Identification of the Citrus Superoxide Dismutase Family and Their Roles in Response to Phytohormones and Citrus Bacterial Canker
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant and Bacterial Materials
2.2. Identification of SOD Genes in the Sweet Orange Genome
2.3. In Silico Characterization of CsSODs
2.4. Xcc and Phytohormone Treatments
2.5. RNA Extraction and qRT-PCR Analysis
2.6. Transient Expression of CsSOD06 and CsSOD08 in Citrus
2.7. Biochemical Indices
2.8. Statistical Analysis
3. Results
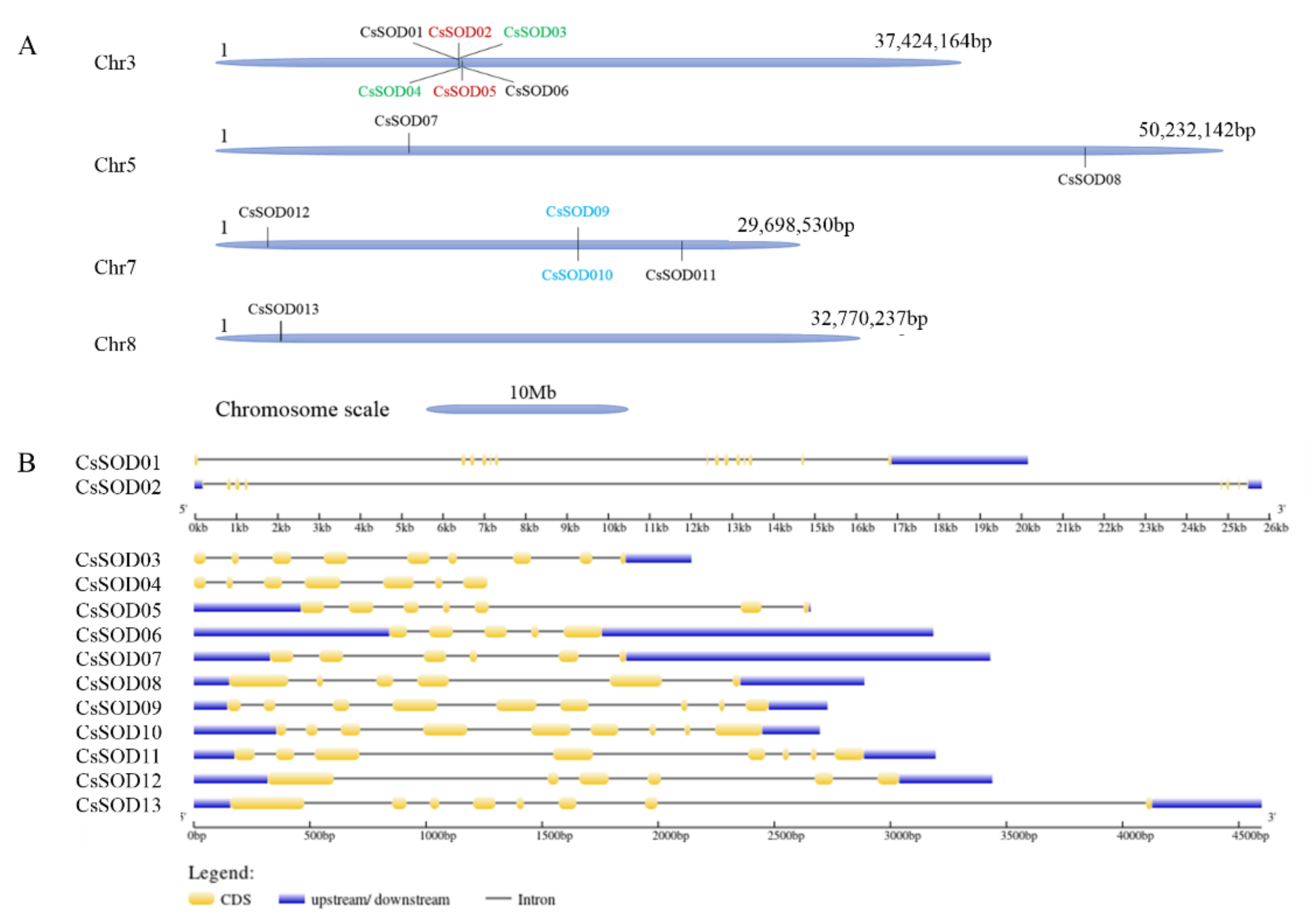
3.1. Genome-Wide Distribution of the Citrus SOD Gene Family
3.2. Chromosomal Distribution and Intron/Exon Configurations of CsSOD Genes
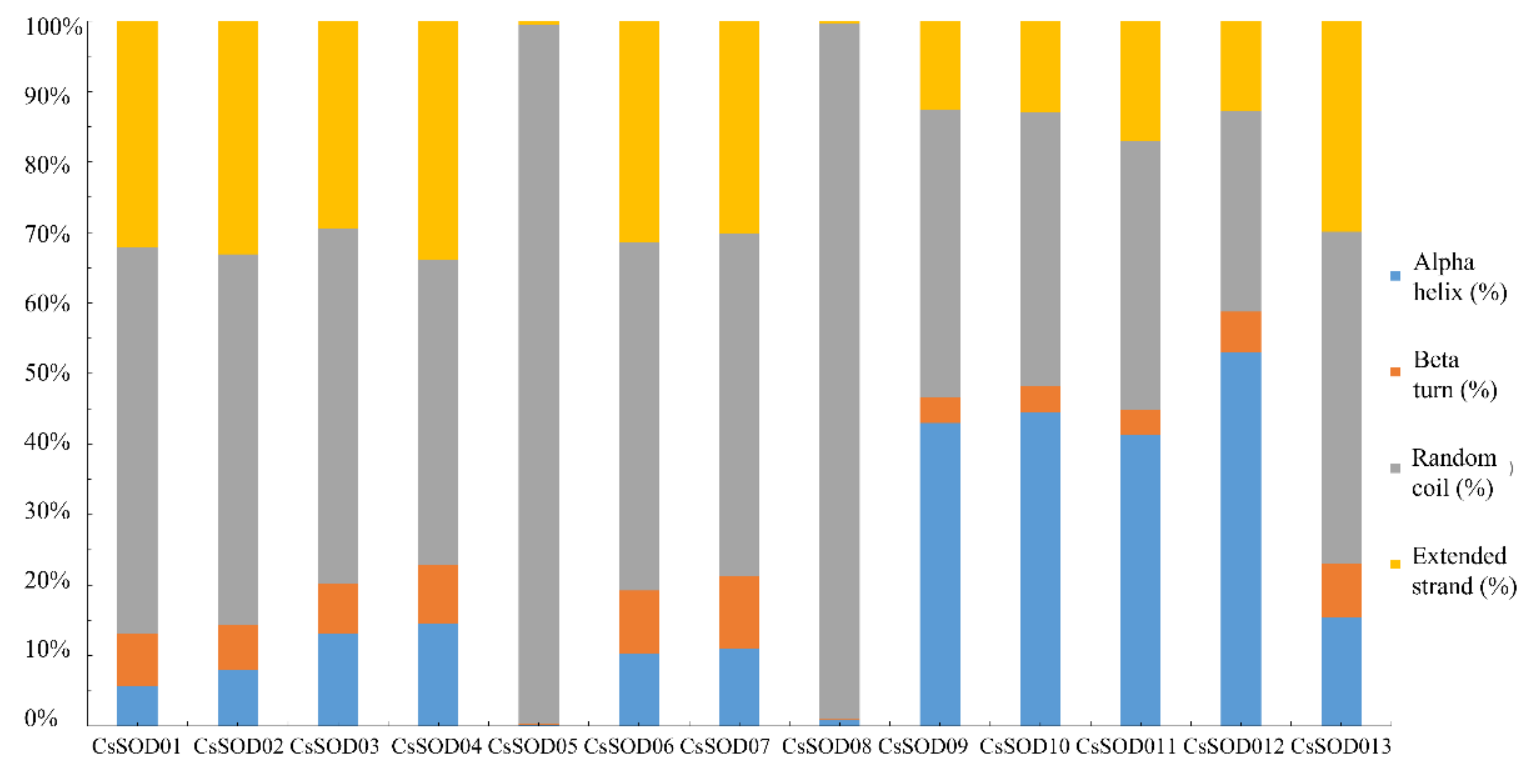
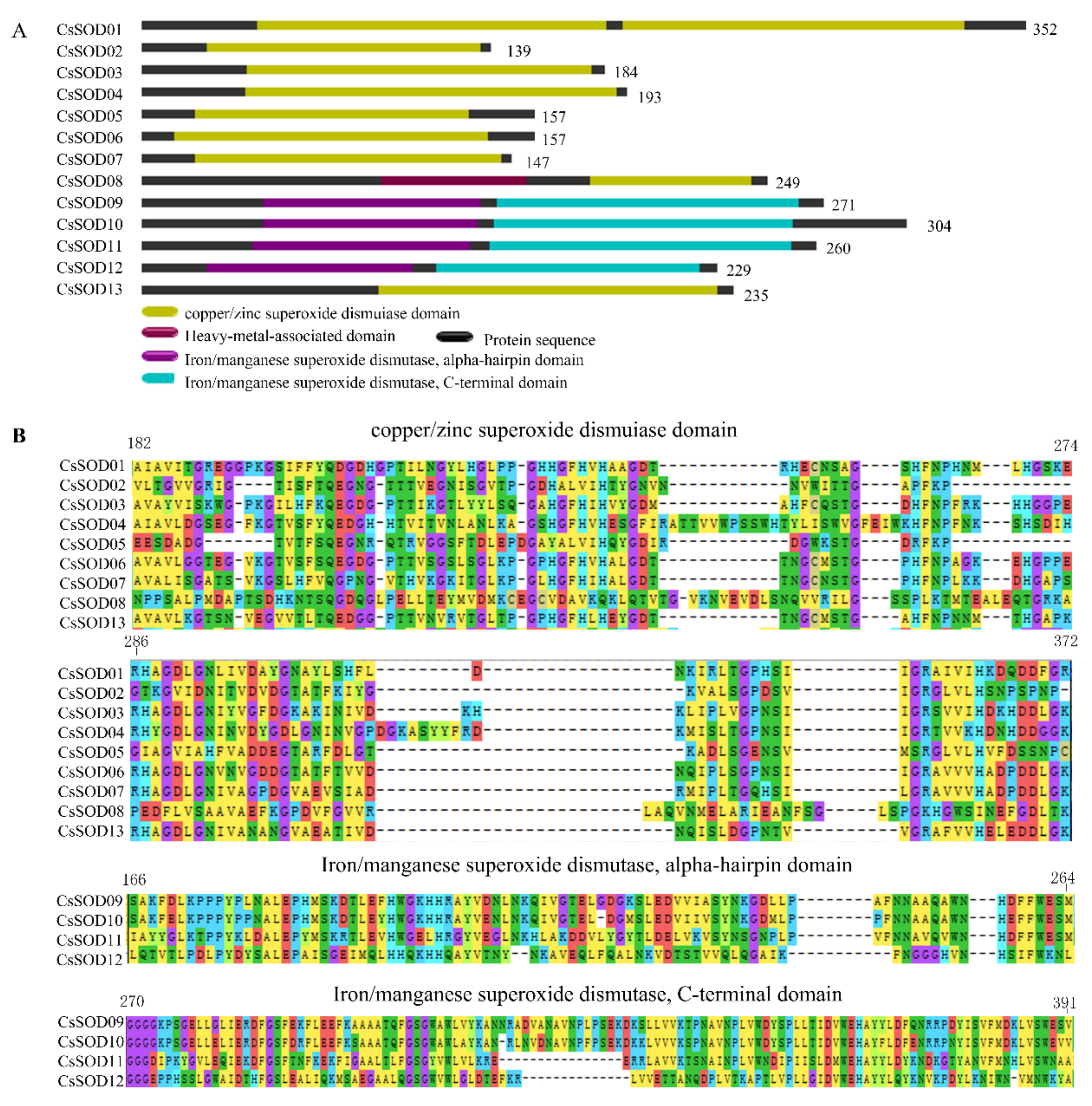
3.3. Functional Domains of CsSODs
3.4. Phylogenetic and Collinearity Analyses of the CsSOD Family in Arabidopsis and Citrus
3.5. Conserved Motif Analysis of CsSOD Family Proteins
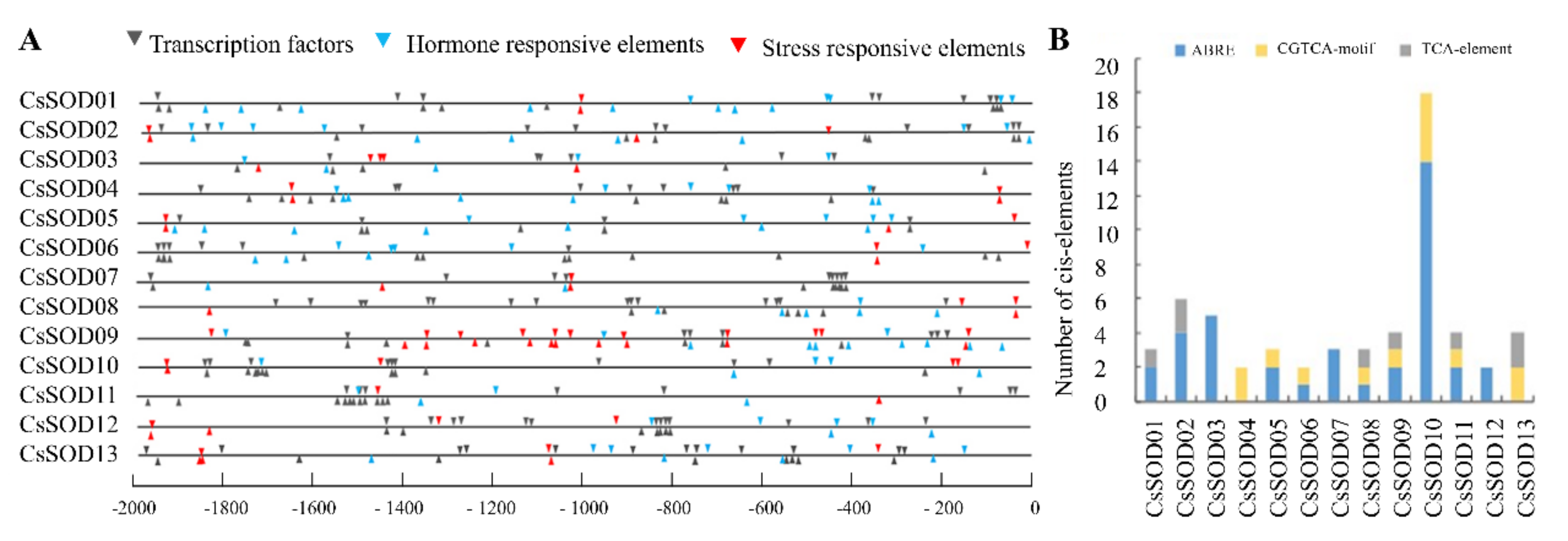
3.6. Analysis of Cis-Elements and Transcription Factor Binding Sites in Putative CsSOD Gene Promoters
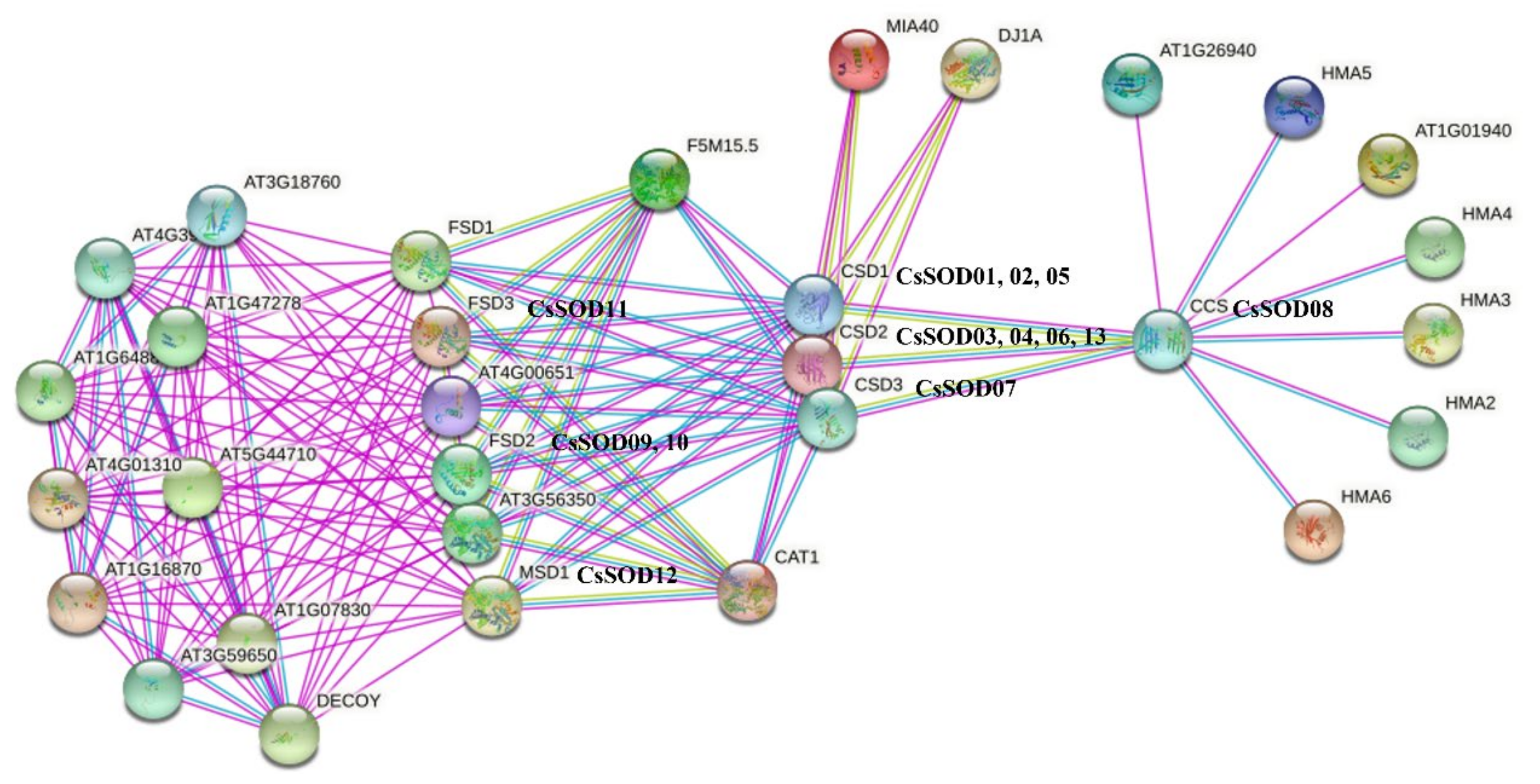
3.7. Predicted Protein–Protein Interaction (PPI) Network of CsSODs
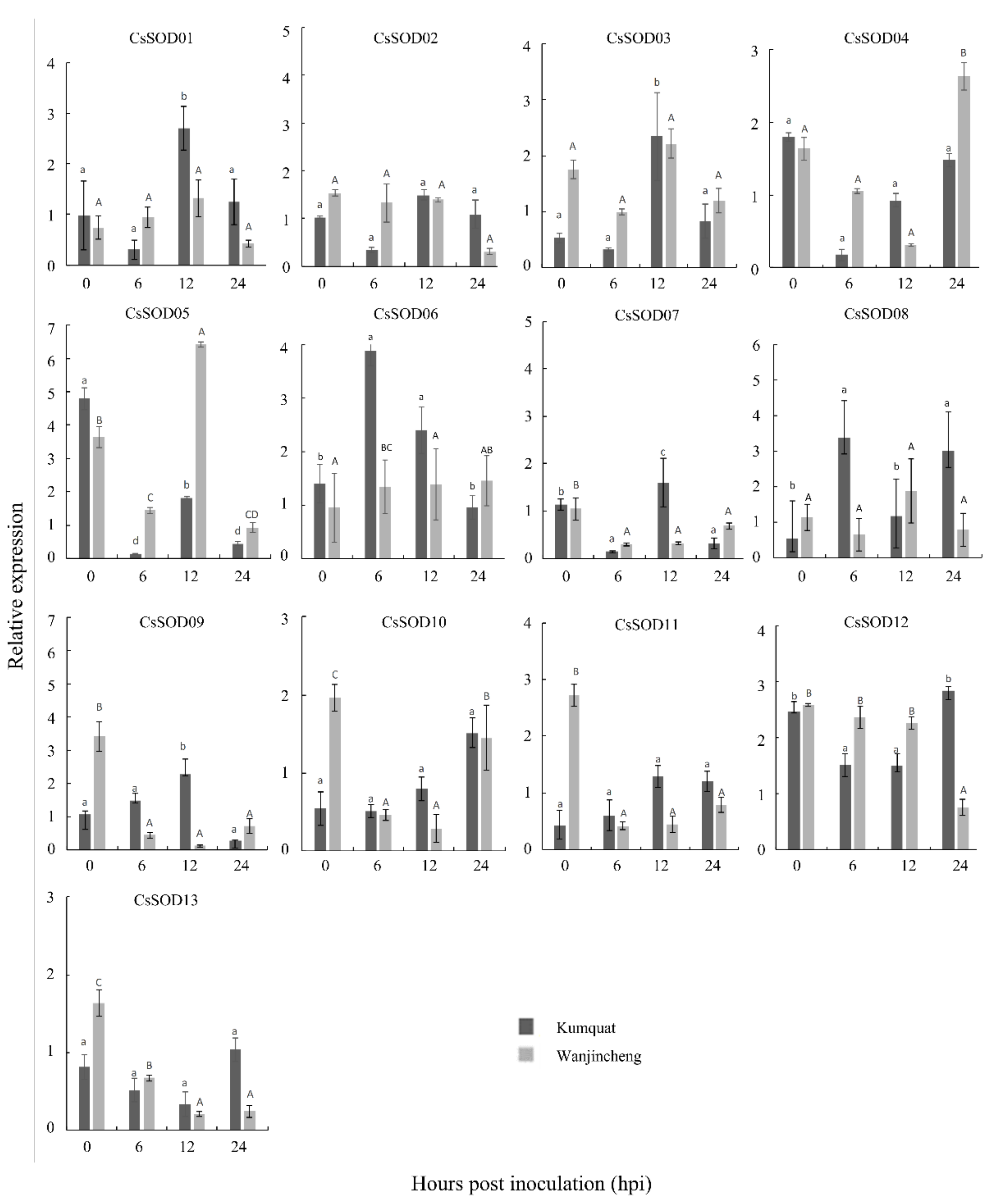
3.8. Expression Patterns of CsSODs Induced by Xcc
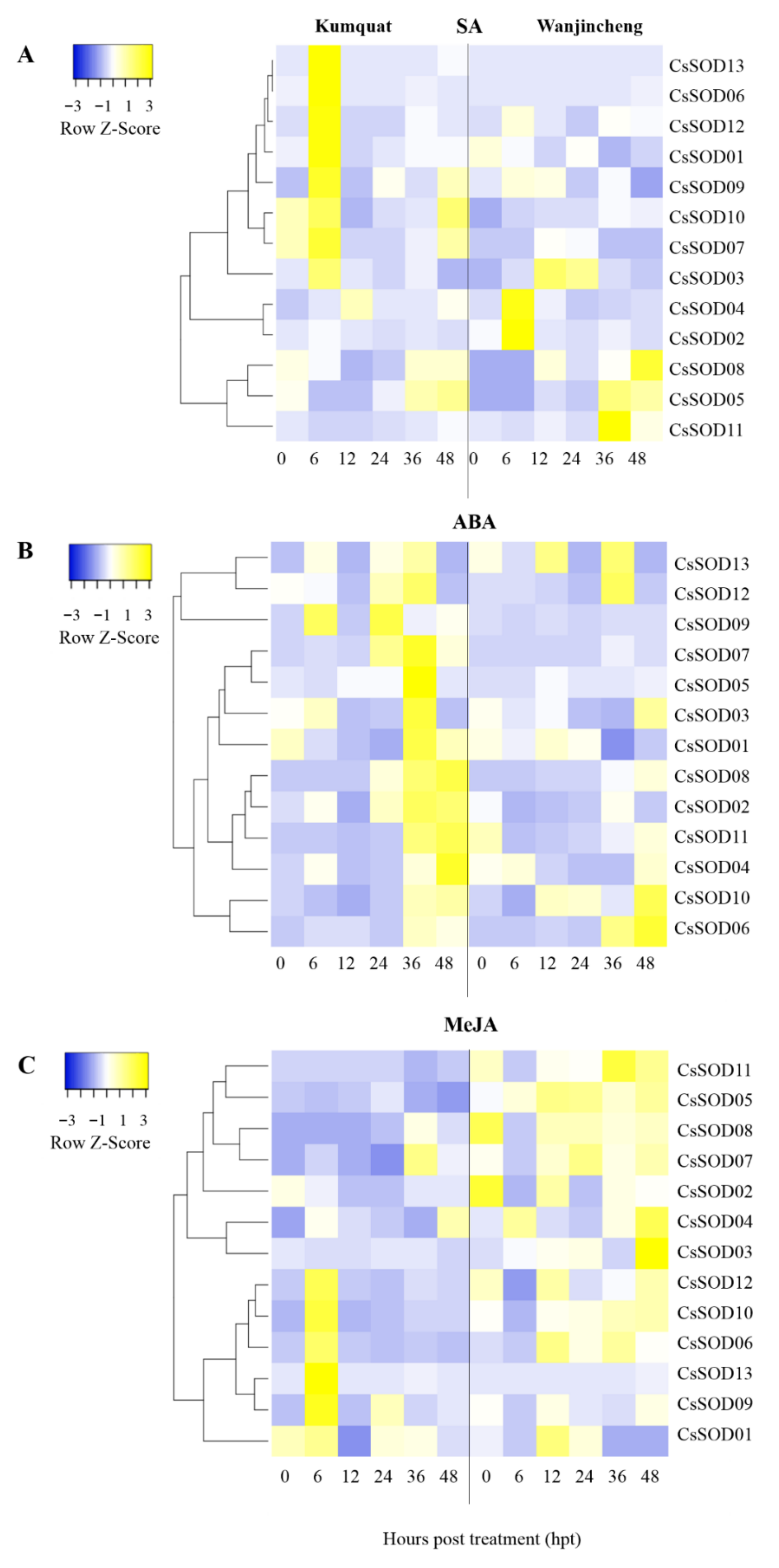
3.9. CsSOD Expression Pattern Induced by Phytohormones
3.10. Transient Expression of CsSOD06 and CsSOD08
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Snowdon, R.J.; Wittkop, B.; Chen, T.W.; Stahl, A. Crop adaptation to climate change as a consequence of long-term breeding. Theor. Appl. Genet. 2021, 134, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Song, H.; Zhang, B.; Lu, Q.; Liu, Z.; Zhang, S.; Guo, R.; Wang, C.; Zhao, Z.; Liu, J.; et al. Genome-wide identification, characterization, and expression analysis of superoxide dismutase (SOD) genes in foxtail millet (Setaria italica L.). 3 Biotech. 2018, 8, 486. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Yu, J.; Cheng, Y.