The Metabolomics Approaches Based on LC-MS/MS for Analysis of Non-Halal Meats in Food Products: A Review
Abstract
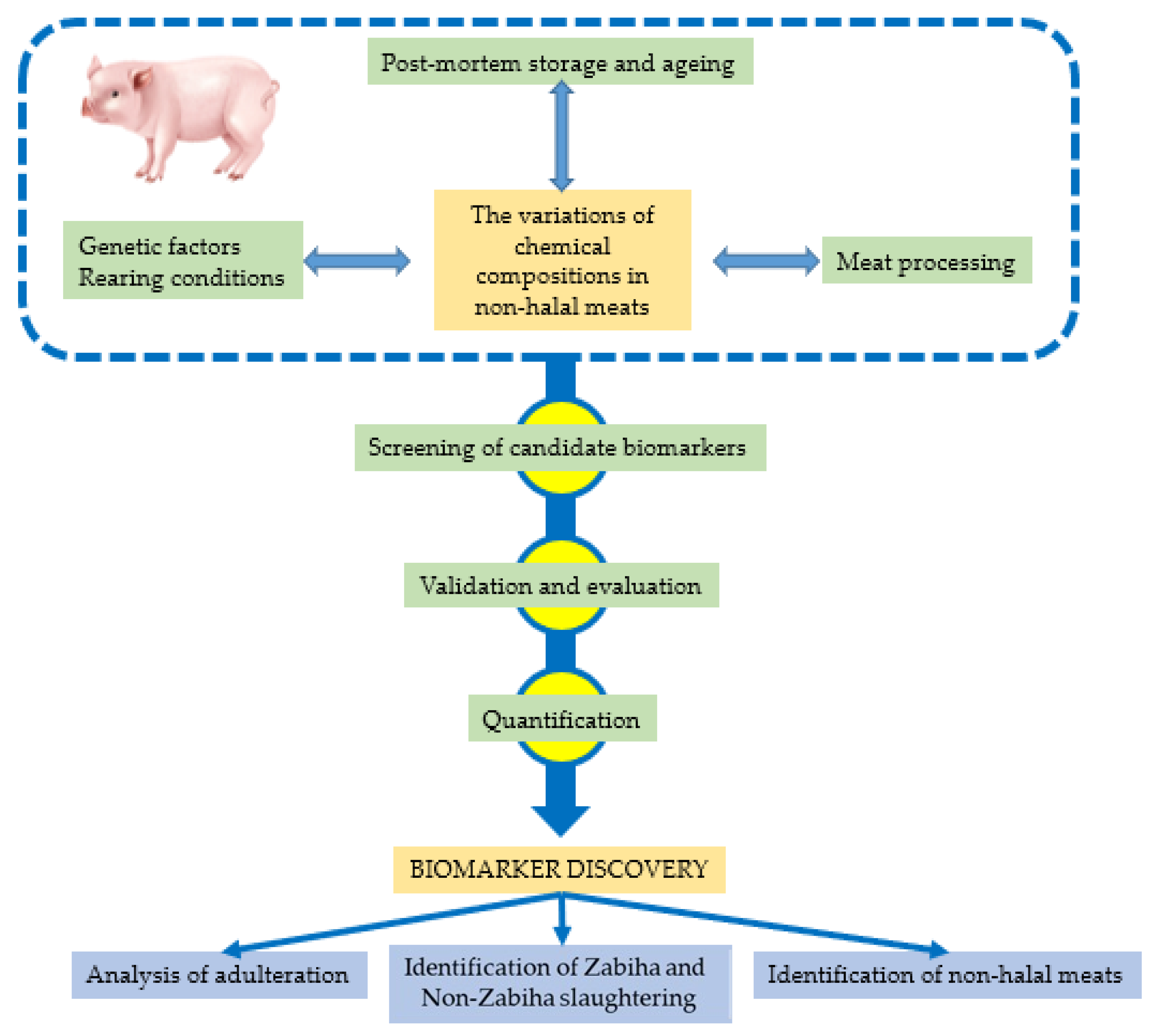
:1. Introduction
2. Methods
3. Metabolomics for Non-Halal Meats’ Analysis
4. LC-MS Technique for Metabolomics Analysis
5. Chemometrics
6. Application of LC-MS for Identification of Non-Halal Meats
7. Analysis of Non-Halal Meats as Adulterants in Halal Meats
8. Application of Metabolomics Studies-Based LC-MS for Analysis of Non-Zabiha Slaughtering
9. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nakyinsige, K.; Man, Y.B.C.; Sazili, A.Q. Halal Authenticity Issues in Meat and Meat Products. Meat Sci. 2012, 91, 207–214. [Google Scholar] [CrossRef] [PubMed]
- El Sheikha, A.F.; Mokhtar, N.F.K.; Amie, C.; Lamasudin, D.U.; Isa, N.M.; Mustafa, S. Authentication Technologies Using DNA-Based Approaches for Meats and Halal Meats Determination. Food Biotechnol. 2017, 31, 281–315. [Google Scholar] [CrossRef]
- Ali, M.E.; Hashim, U.; Mustafa, S.; Che Man, Y.B.; Dhahi, T.S.; Kashif, M.; Uddin, M.K.; Abd Hamid, S.B. Analysis of Pork Adulteration in Commercial Meatballs Targeting Porcine-Specific Mitochondrial Cytochrome b Gene by TaqMan Probe Real-Time Polymerase Chain Reaction. Meat Sci. 2012, 91, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Ridwan, A. Authorization of Halal Certification in Indonesia, Malaysia and Singapore. Int. J. Psychosoc. Rehabil. 2020, 24, 7992–8011. [Google Scholar]
- Demirhan, Y.; Ulca, P.; Senyuva, H.Z. Detection of Porcine DNA in Gelatine and Gelatine-Containing Processed Food Products-Halal/Kosher Authentication. Meat Sci. 2012, 90, 686–689. [Google Scholar] [CrossRef]
- Zia, Q.; Alawami, M.; Mokhtar, N.F.K.; Nhari, R.M.H.R.; Hanish, I. Current Analytical Methods for Porcine Identification in Meat and Meat Products. Food Chem. 2020, 324, 126664. [Google Scholar] [CrossRef]
- Hossain, M.A.M.; Uddin, S.M.K.; Sultana, S.; Wahab, Y.A.; Sagadevan, S.; Johan, M.R.; Ali, M.E. Authentication of Halal and Kosher Meat and Meat Products: Analytical Approaches, Current Progresses and Future Prospects. Crit. Rev. Food Sci. Nutr. 2022, 62, 285–310. [Google Scholar] [CrossRef]
- Rohman, A. The Employment of Fourier Transform Infrared Spectroscopy Coupled with Chemometrics Techniques for Traceability and Authentication of Meat and Meat Products. J. Adv. Vet. Anim. Res. 2019, 6, 9–17. [Google Scholar] [CrossRef]
- Valdés, A.; Beltrán, A.; Mellinas, C.; Jiménez, A.; Garrigós, M.C. Analytical Methods Combined with Multivariate Analysis for Authentication of Animal and Vegetable Food Products with High Fat Content. Trends Food Sci. Technol. 2018, 77, 120–130. [Google Scholar] [CrossRef] [Green Version]
- Rohman, A.; Windarsih, A. The Application of Molecular Spectroscopy in Combination with Chemometrics for Halal Authentication Analysis: A Review. Int. J. Mol. Sci. 2020, 21, 5155. [Google Scholar] [CrossRef]
