Abstract
Jatropha curcas is a woody-shrub species of the Euphorbiaceae family that is widely distributed in tropical and subtropical areas. The great interest in its cultivation lies in the potential for achieving elevated yields of a high-quality oil. Another characteristic that makes J. curcas promising is its ability to produce green energy even in high-salinity soils. For a commercial cultivation to be considered effectively competent to withstand these conditions, it must produce enough to offset production costs. There is no doubt that J. curcas is considered promising, but numerous pilot projects for the commercial planting of J. curcas have failed worldwide, mainly due to a lack of reliable scientific knowledge about the species, its food security, and (mainly) its instability in commercial fruit production. The main goal of this review was to compile published results on tolerance/resistance or sensitivity to salt stress in J. curcas. Updating the knowledge on this theme may allow for researchers to trace strategies for future studies of stress physiology in this promising oil seed species.
1. Introduction
Nine out of ten of the world’s most polluted cities are located in India [1]. The larger the population, the greater the demand for energy, which is commonly obtained from petroleum products or burning coal [2]. In this context, greenhouse gas emissions (GHGs) are the driving force for global climate change. Deforestation accounts for over 20% of the world’s GHG emissions [3]. It is of utmost importance to restore deforested while recycling carbon [3]. Environmental, economic, and social issues have led to the recent trend of using renewable sources of energy worldwide [4], so biomass has increased in popularity and acquired a significant share of the global energy mix over a relatively short time. However, several biomass resources have triggered wide criticism for compromising food resources, agricultural lands, and fresh water to produce energy crops [5], but from environmental and socioeconomic perspectives, biomass is easily available, non-edible, and resilient to climatic variations, thus rendering is a potential biofuel capital for a cleaner and greener future [1]. It was recently projected that the 100.00 (Megatons; MT) of diesel consumed in 2019–2020 [1] could reach 143 and 191 MT in 2025 and 2030, respectively, based on an increase of 6% per annum. Jet fuel is a major product, with an annual production of 253.06 MT, followed by gasoline, fuel gas, and diesel [6]. According to Achten, Verchot [7], Jatropha curcas can absorb 30 tons of carbon per year, and besides substituting biofuels for fossil fuel, the seed shells of J. curcas have a high energy value (~18–19 MJ kg−1). Both husks and shells are not suitable as substrates in biogas digesters due to low digestibility [8]. Unblended 100% J. curcas methyl esters were extensively tested on the road in India with modern Mercedes cars. A total of 80,000 liters were used in these tests, and the overall results were highly satisfactory [8].
J. curcas has emerged as one of the most promising biomass feedstocks for jet biofuel (JBF) production, since it is non-edible and able to grow in non-arable lands with minimal water and energy requirements [6]. However, oil produced by J. curcas seeds remains distant and noncompetitive with conventional Jet-A fuel available in the market of comparable price. Klein-Marcuschamer, Turner [9] investigated the potential of microalgae, Pongamia, and sugarcane in JBF production, and they found minimum selling prices of 2.42, 1.60, and 1.06 $/kg, respectively. Furthermore, Wang [10] reported a minimum JBF selling price of 1.14 $/kg. Moreover, Tao, Milbrandt [11] studied four oil-bearing crops including Jatropha, camelina, pennycress, and castor, among which Jatropha achieved the lowest production cost with a minimum JBF selling price of around 0.40 $ kg−1—well below what IATA [12] presented in its 2021 bulletin (~0.60–0.64 $/kg).
Since the passing of related legislation [13], Brazil has seen a consistent increase in the percent of biodiesel mixed with diesel sourced from fossil fuels, from 2% in 2008 to 20% in 2025 [14]. Nevertheless, the main raw material used for biodiesel production in Brazil is soybean oil (68% in 2019), followed by animal fat (11%) [15]. However, soybean is a versatile crop that is especially important for human and animal nutrition, and J. curcas does not compete with the food market and could be an option to be explored. Considering that J. curcas has a productive life of approximately 50 years [16] and that it attains maximum productivity after 4–5 years, the necessary level of production is achievable [7]. Moreover, this species has shown the ability adapt to different agroclimatic conditions [17,18], including infertile soils, making it suitable for cultivation on degraded soils while protecting them from erosion [19]. Such characteristics make the cultivation of J. curcas suitable for familiar agriculture in marginal lands, where it could improve the standard of living and socio-economic status of farmers, particularly in developing countries [20].