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Li, Z.; Yao, Z.; Yang, Y.; et al. The SOD Gene Family in Tomato: Identification, Phylogenetic Relationships, and Expression Patterns. Front. Plant Sci. 2016, 7, 1279. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Chen, J.; Li, R.; Shang, S.; Tang, X. Genome-wide analysis of the superoxide dismutase (SOD) gene family in Zostera marina and expression profile analysis under temperature stress. PeerJ 2020, 8, e9063. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Deng, F.; Yuan, R.; Shen, F. Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genom. 2017, 18, 376. [Google Scholar] [CrossRef]
- Li, Z.; Han, X.; Song, X.; Zhang, Y.; Jiang, J.; Han, Q.; Liu, M.; Qiao, G.; Zhuo, R. Overexpressing the Sedum alfredii Cu/Zn Superoxide Dismutase Increased Resistance to Oxidative Stress in Transgenic Arabidopsis. Front. Plant Sci. 2017, 8, 1010. [Google Scholar] [CrossRef]
- Huo, C.; He, L.; Yu, T.; Ji, X.; Li, R.; Zhu, S.; Zhang, F.; Xie, H.; Liu, W. The Superoxide Dismutase Gene Family in Nicotiana tabacum: Genome-Wide Identification, Characterization, Expression Profiling and Functional Analysis in Response to Heavy Metal Stress. Front. Plant Sci. 2022, 13, 904105. [Google Scholar] [CrossRef]
- Melicher, P.; Dvorak, P.; Krasylenko, Y.; Shapiguzov, A.; Kangasjarvi, J.; Samaj, J.; Takac, T. Arabidopsis Iron Superoxide Dismutase FSD1 Protects Against Methyl Viologen-Induced Oxidative Stress in a Copper-Dependent Manner. Front. Plant Sci. 2022, 13, 823561. [Google Scholar] [CrossRef]
- Dvorak, P.; Krasylenko, Y.; Ovecka, M.; Basheer, J.; Zapletalova, V.; Samaj, J.; Takac, T. FSD1: Developmentally-regulated plastidial, nuclear and cytoplasmic enzyme with anti-oxidative and osmoprotective role. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Lu, X.; Guan, Q.; Zhu, J. Downregulation of CSD2 by a heat-inducible miR398 is required for thermotolerance in Arabidopsis. Plant Signal. Behav. 2013, 8, e24952. [Google Scholar] [CrossRef][Green Version]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Gallie, D.R.; Chen, Z. Chloroplast-localized iron superoxide dismutases FSD2 and FSD3 are functionally distinct in Arabidopsis. PLoS ONE 2019, 14, e0220078. [Google Scholar] [CrossRef]