- Wakhid, S.; Sarno, R.; Sabilla, S.I. The Effect of Gas Concentration on Detection and Classification of Beef and Pork Mixtures Using E-Nose. Comput. Electron. Agric. 2022, 195, 106838. [Google Scholar] [CrossRef]
- Wu, W.; Zhan, J.; Tang, X.; Li, T.; Duan, S. Characterization and Identification of Pork Flavor Compounds and Their Precursors in Chinese Indigenous Pig Breeds by Volatile Profiling and Multivariate Analysis. Food Chem. 2022, 385, 132543. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, Y.; Du, W.; Jin, L.; Ren, P.; Ren, F.; Xie, J.C. Comparative Analysis of Aroma Compounds in Chinese Traditional Dry-Rendered Fat by HS/GC-IMS, SPME/GC-MS, and SPME/GC-O. J. Food Compos. Anal. 2022, 107, 104378. [Google Scholar] [CrossRef]
- Ellis, D.I.; Muhamadali, H.; Allen, D.P.; Elliott, C.T.; Goodacre, R. A Flavour of Omics Approaches for the Detection of Food Fraud. Curr. Opin. Food Sci. 2016, 10, 7–15. [Google Scholar] [CrossRef]
- Selamat, J.; Rozani, N.A.A.; Murugesu, S. Application of the Metabolomics Approach in Food Authentication. Molecules 2021, 26, 7565. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Q.; Yu, Q.; Chen, Y.; Zhao, Y.; Qie, M. Metabolomics Analysis in Food Authentication; Elsevier: Amsterdam, The Netherlands, 2020; ISBN 9780128163955. [Google Scholar]
- Ballin, N.Z.; Laursen, K.H. To Target or Not to Target? Definitions and Nomenclature for Targeted versus Non-Targeted Analytical Food Authentication. Trends Food Sci. Technol. 2019, 86, 537–543. [Google Scholar] [CrossRef]
- Utpott, M.; Rodrigues, E.; Rios, A.D.O.; Mercali, G.D.; Flôres, S.H. Metabolomics: An Analytical Technique for Food Processing Evaluation. Food Chem. 2022, 366, 130685. [Google Scholar] [CrossRef]
- Ai, Z.; Zhang, Y.; Li, X.; Sun, W.; Liu, Y. Widely Targeted Metabolomics Analysis to Reveal Transformation Mechanism of Cistanche Deserticola Active Compounds During Steaming and Drying Processes. Front. Nutr. 2021, 8, 743. [Google Scholar] [CrossRef]
- Pascale, R.; Onzo, A.; Ciriello, R.; Scrano, L.; Bufo, S.A.; Bianco, G. LC/MS Based Food Metabolomics; Elsevier: Amsterdam, The Netherlands, 2020; ISBN 9780128163955. [Google Scholar]
- Medina, S.; Perestrelo, R.; Silva, P.; Pereira, J.A.M.; Câmara, J.S. Current Trends and Recent Advances on Food Authenticity Technologies and Chemometric Approaches. Trends Food Sci. Technol. 2019, 85, 163–176. [Google Scholar] [CrossRef]
- Gerbig, S.; Neese, S.; Penner, A.; Spengler, B.; Schulz, S. Real-Time Food Authentication Using a Miniature Mass Spectrometer. Anal. Chem. 2017, 89, 10717–10725. [Google Scholar] [CrossRef]
