits that showed high heritability in this study, Bayesian methods performed better than rrBLUP for sugar yield (up to 0.23) and cane yield (up to 0.19). However, rrBLUP outperformed other models for sucrose despite a low prediction accuracy of up to 0.06 (Table 1). The negative correlation values obtained under certain conditions can be explained by the genetic distance between the K-fold genotypes in the training and testing population as well as the systematic downward biased approach using more folds (K = 10 in this study) with a small-size training/testing population [47].
The commonly used rrBLUP method estimates the effect of multiple loci with small effects, and in the process underestimates the effect of major genes [48]. Therefore, alternative approaches model large-effect QTLs segregating in bi-parental mapping populations or markers identified by genome-wide association mapping into rrBLUP as fixed effects. To test this, significant SNPs associated with sucrose content, cane yield, and sugar yield were used as the fixed effect in the rrBLUP model, which yielded mixed results. While there was significant increase in the prediction accuracy for sugar yield and cane yield in most conditions, it decreased in all conditions for the sucrose content except for a slight increase for all combined data. This is possible as the markers from the GWAS study [36] that were used as fixed effects have not yet been validated, and some markers may not have a major effect on the expression of the traits. While prediction accuracy has been reported to increase by using major effect markers as fixed effect covariates [49], no significant difference in the accuracy was observed by the addition of the covariates in corn and sorghum [50]. This further validates that the genetic architecture of a trait, the robustness of the marker effects, and partitioning of the datasets influence the prediction accuracy of a model.
Another analysis calculated the Spearman’s rank correlations in different datasets such as plant cane, ratoon cane, light soil, heavy soil, and combined. The correlations were found to be relatively high (Table 3). This can probably be explained as each genotype accounted for multiple phenotypes based on crop type (plant cane and first, second and third ratoon) or soil type (light, heavy). This led to the probability of having a genotype in both the training and testing population, thereby inflating the rank correlation (prediction accuracy). Likewise, genomic selection with the averaged dataset (over years or crop types or both) as the training dataset and individual data (crop type or year) as the testing dataset also showed a high rank correlation (Figure 1, Supplementary Figure S2) as the testing dataset was derived from the training dataset. In another study, a fixed component due to environment was also introduced into the model. The boxplot results (Figure 2, Supplementary Table S8) showed that the Bayesian models performed better than rrBLUP in most cases. On the other hand, the mean value of genomic prediction accuracy without any fixed effects was very low (Table 1). The prediction accuracy value of each fold, however, fluctuated from very high to very low (Supplementary Table S5). This could be due to various factors such as the number of markers (marker density) used, the size of the training and testing population, the population structure, and relatedness among individuals in the training and testing population.
5. Conclusions
The efficiency of the ever-evolving genomic selection models may vary with breeding populations, trait(s) of interest, genes, or markers governing their expression, etc. Prediction accuracies can be improved in clonally propagated crops such as sugarcane by including non-additive gene actions and large-effect QTLs into the GS models, which contribute significantly to the expression of complex traits in sugarcane. Despite the small size of the training/testing population and marker density, the results from this study and previously reported studies suggest that large and precise datasets will improve the prediction accuracies of the models tested in the present study, which can be effectively utilized in genomic selection as a marker-assisted breeding tool in the Louisiana sugarcane breeding program.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture12091330/s1. Figure S1. Distribution of 3906 SNPs on homeologous chromosomes of sugarcane. Figure S2. Phenotypic distribution of different traits in Sugarcane in five datasets. Table S1. R Packages and functions implemented for genomic prediction in sugarcane. Table S2. Phenotypic values and broad-sense heritability of sucrose percent, cane yield and sugar yield in different datasets. Table S3. Correlations between different combinations of year and crop type datasets. Table S4. W and p values (in parenthesis) from Shapiro-Wilks test for normality. Table S5. Fold-wise information of correlation of actual phenotype and predicted values. Table S6. Details about the training and testing population. Table S7. Rank correlation between actual and predicted value. Table S8. Fold-wise information of rank correlation between actual and predicted value.
Author Contributions
Conceptualization, N.B.; Data curation, M.P.; Formal analysis, S.S. and D.S.; Funding acquisition, S.S. and N.B.; Investigation, N.B.; Methodology, S.S.; Resources, A.H., A.R. and N.B.; Supervision, N.B.; Writing—original draft, S.S. and D.S.; Writing—review & editing, B.B., K.G., C.K., J.T., A.H. and N.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the American Sugar Cane League grant number GR-00008874 and the United States Department of Agriculture National Institute of Food Agriculture Hatch grant number 1018442.