- Wang, F.Z.; Wang, Q.B.; Kwon, S.Y.; Kwak, S.S.; Su, W.A. Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. J. Plant Physiol. 2005, 162, 465–472. [Google Scholar] [CrossRef]
- Shafi, A.; Gill, T.; Sreenivasulu, Y.; Kumar, S.; Ahuja, P.S.; Singh, A.K. Improved callus induction, shoot regeneration, and salt stress tolerance in Arabidopsis overexpressing superoxide dismutase from Potentilla atrosanguinea. Protoplasma 2015, 252, 41–51. [Google Scholar] [CrossRef]
- Wang, Y.C.; Qu, G.Z.; Li, H.Y.; Wu, Y.J.; Wang, C.; Liu, G.F.; Yang, C.P. Enhanced salt tolerance of transgenic poplar plants expressing a manganese superoxide dismutase from Tamarix androssowii. Mol. Biol. Rep. 2010, 37, 1119–1124. [Google Scholar] [CrossRef]
- Lin, K.H.; Sei, S.C.; Su, Y.H.; Chiang, C.M. Overexpression of the Arabidopsis and winter squash superoxide dismutase genes enhances chilling tolerance via ABA-sensitive transcriptional regulation in transgenic Arabidopsis. Plant Signal. Behav. 2019, 14, 1685728. [Google Scholar] [CrossRef]
- Li, Y.; Cao, X.L.; Zhu, Y.; Yang, X.M.; Zhang, K.N.; Xiao, Z.Y.; Wang, H.; Zhao, J.H.; Zhang, L.L.; Li, G.B.; et al. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 2019, 222, 1507–1522. [Google Scholar] [CrossRef]
- Cabrejos, D.A.L.; Alexandrino, A.V.; Pereira, C.M.; Mendonca, D.C.; Pereira, H.D.; Novo-Mansur, M.T.M.; Garratt, R.C.; Goto, L.S. Structural characterization of a pathogenicity-related superoxide dismutase codified by a probably essential gene in Xanthomonas citri subsp. citri. PLoS ONE 2019, 14, e0209988. [Google Scholar] [CrossRef]
- Diaz-Vivancos, P.; Rubio, M.; Mesonero, V.; Periago, P.M.; Barcelo, A.R.; Martinez-Gomez, P.; Hernandez, J.A. The apoplastic antioxidant system in Prunus: Response to long-term plum pox virus infection. J. Exp. Bot. 2006, 57, 3813–3824. [Google Scholar] [CrossRef]
- Fan, W.W.; Wang, L.A.; Chun, H.M.; Wen, W.D.; Yun, C.L.; Zi, H.L.; Yin, S.J.; Geng, J.Y.; Zhang, X.Y. The Influence of the Verticillium dahliae Kleb Infection on the Anti-Enzyme Inside the Body of the Cotton with Different Root Injured Degree. Agric. Sci. China 2007, 6, 816–824. [Google Scholar] [CrossRef]