The production of seeds in J. curcas starts around 6 months after planting [21], and they achieve maximum annual yields of 9–39 tons of seeds ha−1 at 4 years after planting [22]. At the best stage of physiological maturity of the fruits, the seeds are expected to contain 36% moisture (humidity basis) [23] and up to 45% oil in the kernel [24]. Thus, it is possible to estimate the potential productivity of 25 tons of dry seeds ha−1 (64% of 39 tons), and 5.2 tons oil ha−1 year−1 (45% of 15.4 tons when assuming 75% of extraction efficiency) or 5600 L oil ha−1 year−1 (assuming a density of 0.92 kg L−1) [25]. Despite recent results supporting the idea that the productivity of seeds declines six years after planting [22], there have been other estimates of a 50-year productive life of J. curcas, thus strengthening the great production potential of the species under ideal conditions. To put it in perspective, soybean—the most cultivated oilseed crop in world, food purposes included—produces 675 L oil ha−1 on average a year [26].
In April 2013, SG Biofuels (a privately held bioenergy crop company that grows and researches J. curcas for the production of biodiesel, biojet fuel, and specialty chemicals) signed up for more than 101,150 hectares in various field trial and deployment agreements—including an agreement for the trial of J. curcas with Bharat Petroleum in India with 34,803 ha for the first phase commercial deployment following the trials. A similar 30,350 ha deal was set up in Brazil with a consortium including JETBIO, Airbus, the Inter-American Development Bank, Bioventures Brazil, Air BP, and TAM Airlines. Moreover, during the last decade, numerous Jatropha projects have been implemented in Asia, Africa, and Latin America. The National Research Institute for Forestry, Agricultural and Livestock estimated that the potential lands for Jatropha cultivation in Mexico comprised more than 2.6 million hectares. Given this information, in 2007, various southeastern states in Mexico planned to cultivate thousands of hectares (ha), including Veracruz (200,000), Yucatán (6000), Michoacán (6000), and Chiapas (20,000) [27]. The goals of these projects have been diverse, but promoting sustainable rural development, reducing energy dependency, and GHG emissions have been frequent aims. However, doubts have recently been cast on the profitability of J. curcas and the financial viability of its cultivation [27,28].
The present review is a compilation of recent results regarding the effects of salt stress on the physiological, biochemical, and molecular aspects of the energy species J. curcas. Our main objectives were to provide updates on the state of of the issue and to propose new directions for future research on the stress physiology of J. curcas. Why has a plant species with tremendous apparent capacity to generate biodiesel and the possibility of being a world leader in the achievement of green sustainability been the target of so much criticism? Why have many projects worldwide been abandoned before completion? What are the agronomic, physiological, and cellular challenges that we must address for the cultivation of the species? Which strategies are worth investigating and how can molecular biology help to understand the studied processes? This review is intended to clearly present these and other ideas, as well as to unify the extensive published data on the species.
2. Physiological, Biochemical, and Molecular Responses of Jatropha curcas L. to Abiotic Stress
Plants subjected to water deficits usually suffer from both ‘thirst’ (decreased water uptake and transport) and ‘starvation’ (decreased carbon and nutrient uptake). Indeed, due to the fast and efficient stomatal control of transpiration, rather than thirst, starvation is the main cause of drought stress-induced reductions in growth and biomass accumulation in J. curcas. Investigations into the influence of salt-stress deficits on the physiology of J. curcas have reported consistent salt-induced stomatal closure, followed by decreases in transpiration (E) and net photosynthesis (PN) [29,30,31,32,33].
The duration of salt stress determines different plant responses in terms of the velocity and severity of the effects on leaf gas exchange variables and the velocity and degree of recovery after stress release. That was clearly demonstrated by Corte-Real, Miranda [21], who observed the fast and complete or slow and incomplete recovery of leaf gas exchange variables after a cycle of rapid salt-stressor and slow water-deficit treatments. The authors pointed out that rapid salt stress-induced stomatal closure, which kept the leaf water status almost stable, was responsible for the quickly recovery of leaf gas exchange variables when the plants were removed from the saline solution [21].