Data Availability Statement
The data presented in this study are available in Fickett et al. (2019) and Supplementary materials here.
Acknowledgments
S.S. acknowledges the World Bank-funded NAHEP CAAST program for supporting international training at LSU and the Indian Council of Medical Research for the senior research fellowship award. The manuscript was approved for publication by the Louisiana Agricultural Experiment Station as MS# 2022-306-37265.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yadav, S.; Jackson, P.; Wei, X.; Ross, E.M.; Aitken, K.; Deomano, E.; Voss-Fels, K.P. Accelerating Genetic Gain in Sugarcane Breeding Using Genomic Selection. Agronomy 2020, 10, 585. [Google Scholar] [CrossRef]
- United States Department of Agriculture National Agricultural Statistics Service (USDA-NASS). 2021. Available online: https://nassgeodata.gmu.edu/CropScape/ (accessed on 20 July 2022).
- American Sugar Cane League, Louisiana Sugarcane Statistics. Available online: https://www.amscl.org/education/learn/ (accessed on 20 July 2022).
- Goldemberg, J.; Coelho, S.T.; Guardabassi, P. The sustainability of ethanol production from sugarcane. Energy Policy 2008, 36, 2086–2097. [Google Scholar] [CrossRef]
- Gazaffi, R.; Oliveira, K.M.; Souza, A.P.; Garcia, A.A.F. Sugarcane: Breeding Methods and Genetic Mapping. In Sugarcane Bioethanol R &D for Productivity and Sustainability; Cortez, L.A.B., Ed.; Editora Edgard Blucher Publ.: São Paulo, Brazil, 2010; pp. 333–344. [Google Scholar]
- Hale, A.L.; Veremis, J.C.; Tew, T.L.; Burner, D.M.; Legendre, B.; Dunckelman, P. 50 years of sugarcane germplasm enhancement—roadblocks, hurdles, and success. In Proceedings of the International Society of Sugar Cane Technologists 9th Sugarcane Breeding and Germplasm Workshop, Cairns, Australia, 17–21 August 2009. [Google Scholar]
- Khan, N.A.; Bedre, R.; Parco, A.; Bernaola, L.; Hale, A.; Kimbeng, C.; Baisakh, N. Identification of cold-responsive genes in energycane for their use in genetic diversity analysis and future functional marker development. Plant Sci. 2013, 211, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Rousselle, Y.; Chane, A.T.; Anglade, A.; Royaert, S.; Nibouche, S.; Costet, L. Genome wide association mapping of agro-morphological and disease resistance traits in sugarcane. Euphytica 2015, 202, 269–284. [Google Scholar] [CrossRef]
- Matsuoka, S.; Garcia, A.A.F.; Arizono, H. Melhoramento da cana-de-açúcar. Melhor. De Espécies Cultiv. 2005, 2, 205–251. [Google Scholar]
- Landell, M.D.A.; Bressiani, J.A. Melhoramento genético, caracterização e manejo varietal. Cana-De-Açúcar. Camp. Inst. Agronômico 2008, 1, 101–155. [Google Scholar]
- Haley, C.S.; Visscher, P.M. Strategies to utilize marker-quantitative trait loci associations. J. Dairy Sci. 1998, 81, 85–97. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef]
- D’Hont, A.; Ison, D.; Alix, K.; Roux, C.; Glaszmann, J.C. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 1998, 41, 221–225. [Google Scholar] [CrossRef]
- Aitken, K.S. History and development of molecular markers for sugarcane breeding. Sugar Tech 2022, 24, 341–353. [Google Scholar] [CrossRef]
- Costet, L.; Le Cunff, L.; Royaert, S.; Raboin, L.M.; Hervouet, C.; Toubi, L.; D’Hont, A. Haplotype structure around Bru1 reveals a narrow genetic basis for brown rust resistance in modern sugarcane cultivars. Theor. Appl. Genet. 2012, 125, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Glynn, N.C.; Laborde, C.; Davidson, R.W.; Irey, M.S.; Glaz, B.; D’Hont, A.; Comstock, J.C. Utilization of a major brown rust resistance gene in sugarcane breeding. Mol. Breed. 2013, 31, 323–331. [Google Scholar] [CrossRef]