- Li, T.; Huang, M.C.; Zhang, D.D.; Ran, L.; Chen, J.Y.; Sun, W.X.; Qiu, N.W.; Dai, X.F. Extracellular superoxide dismutase VdSOD5 is required for virulence in Verticillium dahliae. J. Integr. Agric. 2021, 20, 1858–1870. [Google Scholar]
- Faize, M.; Faize, L.; Alburquerque, N.; Venisse, J.S.; Burgos, L. Hydrogen peroxide generated by over-expression of cytosolic superoxide dismutase in transgenic plums enhances bacterial canker resistance and modulates plant defence responses. Mol. Biol. Rep. 2020, 47, 5889–5901. [Google Scholar] [CrossRef]
- Coaker, G.L.; Willard, B.; Kinter, M.; Stockinger, E.J.; Francis, D.M. Proteomic analysis of resistance mediated by Rcm 2.0 and Rcm 5.1, two loci controlling resistance to bacterial canker of tomato. Mol. Plant-Microbe Interact. 2004, 17, 1019–1028. [Google Scholar] [CrossRef]
- Sun, L.; Ke, F.; Nie, Z.; Wang, P.; Xu, J. Citrus Genetic Engineering for Disease Resistance: Past, Present and Future. Int. J. Mol. Sci. 2019, 20, 5256. [Google Scholar] [CrossRef]
- Zhang, J.; Huguet-Tapia, J.C.; Hu, Y.; Jones, J.; Wang, N.; Liu, S.; White, F.F. Homologues of CsLOB1 in citrus function as disease susceptibility genes in citrus canker. Mol. Plant Pathol. 2017, 18, 798–810. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, L.L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.B.; Hao, B.H.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Wang, J.; Chen, D.; Lei, Y.; Chang, J.W.; Hao, B.H.; Xing, F.; Li, S.; Xu, Q.; Deng, X.X.; Chen, L.L. Citrus sinensis annotation project (CAP): A comprehensive database for sweet orange genome. PLoS ONE 2014, 9, e87723. [Google Scholar] [CrossRef]
- Reiser, L.; Subramaniam, S.; Li, D.; Huala, E. Using the Arabidopsis Information Resource (TAIR) to Find Information About Arabidopsis Genes. Curr. Protoc. Bioinform. 2017, 60, 1.11.1–1.11.45. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Berhanu Lemma, R.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Perez, N.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Tian, S.; Wang, X.; Li, P.; Wang, H.; Ji, H.; Xie, J.; Qiu, Q.; Shen, D.; Dong, H. Plant Aquaporin AtPIP1;4 Links Apoplastic H2O2 Induction to Disease Immunity Pathways. Plant Physiol. 2016, 171, 1635–1650. [Google Scholar] [CrossRef]