- Chatterjee, N.S.; Chevallier, O.P.; Wielogorska, E.; Black, C.; Elliott, C.T. Simultaneous Authentication of Species Identity and Geographical Origin of Shrimps: Untargeted Metabolomics to Recurrent Biomarker Ions. J. Chromatogr. A 2019, 1599, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Ahmad Ruslan, N.A.S.; Chin, L.X.; Ahmad, M.; Abu Hanifah, S.; Abdullah, Z.; Khor, S.M. Recent Advances in Halal Food Authentication: Challenges and Strategies. J. Food Sci. 2022, 87, 8–35. [Google Scholar] [CrossRef] [PubMed]
- Castro-Puyana, M.; Pérez-Míguez, R.; Montero, L.; Herrero, M. Application of Mass Spectrometry-Based Metabolomics Approaches for Food Safety, Quality and Traceability. TrAC Trends Anal. Chem. 2017, 93, 102–118. [Google Scholar] [CrossRef] [Green Version]
- Esteki, M.; Shahsavari, Z.; Simal-Gandara, J. Use of Spectroscopic Methods in Combination with Linear Discriminant Analysis for Authentication of Food Products. Food Control 2018, 91, 100–112. [Google Scholar] [CrossRef]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The Future of NMR-Based Metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Pranata, A.W.; Yuliana, N.D.; Amalia, L.; Darmawan, N. Volatilomics for Halal and Non-Halal Meatball Authentication Using Solid-Phase Microextraction–Gas Chromatography–Mass Spectrometry. Arab. J. Chem. 2021, 14, 103146. [Google Scholar] [CrossRef]
- Savorani, F.; Khakimov, B.; Viereck, N.; Engelsen, S.B. CHAPTER 8: NMR Foodomics. New Dev. NMR 2018, 2018, 183–245. [Google Scholar] [CrossRef]
- Böhme, K.; Calo-Mata, P.; Barros-Velázquez, J.; Ortea, I. Recent Applications of Omics-Based Technologies to Main Topics in Food Authentication. TrAC Trends Anal. Chem. 2019, 110, 221–232. [Google Scholar] [CrossRef]
- Trivedi, D.K.; Hollywood, K.A.; Rattray, N.J.W.; Ward, H.; Trivedi, D.K.; Greenwood, J.; Ellis, D.I.; Goodacre, R. Meat, the Metabolites: An Integrated Metabolite Profiling and Lipidomics Approach for the Detection of the Adulteration of Beef with Pork. Analyst 2016, 141, 2155–2164. [Google Scholar] [CrossRef] [Green Version]
- Zeki, Ö.C.; Eylem, C.C.; Reçber, T.; Kır, S.; Nemutlu, E. Integration of GC–MS and LC–MS for Untargeted Metabolomics Profiling. J. Pharm. Biomed. Anal. 2020, 190, 113509. [Google Scholar] [CrossRef]
- Windarsih, A.; Suratno; Warmiko, H.D.; Indrianingsih, A.W.; Rohman, A.; Ulumuddin, Y.I. Untargeted Metabolomics and Proteomics Approach Using Liquid Chromatography-Orbitrap High Resolution Mass Spectrometry to Detect Pork Adulteration in Pangasius Hypopthalmus Meat. Food Chem. 2022, 386, 132856. [Google Scholar] [CrossRef] [PubMed]
- Pebriana, R.B.; Rohman, A.; Lukitaningsih, E. Sudjadi Development of FTIR Spectroscopy in Combination with Chemometrics for Analysis of Rat Meat in Beef Sausage Employing Three Lipid Extraction Systems. Int. J. Food Prop. 2017, 20, 1995–2005. [Google Scholar] [CrossRef] [Green Version]
- Bögl, T.; Mlynek, F.; Himmelsbach, M.; Buchberger, W. Comparison of One-Phase and Two-Phase Extraction Methods for Porcine Tissue Lipidomics Applying a Fast and Reliable Tentative Annotation Workflow. Talanta 2022, 236, 122849. [Google Scholar] [CrossRef] [PubMed]
