The Hunt for Mungbean (Vigna radiata (L.) Wilczek) Genotypes and Breeding Lines Resistance to South Indian Bruchid Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Genetic Materials
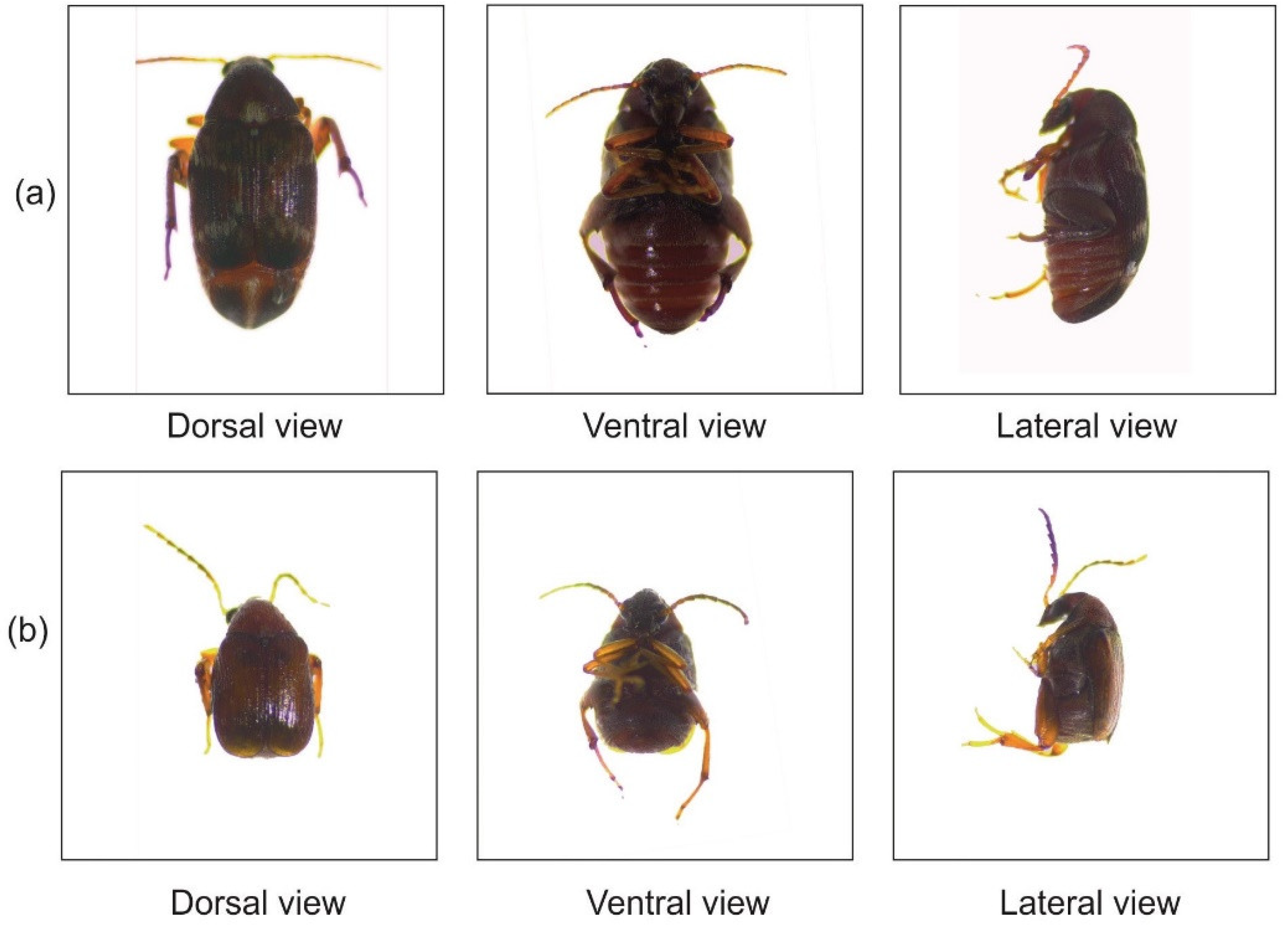
2.2. Source and Identification of Callosobruchus maculatus
2.3. Mass Culturing and Maintenance of Callosobruchus maculatus
2.4. Assay Methodology for Screening Mungbean Genotypes and Resistance Evaluation
- I.
- The number of eggs laid: The total number of eggs laid was counted in each genotype for seven days after adult release;
- II.
- The number of adults emerged: The total number of adults emerged was counted daily to determine the mean developmental period (days) and continued till the cessation of emergence;
- III.
- Adult emergence percentage: (Number of adults emerged/number of eggs laid) × 100;
- IV.
- Female to male ratio;
- V.
- Mean developmental period (MDP) recorded in days.
- Susceptibility Index: log (per cent adult emergence)/mean developmental period.
- Percentage of (%) seed damage: Number of seeds damaged/Number of seeds taken × 100. Based on seed damage percent, the genotypes were categorized as highly resistant (0–10%), resistant (10.1–20%), moderately resistant (20.1–40%), susceptible (40.1–80%), and highly susceptible (80.1–100%) [10].