- Ren, X.; Wang, M.; Wang, Y.; Huang, A. Superoxide anion generation response to wound in Arabidopsis hypocotyl cutting. Plant Signal. Behav. 2021, 16, 1848086. [Google Scholar] [CrossRef]
- Feng, X.; Lai, Z.; Lin, Y.; Lai, G.; Lian, C. Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genom. 2015, 16, 823. [Google Scholar] [CrossRef]
- Gomez, J.M.; Hernandez, J.A.; Jimenez, A.; del Rio, L.A.; Sevilla, F. Differential response of antioxidative enzymes of chloroplasts and mitochondria to long-term NaCl stress of pea plants. Free. Radic. Res. 1999, 31 (Suppl. S1), S11–S18. [Google Scholar] [CrossRef]
- Huang, H.; Wang, H.; Tong, Y.; Wang, Y. Insights into the Superoxide Dismutase Gene Family and Its Roles in Dendrobium catenatum under Abiotic Stresses. Plants 2020, 9, 1452. [Google Scholar] [CrossRef]
- Parker, M.W.; Blake, C.C. Iron-and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 1988, 229, 377–382. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, X.; Wu, Y.; Wang, Y.; He, Y.; Wu, Y. Directly transforming PCR-amplified DNA fragments into plant cells is a versatile system that facilitates the transient expression assay. PLoS ONE 2013, 8, e57171. [Google Scholar] [CrossRef]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genom. 2019, 20, 227. [Google Scholar] [CrossRef]
- Wu, G.; Wilen, R.W.; Robertson, A.J.; Gusta, L.V. Isolation, chromosomal localization, and differential expression of mitochondrial manganese superoxide dismutase and chloroplastic copper/zinc superoxide dismutase genes in wheat. Plant Physiol. 1999, 120, 513–520. [Google Scholar] [CrossRef]
- Baek, K.H.; Skinner, D.Z. Alteration of antioxidant enzyme gene expression during cold acclimation of near-isogenic wheat lines. Plant Sci. 2003, 165, 1221–1227. [Google Scholar] [CrossRef]