- López-Pedrouso, M.; Lorenzo, J.M.; Gagaoua, M.; Franco, D. Application of Proteomic Technologies to Assess the Quality of Raw Pork and Pork Products: An Overview from Farm-to-Fork. Biology 2020, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Amir, S.H.; Yuswan, M.H.; Aizat, W.M.; Mansor, M.K.; Desa, M.N.M.; Yusof, Y.A.; Song, L.K.; Mustafa, S. Comparative Database Search Engine Analysis on Massive Tandem Mass Spectra of Pork-Based Food Products for Halal Proteomics. J. Proteom. 2021, 241, 104240. [Google Scholar] [CrossRef]
- Wadood, S.A.; Boli, G.; Xiaowen, Z.; Hussain, I.; Yimin, W. Recent Development in the Application of Analytical Techniques for the Traceability and Authenticity of Food of Plant Origin. Microchem. J. 2020, 152, 104295. [Google Scholar] [CrossRef]
- Álvarez, G.; Montero, L.; Llorens, L.; Castro-Puyana, M.; Cifuentes, A. Recent Advances in the Application of Capillary Electromigration Methods for Food Analysis and Foodomics. Electrophoresis 2018, 39, 136–159. [Google Scholar] [CrossRef]
- Aszyk, J.; Byliński, H.; Namieśnik, J.; Kot-Wasik, A. Main Strategies, Analytical Trends and Challenges in LC-MS and Ambient Mass Spectrometry–Based Metabolomics. TrAC Trends Anal. Chem. 2018, 108, 278–295. [Google Scholar] [CrossRef]
- López-Ruiz, R.; Romero-González, R.; Garrido Frenich, A. Ultrahigh-Pressure Liquid Chromatography-Mass Spectrometry: An Overview of the Last Decade. TrAC Trends Anal. Chem. 2019, 118, 170–181. [Google Scholar] [CrossRef]
- Kohler, I.; Verhoeven, M.; Haselberg, R.; Gargano, A.F.G. Hydrophilic Interaction Chromatography–Mass Spectrometry for Metabolomics and Proteomics: State-of-the-Art and Current Trends. Microchem. J. 2022, 175, 106986. [Google Scholar] [CrossRef]
- Harrieder, E.M.; Kretschmer, F.; Böcker, S.; Witting, M. Current State-of-the-Art of Separation Methods Used in LC-MS Based Metabolomics and Lipidomics. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2022, 1188, 123069. [Google Scholar] [CrossRef] [PubMed]
- Lucci, P.; Saurina, J.; Núñez, O. Trends in LC-MS and LC-HRMS Analysis and Characterization of Polyphenols in Food. TrAC Trends Anal. Chem. 2017, 88, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Mi, S.; Shang, K.; Jia, W.; Zhang, C.H.; Li, X.; Fan, Y.Q.; Wang, H. Characterization and Discrimination of Taihe Black-Boned Silky Fowl (Gallus Gallus Domesticus Brisson) Muscles Using LC/MS-Based Lipidomics. Food Res. Int. 2018, 109, 187–195. [Google Scholar] [CrossRef]
- Muroya, S.; Ueda, S.; Komatsu, T.; Miyakawa, T.; Ertbjerg, P. Meatabolomics: Muscle and Meat Metabolomics in Domestic Animals. Metabolites 2020, 10, 188. [Google Scholar] [CrossRef]