2.5. Generation of Breeding Population Using Resistant and Susceptible Genotypes
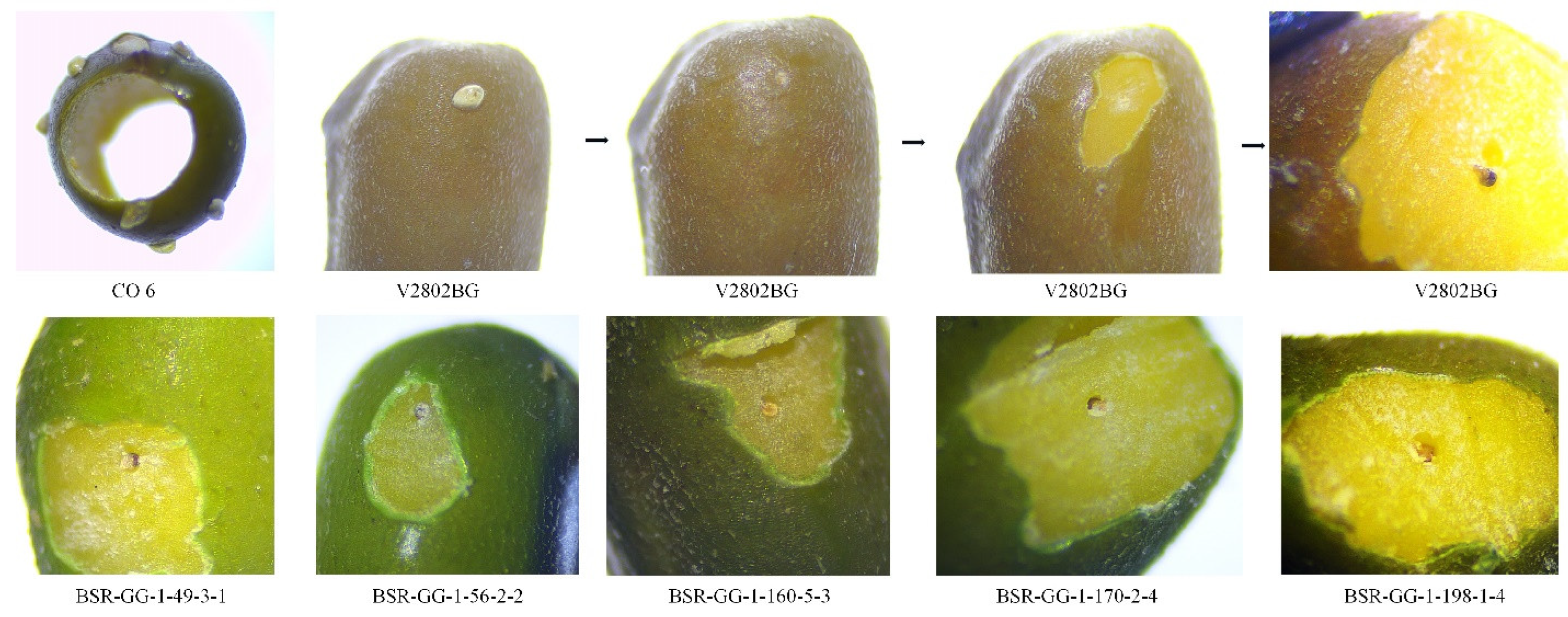
2.6. Assessment of Grub and Morphological Traits
2.7. Statistical Analysis
3. Results
3.1. Bruchid Resistance Determination on 52 Mungbean Genotypes
3.2. Development of Breeding Lines with Bruchid Resistance
3.3. Grub Development in the Resistant Lines
3.4. Agronomic Performance of the Resistant Lines
4. Discussion
4.1. Search for Mungbean Genotypes Resistance to South Indian Bruchid Strain
4.2. Breeding Resistant Lines with Better Agronomic Performances
4.3. Development of Grub and Agronomic Performance of the Resistant Lines
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nair, R.M.; Yang, R.Y.; Easdown, W.J.; Thavarajah, D.; Thavarajah, P.; Hughes, J.A.; Keatinge, J.D.H. Biofortification of mungbean (Vigna radiata) as a whole food to enhance human health. J. Sci. Food Agric. 2013, 93, 1805–1813. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, A.; Shobhana, V.G.; Sudha, M.; Raveendran, M.; Senthil, N.; Pandiyan, M.; Nagarajan, P. Mungbean yellow mosaic virus (MYMV): A threat to green gram (Vigna radiata) production in Asia. Int. J. Pest Manag. 2014, 60, 314–324. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Sudha, M.; Pandiyan, M.; Senthil, N.; Shobhana, V.G.; Nagarajan, P. Screening of MYMV resistant mungbean (Vigna radiata (L.) Wilczek) progenies through Agroinoculation. Int. J. Plant Pathol. 2011, 2, 115–125. [Google Scholar] [CrossRef][Green Version]
- Amin, M.R.; Islam, M.S.; Hasan, M.K.; Sabagh, A.E. Improvement of production and net economic return through intercropping of upland cotton with mungbean. Azarian J. Agric. 2018, 5, 67–75. [Google Scholar]
- He, T.G.; Su, L.R.; Li, Y.R.; Su, T.M.; Qin, F.; Li, Q. Nutrient decomposition rate and sugarcane yield as influenced by mungbean intercropping and crop residue recycling. Sugar Tech. 2018, 20, 154–162. [Google Scholar] [CrossRef]
- Diatta, A.A.; Thomason, W.E.; Abaye, O.; Thompson, T.L.; Battaglia, M.L.; Vaughan, L.J.; Lo, M. Assessment of Nitrogen Fixation by Mungbean Genotypes in Different Soil Textures Using 15 N Natural Abundance Method. J. Soil Sci. Plant Nutr. 2020, 20, 2230–2240. [Google Scholar] [CrossRef]
- Ministry of Agriculture and Farmers Welfare, Department of Agriculture, Cooperation and Farmers Welfare, Government of India. Pulses Revolution from Food to Nutritional Security. 2018. Available online: http://dpd.gov.in (accessed on 20 February 2019).