- Gupta, A.S.; Heinen, J.L.; Holaday, A.S.; Burke, J.J.; Allen, R.D. Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA 1993, 90, 1629–1633. [Google Scholar] [CrossRef]
- Xuan, H.M.; Shi, H.Z.; Guo, Q.S.; Zhang, H.N.; Lei, M.; Wang, C.L. Effects of light intensity on physio-biochemical characteristics of Chrysanthemum indicum. Zhongguo Zhong Yao Za Zhi 2020, 45, 1620–1626. [Google Scholar]
- Wang, S.; Bai, Y.; Shen, C.; Wu, Y.; Zhang, S.; Jiang, D.; Guilfoyle, T.J.; Chen, M.; Qi, Y. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef]
- Giarola, V.; Chen, P.; Dulitz, S.J.; Konig, M.; Manduzio, S.; Bartels, D. The dehydration- and ABA-inducible germin-like protein CpGLP1 from Craterostigma plantagineum has SOD activity and may contribute to cell wall integrity during desiccation. Planta 2020, 252, 84. [Google Scholar] [CrossRef]
- Liu, C.; Wang, H.; Zhang, X.; Ma, F.; Guo, T.; Li, C. Activation of the ABA Signal Pathway Mediated by GABA Improves the Drought Resistance of Apple Seedlings. Int. J. Mol. Sci. 2021, 22, 12676. [Google Scholar] [CrossRef]
- Kurepa, J.; Herouart, D.; Van Montagu, M.; Inze, D. Differential expression of CuZn- and Fe-superoxide dismutase genes of tobacco during development, oxidative stress, and hormonal treatments. Plant Cell Physiol. 1997, 38, 463–470. [Google Scholar] [CrossRef]
- Yang, J.W.; Wang, K.C.; Liang, J.Y.; Wang, J.; Xia, T.S.; Liang, Y.F. Effects of exogenous MeJA, SA and two kinds of endophytic fungi on physiology and total phenols content of seedlings of Bletilla striata. Zhongguo Zhong Yao Za Zhi 2016, 41, 2794–2801. [Google Scholar]
- Fink, R.C.; Scandalios, J.G. Molecular evolution and structure–function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 2002, 399, 19–36. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhou, L.; Liu, J.; Cao, Z.; Du, X.; Huang, F.; Pan, G.; Cheng, F. Involvement of CAT in the detoxification of HT-induced ROS burst in rice anther and its relation to pollen fertility. Plant Cell Rep. 2018, 37, 741–757. [Google Scholar] [CrossRef] [PubMed]