Osmotic adjustment (OA) is a physiological phenomenon observed in some plant species facing osmotic stress caused by salinity or water deficit [34]. OA consists of the active accumulation of organic and inorganic solutes in plant cells (Figure 1), which helps to attract water into the cell, thus maintaining positive turgor pressure [35]. Several types of solutes, such as amino acids, proline, glycine betaine, sugars (e.g., sorbitol, pinitol, and glycerol), and phenolic compounds accumulate in OA under osmotic-limited conditions.
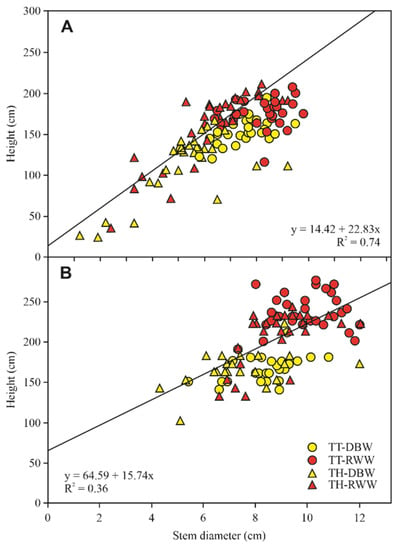
Figure 1.
Stem diameter (A) and height (B) of Jatropha curcas at two different stages of cultivation and under different treatments. Figure adapted from [68].
Many authors [32,36,37,38] have described proline as an osmoregulator and as being promoted by water and salt stress. However, more recent studies [38,39,40,41] have shown that proline, as a key role in plants under drought and salt stresses, not only acts as an osmoprotectant but also functions as an antioxidant. In fact, the synthesis of proline comes from an alternative route of metabolizing glutamate, which is phosphated (by 1 mole of ATP) and reduced to proline from the consumption of 2 moles of NADPH [42]. As a consequence, a stressed plant tends to proportionally reduce the stomatal opening, which limits the entry of CO2 into the system [29,43]. With less CO2 in the system, the Calvin–Benson cycle is compromised and the ATPs and NADPHs that were formed and continue to form through chloroplastidic electron chain transport (ECT) remain in their reduced forms [44,45,46]. Assuming that ATP and NADPH remain in their reduced forms, there would theoretically be no oxidized forms to receive more electrons from ECT; in this case, the electrons would be quickly captured by O2, forming the terrible reactive oxygen species (ROS) [47]. It is in this pathway that proline acts because, as previously reported for the formation of 1 mole of proline, 1 mole of ATP and 2 moles of NADPH are spent and the synthesis of proline enters the system as a stress preventer, not as an osmoregulator. The role of proline is similar to the role of photorespiration, which, in the absence of CO2, can promote the re-oxidation of chloroplast ATPs and NADPHs and (in the same way as explained above) can serve as an escape valve that allows mesophyll cells to continue to promote photosynthesis even with stomatal opening [48] and its consequent compromised CO2 entry.
In addition to proline and soluble sugars, such as pinitol and sorbitol, glycerol can also contribute to osmotic adjustment in plants [49]. The accumulation of potassium (K+) is an effective contribution of the ion to osmotic adjustment. Nevertheless, genotype variation in OA and in osmolyte accumulation has been documented for both R. communis [50] and J. curcas [51], which may be useful in breeding programs.
Tominaga, Inafuku [52] revealed that J. curcas is able to preserve the integrity of the photosystem (PS) II when the stomata are partially [29,32] closed in dry conditions, thus regulating the thermal dissipation and adjusting the quantum efficiency of the PSII to the CO2 fixation capacity. ROS production during abiotic stress is the main cause of protein oxidation and lipid peroxidation, generating toxic molecules, including malondialdehyde (MDA) [18,53] and substances reactive to thiobarbituric acid (TBARS) [54,55,56,57]. In J. curcas leaves, the content of MDA was found to be 1.85-fold higher in 3.5 dS m−1 of electrical conductivity compared to a control; in consequence, the electrolyte leakage was found to be 1.29-times higher in the same treatment [32].