- Gahukar, R.T.; Reddy, G.V. Management of insect pests in the production and storage of minor pulses. Ann. Entomol. Soc. Am. 2018, 111, 172–183. [Google Scholar] [CrossRef]
- Mishra, S.K.; Macedo, M.L.R.; Panda, S.K.; Panigrahi, J. Bruchid pest management in pulses: Past practices, present status and use of modern breeding tools for development of resistant varieties. Ann. App. Biol. 2018, 172, 4–19. [Google Scholar] [CrossRef]
- Mariyammal, I.; Seram, D.; Samyuktha, S.M.; Karthikeyan, A.; Dhasarathan, M.; Murukarthick, J.; Kennedy, J.S.; Malarvizhi, D.; Yang, T.J.; Pandiyan, M.; et al. QTL mapping in Vigna radiata× Vigna umbellata population uncovers major genomic regions associated with bruchid resistance. Mol. Breed. 2019, 39, 1–13. [Google Scholar] [CrossRef]
- Young, N.D.; Kumar, L.; Menancio-Hautea, D.; Danesh, D.; Talekar, N.S.; Shanmugasundarum, S.; Kim, D.H. RFLP mapping of a major bruchid resistance gene in mungbean (Vigna radiata, L. Wilczek). Theor. Appl. Genet. 1992, 84, 839–884. [Google Scholar] [CrossRef]
- Tripathi, A.; Tripathi, D.K.; Chauhan, D.K.; Kumar, N.; Singh, G.S. Paradigms of climate change impacts on some major food sources of the world: A review on current knowledge and future prospects. Agric. Ecosyst. Environ. 2016, 216, 356–373. [Google Scholar] [CrossRef]
- Chawe, K.G.; Venkataramana, P.B.; Ndakidemi, P.A. Assessment of farmers’ indigenous knowledge and preferences: A tool for sustainable lablab bean (Lablab purpureus L. Sweet) improvement and utilization in Northern Tanzania. J. Adv. Biol. Biotechnol. 2019, 21, 1–14. [Google Scholar] [CrossRef]
- Mofunanya, A.A.J.; Namgbe, E.E. Assessment of damage due to Callosobruchus maculatus (Coleoptera: Bruchidae) infestation on germination and nutrient quality of Vigna unguiculata L. (Walp). IOSR-J. Agric. Vet. Sci. 2016, 9, 96–101. [Google Scholar] [CrossRef]
- Hamdi, S.H.; Abidi, S.; Sfayhi, D.; Dhraief, M.Z.; Amri, M.; Boushih, E.; Hedjal-Chebheb, M.; Larbi, K.M.; Jemaa, J.M.B. Nutritional alterations and damages to stored chickpea in relation with the pest status of Callosobruchus maculatus (Chrysomelidae). J. Asia Pac. Entomol. 2017, 20, 1067–1076. [Google Scholar] [CrossRef]
- Sreedhar, M.; Singh, D.V.; Reddy, D.C.; Vasudha, A. Biochemical changes in groundnut pods due to infestation of bruchid Caryedon serratus (Olivier) under stored conditions. J. Stored Prod. Res. 2020, 88, 101678. [Google Scholar] [CrossRef]
- Somta, P.; Ammaranan, C.; Ooi, P.A.C.; Srinives, P. Inheritance of seed resistance to bruchids in cultivated mungbean (Vigna radiata, L. Wilczek). Euphytica 2007, 155, 47–55. [Google Scholar] [CrossRef]
- Soumia, P.S.; Srivastava, C.; Dikshit, H.K.; Pandi, G.G.P. Screening for resistance against pulse beetle, Callosobruchus analis (F.) in greengram (Vigna radiata (L.) Wilczek) accessions. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2015, 87, 551–558. [Google Scholar] [CrossRef]
- Liu, M.S.; Kuo, T.C.Y.; Ko, C.Y.; Wu, D.C.; Li, K.Y.; Lin, W.J.; Lin, C.P.; Wang, Y.W.; Schafleitner, R.; Lo, H.F.; et al. Genomic and transcriptomic comparison of nucleotide variations for insights into bruchid resistance of mungbean (Vigna radiata [L.] R. Wilczek). BMC Plant Biol. 2016, 16, 1–16. [Google Scholar] [CrossRef]
- Singh, C.M.; Singh, P.; Pratap, A.; Pandey, R.; Purwar, S.; Douglas, C.A.; Baek, K.H.; Mishra, A.K. Breeding for enhancing Legumovirus resistance in mungbean: Current understanding and future directions. Agronomy 2019, 9, 622. [Google Scholar] [CrossRef]
- Somta, C.; Somta, P.; Tomooka, N.; Ooi, P.A.C.; Vaughan, D.A.; Srinives, P. Characterization of new sources of mungbean (Vigna radiata (L.) Wilczek) resistance to bruchids, Callosobruchus spp. (Coleoptera: Bruchidae). J. Stored Prod. Res. 2008, 44, 316–321. [Google Scholar] [CrossRef]
- Ponnusamy, D.; Pratap, A.; Singh, S.K.; Gupta, S. Evaluation of screening methods for bruchid beetle (Callosobruchus chinensis) resistance in green gram (Vigna radiata) and blackgram (Vigna mungo) genotypes and influence of seed physical characteristics on its infestation. Vegetos 2014, 27, 60–67. [Google Scholar] [CrossRef]
- Sarkar, S.; Bhattacharyya, S. Screening of green gram genotypes for Bruchid (Callosobruchus chinensis L.) resistance and selection of parental lines for hybridization programme. Legume Res. 2015, 38, 704–706. [Google Scholar] [CrossRef]
- Soumia, P.S.; Srivastava, C.; Pandi, G.; Subramanian, S. Varietal preference of pulse beetle, Callosobruchus maculatus (F.) in greengram. Indian J. Entomol. 2017, 79, 86–91. [Google Scholar] [CrossRef]
- Soumia, P.S.; Chitra, S.; Guru, P.P.G.; Subramanian, S. Screening of green gram accessions against pulse beetle Callosobruchus chinensis (L.). Indian J. Entomol. 2018, 80, 1635–1641. [Google Scholar] [CrossRef]
- Beck, C.W.; Blumer, L.S.; Habib, J. Effects of evolutionary history on adaptation in bean beetles, a model system for inquiry-based laboratories. Evol. Educ. Outreach 2013, 6, 5. [Google Scholar] [CrossRef]
- Seram, D.; Senthil, N.; Pandiyan, M.; Kennedy, J.S. Resistance determination of a South Indian bruchid strain against rice bean landraces of Manipur (India). J. Stored Prod. Res. 2016, 69, 199–206. [Google Scholar] [CrossRef]
- Venkataramana, P.B.; Gowda, R.; Somta, P.; Ramesh, S.; Rao, A.M.; Bhanuprakash, K.; Srinives, P.; Gireesh, C.; Pramila, C.K. Mapping QTL for bruchid resistance in rice bean (Vigna umbellata). Euphytica 2016, 207, 135–147. [Google Scholar] [CrossRef]
- Howe, R. A parameter for experssing the suitability of an environment for insect development. J. Stored Prod. Res. 1971, 7, 63–65. [Google Scholar] [CrossRef]
- IBPGR. Descriptors for Vigna Mungo and Vigna Radiata; International Board for Plant Genetic Resources: Rome, Italy, 1980; pp. 11–15. [Google Scholar]
- Panse, V.; Sukhatme, P. Statistical methods for agricultural research. ICAR New Delhi India 1985, 8, 308–318. [Google Scholar]
- Kempthorne, O. An Introduction to Genetic Statistics; John Willey & Sons. Inc.: New York, NY, USA, 1957. [Google Scholar]
- Manivannan N TNAUSTAT-Statistical Package. 2014. Available online: https://sites.google.com/site/tnaustat (accessed on 10 January 2019).