- Molina, L.; Queme, J.L.; Rosales, F. Comparative analysis between phenotype and Bru1 marker for incidence to brown rust in sugarcane. In Proceedings of the International Society of Sugar Cane Technologists, Townsville, Australia, 16–18 April 2013; Volume 28, pp. 1–6. [Google Scholar]
- Racedo, J.; Perera, M.F.; Bertani, R.; Funes, C.; González, V.; Cuenya, M.I.; Castagnaro, A.P. Bru1 gene and potential alternative sources of resistance to sugarcane brown rust disease. Euphytica 2013, 191, 429–436. [Google Scholar] [CrossRef]
- Parco, A.S.; Avellaneda, M.C.; Hale, A.H.; Hoy, J.W.; Kimbeng, C.A.; Pontif, M.J.; Baisakh, N. Frequency and distribution of the brown rust resistance gene B ru1 and implications for the Louisiana sugarcane breeding programme. Plant Breed. 2014, 133, 654–659. [Google Scholar] [CrossRef]
- Neuber, A.C.; Camilo dos Santos, F.R.; da Costa, J.B.; Volpin, M.; Xavier, M.A.; Perecin, D.; Pinto, L.R. Survey of the Bru1 gene for brown rust resistance in Brazilian local and basic sugarcane germplasm. Plant Breed. 2017, 136, 182–187. [Google Scholar] [CrossRef]
- Li, W.F.; Shan, H.L.; Zhang, R.Y.; Pu, H.C.; Wang, X.Y.; Cang, X.Y.; Huang, Y.K. Identification of field resistance and molecular detection of the brown rust resistance gene Bru1 in new elite sugarcane varieties in China. Crop Prot. 2018, 103, 46–50. [Google Scholar] [CrossRef]
- Hoarau, J.Y.; Grivet, L.; Offmann, B.; Raboin, L.M.; Diorflar, J.P.; Payet, J.; Glaszmann, J.C. Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). II. Detection of QTLs for yield components. Theor. Appl. Genet. 2002, 105, 1027–1037. [Google Scholar] [CrossRef]
- Zhao, Y.; Mette, M.F.; Gowda, M.; Longin, C.F.H.; Reif, J.C. Bridging the gap between marker-assisted and genomic selection of heading time and plant height in hybrid wheat. Heredity 2014, 112, 638–645. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Lorenz, A.J.; Chao, S.; Asoro, F.G.; Heffner, E.L.; Hayashi, T.; Iwata, H.; Jannink, J.L. Genomic selection in plant breeding: Knowledge and prospects. Adv. Agron. 2011, 110, 77–123. [Google Scholar]
- Gouy, M.; Rousselle, Y.; Bastianelli, D.; Lecomte, P.; Bonnal, L.; Roques, D.; Costet, L. Experimental assessment of the accuracy of genomic selection in sugarcane. Theor. Appl. Genet. 2013, 126, 2575–2586. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Hu, Z.; Xu, C. Genomic selection methods for crop improvement: Current status and prospects. Crop J. 2018, 6, 330–340. [Google Scholar] [CrossRef]
- Zeng, J.; Garrick, D.; Dekkers, J.; Fernando, R. A nested mixture model for genomic prediction using whole-genome SNP genotypes. PLoS ONE 2018, 13, e0194683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, C.R. Applications of Linear Models in Animal Breeding; University of Guelph: Guelph, ON, Canada, 1984; p. 462. [Google Scholar]
- Pavlou, M.; Ambler, G.; Seaman, S.R.; Guttmann, O.; Elliott, P.; King, M.; Omar, R.Z. How to develop a more accurate risk prediction model when there are few events. Br. Med. J. 2015, 351, h3868. [Google Scholar] [CrossRef] [PubMed]
- Boutorh, A.; Guessoum, A. Complex diseases SNP selection and classification by hybrid association rule mining and artificial neural network—Based evolutionary algorithms. Eng. Appl. Artif. Intell. 2016, 51, 58–70. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, J.Y.; Ni, Y.L.; Wen, Y.J.; Niu, Y.; Tamba, C.L.; Zhang, Y.M. pLARmEB: Integration of least angle regression with empirical Bayes for multilocus genome-wide association studies. Heredity 2017, 118, 517. [Google Scholar] [CrossRef]
- Mahadevaiah, C.; Appunu, C.; Aitken, K.; Suresha, G.S.; Vignesh, P.; Swamy, H.K.M.; Ram, B. Genomic Selection in Sugarcane: Current Status and Future Prospects. Front. Plant Sci. 2021, 12, 708233. [Google Scholar] [CrossRef]