- Qiao, D.; Zhang, Y.; Xiong, X.; Li, M.; Cai, K.; Luo, H.; Zeng, B. Transcriptome analysis on responses of orchardgrass (Dactylis glomerata L.) leaves to a short term flooding. Hereditas 2020, 157, 20. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kang, Z.; Zhu, K.; Zhao, D.; Yuan, Y.; Yang, S.; Zhen, W.; Hu, X. RBOH1-dependent H2O2 mediates spermine-induced antioxidant enzyme system to enhance tomato seedling tolerance to salinity-alkalinity stress. Plant Physiol. Biochem. 2021, 164, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.Q.; Jung, S. Modulation of chloroplast components and defense responses during programmed cell death in tobacco infected with Pseudomonas syringae. Biochem. Biophys. Res. Commun. 2020, 528, 753–759. [Google Scholar] [CrossRef]
- Kyu, S.Y.; Naing, A.H.; Pe, P.P.W.; Park, K.I.; Kim, C.K. Tomato seeds pretreated with Antifreeze protein type I (AFP I) promotes the germination under cold stress by regulating the genes involved in germination process. Plant Signal. Behav. 2019, 14, 1682796. [Google Scholar] [CrossRef]
- Sandalio, L.M.; Rodriguez-Serrano, M.; Romero-Puertas, M.C.; del Rio, L.A. Role of peroxisomes as a source of reactive oxygen species (ROS) signaling molecules. Subcell. Biochem. 2013, 69, 231–255. [Google Scholar]
- Guan, B.; Lin, Z.; Liu, D.; Li, C.; Zhou, Z.; Mei, F.; Li, J.; Deng, X. Effect of Waterlogging-Induced Autophagy on Programmed Cell Death in Arabidopsis Roots. Front. Plant Sci. 2019, 10, 468. [Google Scholar] [CrossRef]



| Name | CAP ID | Size (aa) | Molecular Weight (Da) | Isoelectric Point (PI) | Grand Average of Hydropathicity | Aliphatic Index | Instability Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| CsSOD01 | Cs_ont_3g017650.1 | 352 | 37,912.86 | 6.92 | −0.572 | 69.80 | 28.48 | Extracellular |
| CsSOD02 | Cs_ont_3g017660.1 | 139 | 14,150.98 | 6.24 | 0.150 | 93.81 | 10.49 | Extracellular/Cytoplasmic |
| CsSOD03 | Cs_ont_3g017710.1 | 183 | 19,859.86 | 9.42 | −0.163 | 88.42 | 22.20 | Extracellular/Cytoplasmic |
| CsSOD04 | Cs_ont_3g017720.1 | 192 | 21,355.90 | 6.72 | −0.320 | 79.64 | 26.48 | Extracellular |
| CsSOD05 | Cs_ont_3g017760.1 | 156 | 16,609.34 | 4.63 | −0.251 | 79.36 | 18.03 | Cytoplasmic |