- Asian Vegetable Research and Development Center [AVRDC]. Progress Report, 1987; AVRDC: Shanhua, Taiwan, 1990; 480p. [Google Scholar]
- Singh, B.B.; Singh., S.R. Breeding for bruchid resistance in cowpea. In Bruchids and Legumes: Economics, Ecology and Coevolution; Springer: Dordrecht, The Netherlands, 1990; pp. 219–228. [Google Scholar] [CrossRef]
- Edde, P.A.; Amatobi, C.I. Seed coat has no value in protecting cowpea seed against attack by Callosobruchus maculatus (F.). J. Stored Prod. Res. 2003, 39, 1–10. [Google Scholar] [CrossRef]
- Bashir, M.A.; Alvi, A.M.; Naz, H. Screening of legume and cereal seeds against Callosobruchus maculatus on the basis of fecundity and longevity. J. Environ. Agric. Sci. 2014, 1, 11. [Google Scholar]
- Sharma, R.; Devi, R.; Soni, A.; Sharma, U.; Yadav, S.; Sharma, R. Growth and developmental responses of Callosobruchus maculatus (F.) on various pulses. Legume Res. 2016, 39, 840–843. [Google Scholar] [CrossRef]
- Kaewwongwal, A.; Liu, C.; Somta, P.; Chen, J.; Tian, J.; Yuan, X.; Chen, X. A second VrPGIP1 allele is associated with bruchid resistance (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) accession ACC41. Mol. Genet. Genom. 2020, 295, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Chotechung, S.; Somta, P.; Chen, J.; Yimram, T.; Chen, X.; Srinives, P. A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. 2016, 129, 1673–1683. [Google Scholar] [CrossRef] [PubMed]
- Kaewwongwal, A.; Chen, J.; Somta, P.; Kongjaimun, A.; Yimram, T.; Chen, X.; Srinives, P. Novel Alleles of Two Tightly Linked Genes Encoding Polygalacturonase-Inhibiting Proteins (VrPGIP1 and VrPGIP2) Associated with the Br Locus That Confer Bruchid (Callosobruchus spp.) Resistance to Mungbean (Vigna radiata) Accession V2709. Front. Plant Sci. 2017, 8, 1692. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Ku, H.; Shafleitner, R.; Bains, T.S.; Kuo, C.G.; Liu, C.; Nair, R.M. The major quantitative trait locus for mungbean yellow mosaic Indian virus resistance is tightly linked in repulsion phase to the major bruchid resistance locus in a cross between mungbean [Vigna radiata (L.) Wilczek] and its wild relative Vigna radiata ssp. sublobata. Euphytica 2013, 192, 205–216. [Google Scholar] [CrossRef]
- Wang, L.; Wu, C.; Zhong, M.; Zhao, D.; Mei, L.; Chen, H.; Wang, S.; Liu, C.; Cheng, X. Construction of an integrated map and location of a bruchid resistance gene in mungbean. Crop J. 2016, 4, 60–366. [Google Scholar] [CrossRef][Green Version]
- Somta, P.; Kaga, A.; Tomooka, N.; Kashiwaba, K.; Isemura, T.; Chaitieng, B.; Srinives, P.; Vaughan, D.A. Development of an interspecific Vigna linkage map between Vigna umbellata (Thunb.) Ohwi & Ohashi and V. nakashimae (Ohwi) Ohwi & Ohashi and its use in analysis of bruchid resistance and comparative genomics. Plant Breed. 2006, 125, 77–84. [Google Scholar]
- Smith, C.M. Plant Resistance to Arthropods: Molecular and Conventional Approaches; Springer: Dordrecht, The Netherlands, 2005. [Google Scholar]
- Smith, C.M.; Clement, S.L. Molecular bases of plant resistance to arthropods. Annu. Rev. Entomol. 2012, 57, 309–328. [Google Scholar] [CrossRef]
- Sadasivam, S.; Thayumanayan, B. Molecular Host Plant Resistance to Pests; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar] [CrossRef]
- Edwards, O.; Singh, K.B. Resistance to insect pests: What do legumes have to offer? Euphytica 2006, 147, 273–285. [Google Scholar] [CrossRef]
- Eduardo, W.I.; Junior, A.L.B.; de Moraes, R.F.O.; Chiorato, A.F.; Perlatti, B.; Forim, M.R. Antibiosis levels of common bean genotypes toward Zabrotes subfasciatus (Boheman) (Coleoptera: Bruchidae) and its correlation with flavonoids. J. Stored Prod. Res. 2016, 67, 63–70. [Google Scholar] [CrossRef]
- Devi, M.B.; Devi, N.V. Biology and morphometric measurement of cowpea weevil, Callosobruchus maculatus Fab. (Coleoptera: Chrysomelidae) in green gram. J. Entomol. Zool. Stud. 2014, 2, 74–76. [Google Scholar]
- Dick, K.M. Bionomic Variation among Three Populations of the Southern Cowpea Weevil, Callosobruchus maculatus, and Their Responses to Resistant Varieties of the Primary Host. Ph.D. Thesis, University of London, Bedford College, Bedford, UK, 1984. [Google Scholar]
- Dick, K.M.; Credland, P.F. Changes in the response of Callosobruchus maculatus (Coleoptera: Bruchidae) to a resistant variety of cowpea. J. Stored Prod. Res. 1986, 22, 227–233. [Google Scholar] [CrossRef]