- Meher, P.K.; Kumar, A.; Pradhan, S.K. Genomic Selection Using Bayesian Methods: Models, Software, and Application. In Genomics of Cereal Crops; Humana: New York, NY, USA, 2022; pp. 259–269. [Google Scholar]
- Avellaneda, M.C.; Parco, A.P.; Hoy, J.W.; Baisakh, N. Putative resistance-associated genes induced in sugarcane in response to the brown rust fungus, Puccinia melanocephala and their use in genetic diversity analysis of Louisiana sugarcane clones. Plant Gene 2018, 14, 20–28. [Google Scholar] [CrossRef]
- Fickett, N.; Gutierrez, A.; Verma, M.; Pontif, M.; Hale, A.; Kimbeng, C.; Baisakh, N. Genome-wide association mapping identifies markers associated with cane yield components and sucrose traits in the Louisiana sugarcane core collection. Genomics 2019, 111, 1794–1801. [Google Scholar] [CrossRef]
- de Los Campos, G.; Sorensen, D.; Gianola, D. Genomic heritability: What is it? PLoS Genet. 2015, 11, e1005048. [Google Scholar] [CrossRef]
- Hoerl, A.E.; Kennard, R.W. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics 1970, 12, 55–67. [Google Scholar] [CrossRef]
- Park, T.; Casella, G. The bayesian lasso. J. Am. Stat. Assoc. 2008, 103, 681–686. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the Bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef] [Green Version]
- Deomano, E.; Jackson, P.; Wei, X.; Aitken, K.; Kota, R.; Pérez-Rodríguez, P. Genomic prediction of sugar content and cane yield in sugar cane clones in different stages of selection in a breeding program, with and without pedigree information. Mol. Breed. 2020, 40, 38. [Google Scholar] [CrossRef]
- Hayes, B.J.; Wei, X.; Joyce, P.; Atkin, F.; Deomano, E.; Yue, J.; Voss-Fels, K.P. Accuracy of genomic prediction of complex traits in sugarcane. Theor. Appl. Genet. 2021, 134, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; McCord, P.H.; Olatoye, M.O.; Qin, L.; Sood, S.; Lipka, A.E.; Todd, J.R. Experimental evaluation of genomic selection prediction for rust resistance in sugarcane. Plant Genome 2021, 14, e20148. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Wei, X.; Ross, E.M.; Frisch, M.; Aitken, K.S.; Cooper, M.; Hayes, B.J. Strategies and considerations for implementing genomic selection to improve traits with additive and non-additive genetic architectures in sugarcane breeding. Theor. Appl. Genet. 2021, 134, 1493–1511. [Google Scholar] [CrossRef]
- Yadav, S.; Wei, X.; Joyce, P.; Atkin, F.; Deomano, E.; Sun, Y.; Voss-Fels, K.P. Improved genomic prediction of clonal performance in sugarcane by exploiting non-additive genetic effects. Theor. Appl. Genet. 2021, 134, 2235–2252. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Dekkers, J. The impact of genetic relationship information on genome-assisted breeding values. Genetics 2007, 177, 2389–2397. [Google Scholar] [CrossRef]
- Zhou, Y.; Vales, M.I.; Wang, A.; Zhang, Z. Systematic bias of correlation coefficient may explain negative accuracy of genomic prediction. Brief. Bioinform. 2017, 18, 744–753. [Google Scholar] [CrossRef]
- Bernardo, R. Genomewide selection when major genes are known. Crop Sci. 2014, 54, 68–75. [Google Scholar] [CrossRef]
- Sarinelli, J.M.; Murphy, J.P.; Tyagi, P.; Holland, J.B.; Johnson, J.W.; Mergoum, M.; Brown-Guedira, G. Training population selection and use of fixed effects to optimize genomic predictions in a historical USA winter wheat panel. Theor. Appl. Genet. 2019, 132, 1247–1261. [Google Scholar] [CrossRef] [PubMed]
- Rice, B.; Lipka, A.E. Evaluation of RR-BLUP genomic selection models that incorporate peak genome-wide association study signals in maize and sorghum. Plant Genome 2019, 12, 180052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).