| CsSOD06 | Cs_ont_3g017770.1 | 156 | 16,150.05 | 5.50 | −0.128 | 77.37 | 16.70 | Cytoplasmic |
| CsSOD07 | Cs_ont_5g014800.1 | 146 | 14,885.84 | 6.78 | −0.112 | 90.82 | 3.40 | Cytoplasmic |
| CsSOD08 | Cs_ont_5g041000.1 | 248 | 26,520.07 | 7.76 | −0.150 | 79.80 | 36.60 | Chloroplast |
| CsSOD09 | Cs_ont_7g013920.1 | 270 | 30,095.09 | 8.57 | −0.439 | 74.52 | 36.92 | Chloroplast |
| CsSOD10 | Cs_ont_7g013930.1 | 303 | 34,616.86 | 5.22 | −0.674 | 67.29 | 47.65 | Cytoplasmic/Chloroplast |
| CsSOD11 | Cs_ont_7g019080.1 | 259 | 29,467.85 | 8.66 | −0.275 | 85.83 | 31.53 | Mitochondrial/Chloroplast |
| CsSOD12 | Cs_ont_7g002980.1 | 228 | 25,288.88 | 6.79 | −0.212 | 92.81 | 36.56 | Mitochondrial |
| CsSOD13 | Cs_ont_8g006180.1 | 234 | 23,705.69 | 6.66 | 0.037 | 89.19 | 27.29 | Chloroplast |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Fu, J.; Huang, X.; Fan, J.; Qin, X.; Yu, Q.; Zhang, C.; Xian, B.; Chen, S.; He, Y.; et al. Identification of the Citrus Superoxide Dismutase Family and Their Roles in Response to Phytohormones and Citrus Bacterial Canker. Agriculture 2022, 12, 1254. https://doi.org/10.3390/agriculture12081254
Yang W, Fu J, Huang X, Fan J, Qin X, Yu Q, Zhang C, Xian B, Chen S, He Y, et al. Identification of the Citrus Superoxide Dismutase Family and Their Roles in Response to Phytohormones and Citrus Bacterial Canker. Agriculture. 2022; 12(8):1254. https://doi.org/10.3390/agriculture12081254
Chicago/Turabian StyleYang, Wen, Jia Fu, Xin Huang, Jie Fan, Xiujuan Qin, Qiyuan Yu, Chenxi Zhang, Baohang Xian, Shanchun Chen, Yongrui He, and et al. 2022. "Identification of the Citrus Superoxide Dismutase Family and Their Roles in Response to Phytohormones and Citrus Bacterial Canker" Agriculture 12, no. 8: 1254. https://doi.org/10.3390/agriculture12081254
APA StyleYang, W., Fu, J., Huang, X., Fan, J., Qin, X., Yu, Q., Zhang, C., Xian, B., Chen, S., He, Y., & Li, Q. (2022). Identification of the Citrus Superoxide Dismutase Family and Their Roles in Response to Phytohormones and Citrus Bacterial Canker. Agriculture, 12(8), 1254. https://doi.org/10.3390/agriculture12081254