- Edde, P.A.; Amatobi, C.I. Relative resistance of some cowpea varieties to Callosobruchus maculatus (F.) (Coleoptera: Bruchidae). J. Sustain. Agric. 2001, 17, 67–77. [Google Scholar] [CrossRef]
- Asian Vegetable Research and Development Center [AVRDC]. Progress Report for 1991; AVRDC: Shanhua, Taiwan, 1992; p. 410. [Google Scholar]
- Talekar, N.S.; Lin, C.P. Characterization of Callosobruchus chinensis (Coleoptera: Bruchidae) resistance in mungbean. J. Econ. Entomol. 1992, 85, 1150–1153. [Google Scholar] [CrossRef]
- Souframanien, J.; Gopalakrishna, T. Source for bruchid resistance and its inheritance in Trombay wild urdbean Vigna. J. Food Legumes 2007, 20, 19–21. [Google Scholar]
- Castro, M.D.; Baldin, E.L.; Cruz, P.L.; Souza, C.M.; Silva, P.H. Characterization of cowpea genotype resistance to Callosobruchus maculatus. Pesqui. Agropecu. Bras. 2013, 48, 1201–1209. [Google Scholar] [CrossRef]
- Kaur, H.; Gill, R.S.; Kaur, S. Antinutritional factors in rice bean, Vigna umbellata Thunb. (Ohwi and Ohashi) against Callosobruchus maculatus F. (Coleoptera: Bruchidae). Phytoparasitica 2016, 44, 609–614. [Google Scholar] [CrossRef]
- Miesho, B.; Msiska, U.; Hailay, M.; Malinga, G.; Odong, T.; Edema, R.; Gibson, P.; Rubaihayo, P.; Kyamanywa, S. Biochemical basis of cowpea resistance to bruchid, Callosobruchus maculatus (F.). IJACR 2017, 5, 219–227. [Google Scholar]
- Grazziotin, M.A.; Cabral, G.B.; Ibrahim, A.B.; Machado, R.B.; Aragao, F.J. Expression of the Arcelin 1 gene from Phaseolus vulgaris L. in cowpea seeds (Vigna unguiculata L.) confers bruchid resistance. Ann. App. Biol. 2020, 176, 268–274. [Google Scholar] [CrossRef]
- Jaba, J.; Bhandi, S.; Deshmukh, S.; Pallipparambil, G.R.; Mishra, S.P.; Arora, N. Identification, evaluation and utilization of resistance to insect pests in grain legumes: Advancement and restrictions. In Genetic Enhancement in Major Food Legumes; Springer: Cham, Germany, 2021; pp. 197–230. [Google Scholar]
- Caroline, N.M.; Deogracious, P.M.; George, M.T.; James, R.M.; Joel, W.D.; Paul, M.K. Identification of potential seed storage protein responsible for bruchid resistance in common bean landraces from Tanzania and Malawi. Afr. J. Biotechnol. 2022, 21, 35–45. [Google Scholar] [CrossRef]
- Krisnawati, A.; Adie, M.M.; Soegianto, A.; Waluyo, B. Pod shattering resistance and agronomic traits in F5 segregating populations of soybean. SABRAO J. Breed. Genet. 2019, 51, 266–280. [Google Scholar]


| S.No. | Genotypes | No.of Eggs Laid | No. of Adults Emerged | Adult Emergence Percentage | Mean Developmental Period | No. of Males Emerged | No. of Females Emerged | Female to Male Ratio | Susceptibility Index | Damage Percentage | Category |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NM94 | 52 ± 3.28 h | 20 | 38.46 pq | 25.75 ± 0.65 qrstuv | 14 ± 0.88 b | 6 ± 0.88 j | 0.43 j | 0.062 defghijklmnopq | 100 | HS |
| 2 | Binamung 2 | 30 ± 3.76 x | 20 | 66.67 a | 23.35 ± 0.63 y | 12 ± 1.15 d | 8 ± 1.15 h | 0.67 h | 0.078 a | 100 | HS |
| 3 | Barimung 7 | 50 ± 5.29 i | 20 | 40.00 op | 25.75 ± 0.88 qrstuv | 11 ± 1.45 e | 9 ± 1.45 g | 0.82 g | 0.062 cdefghijklmnop | 100 | HS |
| 4 | Barimung 4 | 63 ± 2.96 c | 20 | 31.75 t | 24.85 ± 0.49 vwx | 11 ± 0.88 e | 9 ± 0.88 g | 0.82 g | 0.060 ghijklmnopq | 100 | HS |
| 5 | Binamung 7 | 36 ± 4.33 u | 20 | 55.56 d | 25.80 ± 0.28 pqrstu | 6 ± 0.33 j | 14 ± 0.33 b | 2.33 b | 0.068 bcdefg | 100 | HS |
| 6 | Barimung 5 | 48 ± 2.73 j | 20 | 41.67 no | 28.95 ± 0.58 cdefg | 10 ± 0.58 f | 10 ± 0.58 f | 1.00 f | 0.056 mnopqrs | 100 | HS |
| 7 | Nigerian variety | 40 ± 2.19 qr | 20 | 50.00 gh | 26.80 ± 0.65 jklmnopq | 13 ± 0.88 c | 7 ± 0.88 i | 0.54 i | 0.063 bcdefjklm | 100 | HS |
| 8 | DM 2 | 58 ± 2.91 d | 20 | 34.48 s | 26.60 ± 0.62 y | 7 ± 0.58 i | 13 ± 0.58 c | 1.86 c | 0.058 bcdefghi | 100 | HS |
| 9 | Ilangai 2 | 36 ± 3.18 u | 20 | 55.56 d | 25.15 ± 0.82 tuvw | 13 ± 0.88 c | 7 ± 0.88 i | 0.54 i | 0.069 bc | 100 | HS |
| 10 | Ilangai 1 | 32 ± 3.18 w | 20 | 62.50 b | 25.45 ± 0.85 rstuvw | 10 ± 1.33 f | 10 ± 1.33 f | 1.00 f | 0.071 b | 100 | HS |
| 11 | EC 396097 | 42 ± 6.08 op | 20 | 47.62 ij | 27.95 ± 0.69 fghij | 13 ± 1.53 c | 7 ± 1.53 i | 0.54 i | 0.060 hijklmnopq | 100 | HS |
| 12 | HUM 2 | 41 ± 3.79 pq | 20 | 48.78 hi | 26.20 ± 0.70 nopqrst | 7 ± 0.88 i | 13 ± 0.88 c | 1.86 c | 0.064 bcdefghijk | 100 | HS |
| 13 | EC 396099 | 41 ± 2.91 pq | 20 | 48.78 hi | 29.05 ± 0.66 cdef | 12 ± 1.86 d | 8 ± 1.86 h | 0.67 h | 0.058 jklmnopqr | 100 | HS |