Water-stressed plants have developed other mechanisms, including antioxidant defense systems, with enzymes such as superoxide dismutase (SOD; EC 1.15.1.1), peroxidase (POD; EC 1.11.1.7), and catalase (CAT; EC 1.11.1.6). While SOD acts on reduced oxygen derivatives (e.g., superoxide radical, O2•-, singlet oxygen, and 1O2 and hydroxyl radical HO) to form H2O2, CAT and APX enzymes act in the dismutation of H2O2 into H2O and O2. In addition, plants can use non-enzymatic antioxidants consisting of low molecular-weight metabolites such as carotenoids, ascorbic acid, glutathione, tocopherols, and phenolic compounds [58], which increases their ability to grow and develop in significant salinity conditions. The balance between ROS production and antioxidant activity will determine different responses in plant cells, such as signaling and oxidative stress [59]. Increases in SOD, CAT, and ascorbate peroxidase (APX) activities would demonstrate the strong participation of the enzyme system in J. curcas induced by water deficits for 4, 8, and 18 days, in addition to aiding the process of recovery from drought (Pompelli et al., 2010). In turn, the maintenance of redox homeostasis can contribute to plant drought tolerance [60].
3. How Does Salinity Modulate Physiological, Biochemical and Molecular Responses in J. curcas?
As soil salinity increases, the extraction of water from the soil becomes increasingly difficult for plants [61]. Surprisingly, an estimated one to two percent reduction in the world’s arable area is observed yearly as an effect of salinization, indicating the severity and importance of secondary salinization. In addition, salinity has often been associated with the decreased yield and stability of some crop species [62].
3.1. Advantages and Disadvantages of Wastewater in Agriculture
In the global context, it is evident that recycled urban wastewater (RWW) in agriculture will increase in water-scarce countries in the near future, particularly in the vicinity of cities [63,64]. Irrigation with RWW has been used for three purposes: (i) as a complementary treatment method for wastewater; (ii) as an available water source for agriculture—a sector demanding ~70% of the consumptive water use in Brazil; and (iii) as a nutrient source associated with mineral fertilizer savings and high crop yields [16,65]. For this role, Brazil deserves special attention, since a record export of agricultural products such as soybeans, corn, cellulose, sugarcane, and coffee that should exceed 80.87 billion tons is expected for 2022 [66]. Thus, if Brazil could replace 10% of freshwater with wastewater in agriculture, a few billion mm3 of freshwater would be spared, thus making Brazil the focus of attention for the gradual use of RWW instead of freshwater, not for its ecological disasters [64]. Various studies have shown that the soil application of RWW as a water and nutrient source for agricultural irrigation represents a low-cost alternative [67] and is applicable in both dry and humid regions. Hence, irrigation with RWW has practical importance regarding nutrient availability, especially in the acidic soils of Brazil that have inherently low natural fertility [65]. In Morocco, 700 mil m3 of RWW was discharged without treatment, an amount that is predicted to increase to 900 mil m3 in 2020 [63], and just about 70 mil m3 are being re-used each year. The benefits of RWW use additionally include the conservation of fresh water reserves, reductions in the discharge of pollutants into water bodies, potential for the recycling of both water and nutrients that otherwise would have been disposed of into the environment and consequently contaminate natural water bodies, and the supply of nutrients to crops due to the organic matter and nutrients present in this water [68,69].
Clay-loam soils, generally with higher concentrations of nutrients than sandy soils, may be more suitable for J. curcas cultivation [70]. Many scholars have also noted the preference of J. curcas for sandy soils, which are more favorable to root development than clay soils [16,65]. Dorta-Santos, Tejedor [68] cultivated J. curcas plants in two soil types, sandy-loam soil (TT) and clay-loam soil (TH), plus two irrigation forms, RWW and desalinated brackish groundwater (DBW) (Figure 1). These authors showed that relationship between the base diameter and height of the plants indicated that both morphometrical variables were significantly affected by the soil and water factors but not by the interaction between them. Thus, for the same soil type, irrigation using RWW proved better for plant growth than DBW irrigation, and for the same type of water, the TT proved more favorable than TH. Regarding seed production, these authors reported that production under RWW irrigation increased 12-fold with respect to that obtained using DBW (mean ~2258 versus 197 kg·ha−1). Additionally, due to the plant’s relatively wild nature and the enormous gaps in knowledge about its genetic background and optimized agronomic practices, many J. curcas projects have failed to reach commercially viable yields [28,71] and were subsequently abandoned.