- Perez de Souza, L.; Alseekh, S.; Scossa, F.; Fernie, A.R. Ultra-High-Performance Liquid Chromatography High-Resolution Mass Spectrometry Variants for Metabolomics Research. Nat. Methods 2021, 18, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Källsten, M.; Pijnappel, M.; Hartmann, R.; Lehmann, F.; Kovac, L.; Lind, S.B.; Bergquist, J. Application of Triple Quadrupole Mass Spectrometry for the Characterization of Antibody–Drug Conjugates. Anal. Bioanal. Chem. 2019, 411, 2569–2576. [Google Scholar] [CrossRef] [Green Version]
- Boesl, U. Time-of-Flight Mass Spectrometry: Introduction to the Basics. Mass Spectrom. Rev. 2017, 36, 86–109. [Google Scholar] [CrossRef]
- Špánik, I.; Machyňáková, A. Recent Applications of Gas Chromatography with High-Resolution Mass Spectrometry. J. Sep. Sci. 2018, 41, 163–179. [Google Scholar] [CrossRef]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass Spectrometry-Based Metabolomics: A Guide for Annotation, Quantification and Best Reporting Practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef]
- Rubert, J.; Zachariasova, M.; Hajslova, J. Advances in High-Resolution Mass Spectrometry Based on Metabolomics Studies for Food—A Review. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2015, 32, 1685–1708. [Google Scholar] [CrossRef]
- Lacalle-Bergeron, L.; Izquierdo-Sandoval, D.; Sancho, J.V.; López, F.J.; Hernández, F.; Portolés, T. Chromatography Hyphenated to High Resolution Mass Spectrometry in Untargeted Metabolomics for Investigation of Food (Bio)Markers. TrAC Trends Anal. Chem. 2021, 135, 116161. [Google Scholar] [CrossRef]
- Andjelković, U.; Gajdošik, M.Š.; Gašo-Sokač, D.; Martinović, T.; Josić, D. Foodomics and Food Safety: Where We Are. Food Technol. Biotechnol. 2017, 55, 290–307. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; de Boves Harrington, P. Chemometric Applications in Metabolomic Studies Using Chromatography-Mass Spectrometry. TrAC Trends Anal. Chem. 2021, 135, 116165. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar] [CrossRef]
- Cao, M.; Han, Q.; Zhang, J.; Zhang, R.; Wang, J.; Gu, W.; Kang, W.; Lian, K.; Ai, L. An Untargeted and Pseudotargeted Metabolomic Combination Approach to Identify Differential Markers to Distinguish Live from Dead Pork Meat by Liquid Chromatography–Mass Spectrometry. J. Chromatogr. A 2020, 1610, 460553. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. PCA as a Practical Indicator of OPLS-DA Model Reliability. Curr. Metab. 2016, 4, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, L.; Johansson, E.; Kettenah-Wold, N.; Trygg, J.; Wikström, C.; Wold, S. Multi- and Megavariate Data Analysis; Umetrics AB: Umeå, Sweden, 2006; ISBN 9789197373029. [Google Scholar]
- Xu, Y.; Muhamadali, H.; Sayqal, A.; Dixon, N.; Goodacre, R. Partial Least Squares with Structured Output for Modelling the Metabolomics Data Obtained from Complex Experimental Designs: A Study into the Y-Block Coding. Metabolites 2016, 6, 38. [Google Scholar] [CrossRef]