| 14 | EC 396103 | 38 ± 3.84 st | 20 | 52.63 ef | 28.15 ± 0.17 efghi | 8 ± 0.88 h | 12 ± 0.88 d | 1.50 d | 0.061 efghijklmnopq | 100 | HS |
| 15 | EC 396107 | 32 ± 5.04 w | 20 | 62.50 b | 27.75 ± 0.88 ghijk | 12 ± 1.53 d | 8 ± 1.53 h | 0.67 h | 0.065 bcdefghijk | 100 | HS |
| 16 | EC 396104 | 48 ± 4.33 j | 20 | 41.67 no | 29.25 ± 0.69 bcde | 11 ± 0.58 e | 9 ± 0.58 g | 0.82 g | 0.055 opqrs | 100 | HS |
| 17 | EC 396114 | 51 ± 3.18 hi | 20 | 39.22 p | 26.15 ± 0.89 nopqrst | 8 ± 0.88 h | 12 ± 0.88 d | 1.50 d | 0.061 fghijklmnopq | 100 | HS |
| 18 | EC 396115 | 48 ± 2.73 j | 20 | 41.67 no | 29.05 ± 0.81 cdef | 12 ± 1.45 d | 8 ± 1.45 h | 0.67 h | 0.056 nopqrs | 100 | HS |
| 19 | EC 396126 | 39 ± 2.85 rs | 20 | 51.28 fg | 27.65 ± 0.69 hijkl | 7 ± 0.33 i | 13 ± 0.33 c | 1.86 c | 0.062 defghijklmnopq | 100 | HS |
| 20 | EC 396100 | 41 ± 2.65 pq | 20 | 48.78 hi | 26.40 ± 0.32 mnopqrs | 11 ± 1.45 e | 9 ± 1.45 g | 0.82 g | 0.064 bcdefghijk | 100 | HS |
| 21 | EC 396121 | 69 ± 1.73 a | 20 | 28.99 u | 25.05 ± 0.92 tuvw | 15 ± 0.58 a | 5 ± 0.58 k | 0.33 k | 0.058 ijklmnopqr | 100 | HS |
| 22 | BDYR 3 | 34 ± 1.53 v | 20 | 58.82 c | 26.05 ± 0.45 opqrstu | 9 ± 1.53 g | 11 ± 1.53 e | 1.22 e | 0.068 bcdef | 100 | HS |
| 23 | EC 396106 | 62 ± 0.88 c | 20 | 32.26 t | 27.75 ± 0.37 ghijk | 12 ± 1.20 d | 8 ± 1.20 h | 0.67 h | 0.054 qrs | 100 | HS |
| 24 | EC 396110 | 46 ± 3.76 kl | 20 | 43.48 mn | 28.45 ± 0.91 defgh | 13 ± 1.33 c | 7 ± 1.33 i | 0.54 i | 0.058 klmnopqr | 100 | HS |
| 25 | EC 396108 | 41 ± 1.15 pq | 20 | 48.78 hi | 28.05 ± 0.50 efghi | 8 ± 1.45 h | 12 ± 1.45 d | 1.50 d | 0.060 ghijklmnopq | 100 | HS |
| 26 | EC 396105 | 56 ± 4.62 ef | 20 | 35.71 rs | 27.00 ± 0.18 ijklmnop | 11 ± 1.20 e | 9 ± 1.20 g | 0.82 g | 0.058 klmnopqr | 100 | HS |
| 27 | EC 396118 | 41 ± 1.76 p | 20 | 48.78 hi | 30.30 ± 0.23 ab | 10 ± 1.45 f | 10 ± 1.45 f | 1.00 f | 0.056 t | 100 | HS |
| 28 | EC 396120 | 57 ± 2.96 de | 20 | 35.09 gs | 26.45 ± 0.42 lmnopqrs | 8 ± 0.88 h | 12 ± 0.88 d | 1.50 d | 0.058 ikmnopqr | 100 | HS |
| 29 | EC 118889 | 39 ± 3.76 rs | 20 | 51.28 f | 26.10 ± 0.56 opqrst | 14 ± 0.33 b | 6 ± 0.33 j | 0.43 j | 0.066 bcdefghij | 100 | HS |
| 30 | AVRDC 1785/5 | 38 ± 1.15 st | 20 | 52.63 ef | 24.40 ± 0.49 wxy | 5 ± 0.33 k | 15 ± 0.33 a | 3.00 a | 0.071 b | 100 | HS |
| 31 | BDYR 2 | 45 ± 1.76 lm | 20 | 44.44 lm | 25.05 ± 0.42 tuvw | 12 ± 0.88 d | 8 ± 0.88 h | 0.67 h | 0.066 bcdefghi | 100 | HS |
| 32 | EC 396101 | 54 ± 3.18 g | 20 | 37.04 q | 26.30 ± 0.56 vwx | 13 ± 0.88 c | 7 ± 0.88 i | 0.54 i | 0.060 bcdefghijklmn | 100 | HS |
| 33 | EC 396102 | 44 ± 1.73 mn | 20 | 45.45 kl | 29.45 ± 0.75 bcd | 6 ± 0.67 j | 14 ± 0.67 b | 2.33 b | 0.056 lmnopqrs | 100 | HS |
| 34 | EC 396111 | 65 ± 2.03 b | 20 | 30.77 tu | 30.00 ± 0.22 bc | 14 ± 0.33 b | 6 ± 0.33 j | 0.43 j | 0.050 s | 100 | HS |
| 35 | EC 396116 | 38 ± 1.45 st | 20 | 52.63 ef | 27.45 ± 0.86 gijklm | 13 ± 1.15 c | 7 ± 1.15 i | 0.54 i | 0.063 cdefijklmno | 100 | HS |
| 36 | EC 396117 | 37 ± 1.45 tu | 20 | 54.05 de | 31.50 ± 0.71 a | 12 ± 1.15 d | 8 ± 1.15 h | 0.67 h | 0.055 pqrs | 100 | HS |
| 37 | EC 396125 | 33 ± 2.03 vw | 20 | 60.61 bc | 27.10 ± 0.26 ijklmno | 11 ± 0.88 e | 9 ± 0.88 g | 0.82 g | 0.066 bcdefghi | 100 | HS |
| 38 | EC 396113 | 63 ± 2.03 c | 20 | 31.75 t | 25.75 ± 0.71 qrstuv | 7 ± 0.88 i | 13 ± 0.88 c | 1.86 c | 0.058 ijklpqr | 100 | HS |
| 39 | EC 396123 | 47 ± 1.45 jk | 20 | 42.55 mn | 23.75 ± 0.20 xy | 10 ± 1.20 f | 10 ± 1.20 f | 1.00 f | 0.069 bcde | 100 | HS |
| 40 | EC 396122 | 42 ± 3.48 op | 20 | 47.62 ij | 25.50 ± 0.45 rstuvw | 13 ± 0.33 c | 7 ± 0.33 i | 0.54 i | 0.066 bcdefghi | 100 | HS |
| 41 | BDYR 1 | 55 ± 2.96 fg | 20 | 36.36 rs | 30.15 ± 0.76 bc | 14 ± 0.88 b | 6 ± 0.88 j | 0.43 j | 0.052 rs | 100 | HS |
| 42 | V2709 | 25 ± 2.40 y | 0 | 0.00 v | 0.00 ± 0.00 z | 0 ± 0.00 l | 0 ± 0.00 l | 0.00 l | 0.000 u | 0 | HR |
| 43 | HG 22 | 32 ± 4.36 w | 20 | 62.50 b | 26.10 ± 0.45 opqrst | 12 ± 0.58 d | 8 ± 0.58 h | 0.67 h | 0.069 bcd | 100 | HS |
| 44 | ML 818 | 42 ± 6.06 op | 20 | 47.62 ij | 27.35 ± 0.75 gijklmn | 13 ± 1.45 c | 7 ± 1.45 i | 0.54 i | 0.061 defghijklmnopq | 100 | HS |
| 45 | VGGRU 1 | 58 ± 3.06 d | 20 | 34.48 s | 25.60 ± 0.19 qrstuvw | 9 ± 1.45 g | 11 ± 1.45 e | 1.22 e | 0.060 hijklmnopq | 100 | HS |
| 46 | ML 1108 | 62 ± 5.13 c | 20 | 32.26 t | 26.15 ± 0.51 nopqrst | 12 ± 0.88 d | 8 ± 0.88 h | 0.67 h | 0.058 klmnopqr | 100 | HS |
| 47 | Basanti | 34 ± 3.21 v | 20 | 58.82 c | 26.55 ± 0.42 klmnopqr | 5 ± 0.58 k | 15 ± 0.58 a | 3.00 a | 0.067 bcdefgh | 100 | HS |
| 48 | KMG 189 | 39 ± 6.06 rs | 20 | 51.28 fg | 25.30 ± 0.92 stuvw | 12 ± 1.15 d | 8 ± 1.15 h | 0.67 h | 0.068 bcdef | 100 | HS |
| 49 | EC 396098 | 40 ± 6.11 qr | 20 | 50.00 gh | 26.05 ± 1.08 opqrstu | 6 ± 0.33 j | 14 ± 0.33 b | 2.33 b | 0.065 bcdefghij | 100 | HS |
| 50 | LM 469 | 43 ± 5.49 no | 20 | 46.51 hjk | 26.20 ± 0.78 nopqrst | 13 ± 1.20 c | 7 ± 1.20 i | 0.54 i | 0.064 bcdefghijkl | 100 | HS |
| 51 | T 1 | 41 ± 6.17 pq | 20 | 48.78 i | 25.75 ± 1.22 qrstuv | 9 ± 0.58 g | 11 ± 0.58 e | 1.22 e | 0.066 bcdefghij | 100 | HS |
| 52 | V2802 BG | 22 ± 3.46 z | 0 | 0.00 v | 0.00 ± 0.00 z | 0 ± 0.00 l | 0 ± 0.00 l | 0.00 l | 0.000 u | 0 | HR |
| Mean | 44.42 | 19.23 | 44.44 | 25.73 | 10.17 | 9.06 | 1.01 | 0.059 | 96.15 | - | |
| SEd | 0.91 | - | 0.96 | 0.61 | 0.24 | 0.19 | 0.02 | 0.004 | - | - | |
| CD (p = 0.05) | 1.81 | - | 1.91 | 1.21 | 0.48 | 0.38 | 0.04 | 0.008 | - | - | |
| Parents | Plant Height | Days to 50% Flowering | No. of Pods/Plant | Pod Length | No. of Seeds/Pod | Hundred Seed Weight | Single Plant Yield |
|---|---|---|---|---|---|---|---|
| Lines | |||||||
| CO 5 | 6.28 ** | 0.31 | 3.44 | 0.15 | 0.03 | −0.14 ** | 0.82 |
| CO 6 | 10.83 ** | −0.03 | 7.11 ** | 0.95 ** | 0.86 ** | 0.09 ** | 5.63 ** |
| CO 7 | 2.31 ** | −0.53 * | −3.56 | 0.32 ** | 0.86 ** | 0.30 ** | 2.24 |
| CO 8 | −4.75 ** | −0.19 | −4.89 * | −0.85 ** | 0.19 | −0.21 ** | −3.50 ** |
| VBN 2 | −8.72 ** | −1.53 ** | 6.11 * | −0.12 | −0.47 | 0.09 ** | 1.70 |
| VBN 3 | −5.95 ** | 1.97 ** | −8.22 ** | −0.44 ** | −1.47 | −0.13 ** | −6.89 ** |
| Testers | |||||||
| V2802BG | 0.03 | −0.36 ** | 0.83 | 0.12 * | −0.25 | 0.01 | 0.05 |
| V2709 | −0.03 | 0.36 ** | −0.83 | −0.12 * | 0.25 | −0.01 | −0.05 |
| SE (Lines) | 1.06 | 0.21 | 2.24 | 0.08 | 0.28 | 0.02 | 1.17 |
| SE (Testers) | 0.61 | 0.11 | 1.29 | 0.05 | 0.16 | 0.02 | 0.67 |
| S.No. | Genotypes | No. of Eggs Laid | Adult Emergence | Adult Emergence Percentage | Mean Developmental Period | No. of Males Emerged | No. of Females Emerged | Female to Male Ratio | Susceptibility Index | Damage Percentage | Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | BSR-GG-1-42-1 | 23 q | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 2 | BSR-GG-1-42-2 | 28 m | 1 f | 3.57 d | 31.00 a | 1 e | 0 e | 0.00 e | 0.018 d | 5.00 e | HR |
| 3 | BSR-GG-1-42-3 | 78 a | 3 d | 3.85 g | 31.67 a | 3 c | 0 c | 0.00 e | 0.019 d | 15.00 d | R |
| 4 | BSR-GG-1-42-4 | 50 f | 7 b | 14.00 f | 31.71 a | 4 b | 3 c | 0.75 c | 0.036 c | 35.00 b | MR |
| 5 | BSR-GG-1-42-5 | 48 g | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 6 | BSR-GG-1-49-1 | 21 rs | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 7 | BSR-GG-1-49-2 | 26 op | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 8 | BSR-GG-1-49-3 | 51 f | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 9 | BSR-GG-1-49-4 | 22 qr | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 10 | BSR-GG-1-49-5 | 23 q | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 11 | BSR-GG-1-56-1 | 32 j | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 12 | BSR-GG-1-56-2 | 28 m | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 13 | BSR-GG-1-56-3 | 36 i | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 14 | BSR-GG-1-56-4 | 67 b | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 15 | BSR-GG-1-56-5 | 60 c | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 16 | BSR-GG-1-97-1 | 27 o | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 17 | BSR-GG-1-97-2 | 27 o | 5 c | 18.52 b | 26.20 c | 3 c | 1 d | 0.33 d | 0.048 b | 25.00 c | MR |
| 18 | BSR-GG-1-97-3 | 38 h | 1 e | 2.63 e | 25.00 d | 1 e | 0 e | 0.00 e | 0.017 d | 5.00 e | HR |
| 19 | BSR-GG-1-97-4 | 58 d | 7 b | 12.07 c | 29.28 b | 2 d | 5 b | 2.50 a | 0.037 c | 35.00 b | MR |