- Leng, T.; Li, F.; Xiong, L.; Xiong, Q.; Zhu, M.; Chen, Y. Quantitative Detection of Binary and Ternary Adulteration of Minced Beef Meat with Pork and Duck Meat by NIR Combined with Chemometrics. Food Control 2020, 113, 107203. [Google Scholar] [CrossRef]
- Ali, M.E.; Hashim, U.; Dhahi, T.S.; Mustafa, S.; Man, Y.B.C.; Latif, M.A. Analysis of Pork Adulteration in Commercial Burgers Targeting Porcine-Specific Mitochondrial Cytochrome B Gene by TaqMan Probe Real-Time Polymerase Chain Reaction. Food Anal. Methods 2012, 5, 784–794. [Google Scholar] [CrossRef]
- Sarah, S.A.; Faradalila, W.N.; Salwani, M.S.; Amin, I.; Karsani, S.A.; Sazili, A.Q. LC-QTOF-MS Identification of Porcine-Specific Peptide in Heat Treated Pork Identifies Candidate Markers for Meat Species Determination. Food Chem. 2016, 199, 157–164. [Google Scholar] [CrossRef]
- Yuswan, M.H.; Aizat, W.M.; Lokman, A.A.; Desa, M.N.M.; Mustafa, S.; Junoh, N.M.; Yusof, Z.N.B.; Mohamed, R.; Mohmad, Z.; Lamasudin, D.U. Chemometrics-Assisted Shotgun Proteomics for Establishment of Potential Peptide Markers of Non-Halal Pork (Sus Scrofa) among Halal Beef and Chicken. Food Anal. Methods 2018, 11, 3505–3515. [Google Scholar] [CrossRef]
- Mi, S.; Shang, K.; Li, X.; Zhang, C.H.; Liu, J.Q.; Huang, D.Q. Characterization and Discrimination of Selected China’s Domestic Pork Using an LC-MS-Based Lipidomics Approach. Food Control 2019, 100, 305–314. [Google Scholar] [CrossRef]
- Yuswan, M.H.; Aizat, W.M.; Desa, M.N.M.; Hashim, A.M.; Rahim, N.A.; Mustafa, S.; Mohamed, R.; Lamasudin, D.U. Improved Gel-Enhanced Liquid Chromatography-Mass Spectrometry by Chemometrics for Halal Proteomics. Chemom. Intell. Lab. Syst. 2019, 192, 103825. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Y.; Liu, C.; Qin, R.; Gong, D.; Wang, R.; Zhang, D.; Che, L.; Chen, D.; Xin, G.; et al. Multi-Omics Profiling Highlights Lipid Metabolism Alterations in Pigs Fed Low-Dose Antibiotics. BMC Genet. 2020, 21, 112. [Google Scholar] [CrossRef] [PubMed]
- Hye, L.J.; Min, A.J.; Jin, K.D.; Jin, K.H.; Hun, L.S. Use of LC-Orbitrap MS and FT-NIRS with Multivariate Analysis to Determine Geographic Origin of Boston Butt Pork. Int. J. Food Prop. 2022, 25, 128–143. [Google Scholar] [CrossRef]
- Von Bargen, C.; Dojahn, J.; Waidelich, D.; Humpf, H.U.; Brockmeyer, J. New Sensitive High-Performance Liquid Chromatography-Tandem Mass Spectrometry Method for the Detection of Horse and Pork in Halal Beef. J. Agric. Food Chem. 2013, 61, 11986–11994. [Google Scholar] [CrossRef] [PubMed]
- Prandi, B.; Lambertini, F.; Faccini, A.; Suman, M.; Leporati, A.; Tedeschi, T.; Sforza, S. Mass Spectrometry Quantification of Beef and Pork Meat in Highly Processed Food: Application on Bolognese Sauce. Food Control. 2017, 74, 61–69. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Kang, C.; Zhao, W.; Li, S.; Wang, S. Assessment of Carbonic Anhydrase 3 as a Marker for Meat Authenticity and Performance of LC-MS/MS for Pork Content. Food Chem. 2021, 342, 128240. [Google Scholar] [CrossRef]