| 20 | BSR-GG-1-97-5 | 30 kl | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 21 | BSR-GG-1-160-1 | 25 p | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 22 | BSR-GG-1-160-2 | 20 s | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 23 | BSR-GG-1-160-3 | 31 jk | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 24 | BSR-GG-1-160-4 | 55 e | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 25 | BSR-GG-1-160-5 | 48 g | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 26 | BSR-GG-1-170-1 | 23 q | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 27 | BSR-GG-1-170-2 | 25 p | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 28 | BSR-GG-1-170-3 | 26 op | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 29 | BSR-GG-1-170-4 | 32 j | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 30 | BSR-GG-1-170-5 | 30 kl | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 31 | BSR-GG-1-198-1 | 39 h | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 32 | BSR-GG-1-198-2 | 29 lm | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 33 | BSR-GG-1-198-3 | 27 o | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 34 | BSR-GG-1-198-4 | 60 c | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| 35 | BSR-GG-1-198-5 | 32 j | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| Parents | |||||||||||
| 36 | CO6 | 48 g | 20 a | 41.67 a | 26.10 c | 9 a | 11 a | 1.22 b | 0.062 a | 100.00 a | HS |
| 37 | V2802 BG | 16 t | 0 f | 0.00 h | 0.00 e | 0 f | 0 e | 0.00 e | 0.000 e | 0.00 f | HR |
| Mean | 36.33 | 1.00 | 2.21 | 5.72 | 0.49 | 0.46 | 0.11 | 0.01 | 4.86 | - | |
| SEd | 0.91 | 0.11 | 0.07 | 0.22 | 0.02 | 0.03 | 0.01 | 0.00 | 0.57 | - | |
| CD (p = 0.05) | 1.82 | 0.22 | 0.13 | 0.44 | 0.05 | 0.05 | 0.02 | 0.00 | 1.13 | - | |
| S.No. | Parents and Resistant Lines | Length (mm) | Breadth (mm) |
|---|---|---|---|
| 1. | BSR-GG-1-49-3-1 | 0.19 b | 0.12 b |
| 2. | BSR-GG-1-56-2-2 | 0.20 b | 0.14 b |
| 3. | BSR-GG-1-160-5-3 | 0.20 b | 0.11 b |
| 4. | BSR-GG-1-170-2-4 | 0.22 b | 0.13 b |
| 5. | BSR-GG-1-198-1-4 | 0.21 b | 0.13 b |
| 6. | CO 6 | 4.05 a | 2.65 a |
| 7. | V2802 BG | 0.21 b | 0.13 b |
| Mean | 0.75 | 0.49 | |
| SEd | 0.04 | 0.01 | |
| CV% | 6.41 | 3.56 | |
| S.No. | Genotypes | PH | DFPF | PPP | PL | SPP | HSW | SPY |
|---|---|---|---|---|---|---|---|---|
| 1 | BSR-GG-1-49-3-1 | 64.23 ± 6.31 | 35.33 ± 0.33 | 41.33 ± 1.76 | 8.27 ± 0.15 | 13.00 ± 0.58 | 3.95 ± 0.02 | 16.71 ± 0.66 |
| 2 | BSR-GG-1-56-2-2 | 60.17 ± 4.13 | 35.67 ± 0.33 | 43.67 ± 3.38 | 7.83 ± 0.12 | 12.67 ± 0.33 | 4.04 ± 0.05 | 18.18 ± 0.83 |
| 3 | BSR-GG-1-160-5-3 | 62.40 ± 2.15 | 34.67 ± 0.33 | 45.67 ± 2.73 | 8.53 ± 0.06 * | 13.00 ± 0.58 | 4.02 ± 0.06 | 19.70 ± 1.41 |
| 4 | BSR-GG-1-170-2-4 | 61.97 ± 2.44 | 35.67 ± 0.33 | 44.00 ± 2.52 | 7.73 ± 0.15 | 12.33 ± 0.33 | 3.98 ± 0.03 | 16.31 ± 0.50 |
| 5 | BSR-GG-1-198-1-4 | 62.73 ± 3.46 | 36.33 ± 0.33 | 48.33 ± 2.60 | 8.23 ± 0.15 | 12.67 ± 0.33 | 3.90 ± 0.04 | 18.72 ± 1.00 |
| 6 | CO 6 | 60.62 ± 2.66 | 35.67 ± 0.33 | 42.33 ±2.03 | 7.33 ± 0.06 | 12.33 ± 0.33 | 3.79 ± 0.04 | 15.99 ± 0.34 |
| Mean | 62.02 | 35.56 | 44.22 | 7.99 | 12.67 | 3.95 | 17.60 | |
| SEd | 5.45 | 0.41 | 3.56 | 0.11 | 0.62 | 0.06 | 1.31 | |
| CD (p = 0.05) | 12.15 | 0.92 | 7.93 | 0.24 | 1.37 | 0.14 | 2.91 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samyuktha, S.M.; Malarvizhi, D.; Mariyammal, I.; Karthikeyan, A.; Seram, D.; Dhasarathan, M.; Juliet Hepziba, S.; Sheela, V.; Thanga Hemavathy, A.; Kavithamani, D.; et al. The Hunt for Mungbean (Vigna radiata (L.) Wilczek) Genotypes and Breeding Lines Resistance to South Indian Bruchid Strain. Agriculture 2022, 12, 1050. https://doi.org/10.3390/agriculture12071050
Samyuktha SM, Malarvizhi D, Mariyammal I, Karthikeyan A, Seram D, Dhasarathan M, Juliet Hepziba S, Sheela V, Thanga Hemavathy A, Kavithamani D, et al. The Hunt for Mungbean (Vigna radiata (L.) Wilczek) Genotypes and Breeding Lines Resistance to South Indian Bruchid Strain. Agriculture. 2022; 12(7):1050. https://doi.org/10.3390/agriculture12071050
Chicago/Turabian StyleSamyuktha, Santhi Madhavan, Devarajan Malarvizhi, Irulappan Mariyammal, Adhimoolam Karthikeyan, Devina Seram, Manickam Dhasarathan, Sundarrajan Juliet Hepziba, Venugopal Sheela, Arumugam Thanga Hemavathy, Duraisamy Kavithamani, and et al. 2022. "The Hunt for Mungbean (Vigna radiata (L.) Wilczek) Genotypes and Breeding Lines Resistance to South Indian Bruchid Strain" Agriculture 12, no. 7: 1050. https://doi.org/10.3390/agriculture12071050
APA StyleSamyuktha, S. M., Malarvizhi, D., Mariyammal, I., Karthikeyan, A., Seram, D., Dhasarathan, M., Juliet Hepziba, S., Sheela, V., Thanga Hemavathy, A., Kavithamani, D., Kavitha, S., & Senthil, N. (2022). The Hunt for Mungbean (Vigna radiata (L.) Wilczek) Genotypes and Breeding Lines Resistance to South Indian Bruchid Strain. Agriculture, 12(7), 1050. https://doi.org/10.3390/agriculture12071050