- Abbas, N.; Ali, A.; Kumari, S.; Iqbal, A.; Husain, A.; Saeed, T.; AbdulAmer Al-Ballam, Z.; Ahmed, N.; El-Seedi, H.R.; Musharraf, S.G. Untargeted-Metabolomics Differentiation between Poultry Samples Slaughtered with and without Detaching Spinal Cord. Arab. J. Chem. 2020, 13, 9081–9089. [Google Scholar] [CrossRef]
- Ali, N.S.M.; Zabidi, A.R.; Manap, M.N.A.; Zahari, S.M.S.N.S.; Yahaya, N. Effect of Different Slaughtering Methods on Metabolites of Broiler Chickens Using Ultra High-Performance Liquid Chromatography-Time of Flight-Mass Spectrometry (UHPLC-TOF-MS). Food Res. 2020, 4, 133–138. [Google Scholar] [CrossRef]
- Sidwick, K.L.; Johnson, A.E.; Adam, C.D.; Pereira, L.; Thompson, D.F. Use of Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry and Metabonomic Profiling to Differentiate between Normally Slaughtered and Dead on Arrival Poultry Meat. Anal. Chem. 2017, 89, 12131–12136. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.N.; Miller, J.C.; Miller, R.D. Statistics and Chemometrics for Analytical Chemistry, 7th ed.; Pearson Education Limited: Harlow, UK, 2018. [Google Scholar]
- Nair, M.S.; Yao, D.; Chen, C.; Pieters, M. Serum Metabolite Markers of Early Mycoplasma Hyopneumoniae Infection in Pigs. Vet. Res. 2019, 50, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babu, A.F.; Koistinen, V.M.; Turunen, S.; Solano-Aguilar, G.; Urban, J.F.; Zarei, I.; Hanhineva, K. Identification and Distribution of Sterols, Bile Acids, and Acylcarnitines by LC–MS/MS in Humans, Mice, and Pigs—A Qualitative Analysis. Metabolites 2022, 12, 49. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Windarsih, A.; Rohman, A.; Riswanto, F.D.O.; Dachriyanus; Yuliana, N.D.; Bakar, N.K.A. The Metabolomics Approaches Based on LC-MS/MS for Analysis of Non-Halal Meats in Food Products: A Review. Agriculture 2022, 12, 984. https://doi.org/10.3390/agriculture12070984
Windarsih A, Rohman A, Riswanto FDO, Dachriyanus, Yuliana ND, Bakar NKA. The Metabolomics Approaches Based on LC-MS/MS for Analysis of Non-Halal Meats in Food Products: A Review. Agriculture. 2022; 12(7):984. https://doi.org/10.3390/agriculture12070984
Chicago/Turabian StyleWindarsih, Anjar, Abdul Rohman, Florentinus Dika Octa Riswanto, Dachriyanus, Nancy Dewi Yuliana, and Nor Kartini Abu Bakar. 2022. "The Metabolomics Approaches Based on LC-MS/MS for Analysis of Non-Halal Meats in Food Products: A Review" Agriculture 12, no. 7: 984. https://doi.org/10.3390/agriculture12070984
APA StyleWindarsih, A., Rohman, A., Riswanto, F. D. O., Dachriyanus, Yuliana, N. D., & Bakar, N. K. A. (2022). The Metabolomics Approaches Based on LC-MS/MS for Analysis of Non-Halal Meats in Food Products: A Review. Agriculture, 12(7), 984. https://doi.org/10.3390/agriculture12070984