3.2. Sodium (Na+) and Chloride (Cl−) Compartmentalization Related to Salt Stress Tolerance
Concerning appropriate soil characteristics for growing J. curcas, the data published by different authors have often proven contradictory. Some authors have considered J. curcas to be a salt-sensitive plant [21,37,63,72], while others have considered J. curcas to be moderately sensitive [73] or moderately tolerant to salinity [63,68,74,75,76,77]. Accordingly, the potential of J. curcas in terms of tolerance to salinity remains controversial. This divergence may be related to the diversity of physiological responses among genotypes in different studies and a lack of high yielding variety. An understanding of J. curcas’s response mechanisms under salt stress would thus provide valuable information for improving stress tolerance. It seems plausible that understanding the physiological and molecular mechanisms underlying salt stress resistance (SSR) is a promising and cost-effective strategy for developing highly resistant cultivars.
The adverse effects of salinity are the results of three primary factors: first, the inhibition of water uptake caused by the low water potential of the soil; second, the toxic effects of the Na+ and Cl− ions at the cellular level; and third, the alteration of the nutritional balance, resulting in high ratios of Na+/Ca2+, Na+/K+, Na+/Mg2+, Cl−/NO3−, and Cl−/H2PO4−. Plants have developed complex mechanisms for the avoidance of salinity stress. These mechanisms include (i) the accumulation of significant amounts of ions to ensure sufficient water uptake (a process known as osmotic adjustment; Figure 2A), (ii) the ability to avoid ion translocation from the root to the aerial part, (iii) the compartmentalization of toxic ions within the cell, and (iv) a mechanism of Na+ exclusion via export across the plasma membranes [35,78,79,80]. In seedlings, increasing salt levels were found to result in a consistent decrease in total water loss associated with an increase in Na+ concentration in leaves, while in 3-year-old J. curcas, the total water loss of plants treated with 25 mmol of NaCl was higher than that of control plants and then decreased at higher NaCl levels until 100 mmol. The total water loss values at high NaCl treatments (100, 150, and 200 mmol of NaCl) were lowest and of similar magnitude [63].
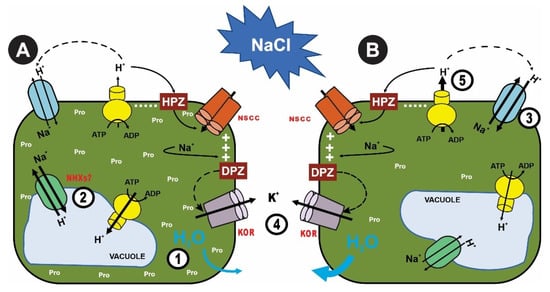
Figure 2.
(A) Osmotic adjustment through the accumulation of compatible solutes such as proline in leaves and roots to prevent the loss of water (1). Na+ compartmentalization in vacuole-like structures suggests the involvement of vacuolar and endosomal Na+/H+ antiporters (NHX) (2). (3) Na+ extrusion or efflux from the cytosol to the apoplast by SOS1 exchangers (3). (B) Regulation of K+ homeostasis through K+ retention in roots is crucial in plants (4). (5) Increase in the H+-ATPase activity to cell membrane repolarization or Na+ extrusion. KOR: outward-rectifying K+ channels (5). HPZ is hyperpolarization, and DPZ is depolarization. Modified from the work of Quintal, Velarde-Buerdía et al. (2014).
The progressive accumulation of salt in photosynthetically active mesophyll tissues leads to the further inhibition of CO2 assimilation because it affects the processes of photosynthesis in chloroplasts. This involves suppressing the activity of photosynthetic enzymes of the Calvin cycle, disturbing chlorophyll biosynthesis, and reducing the efficiency of the operation and structural integrity of the photosynthetic apparatus and thylakoid membranes (revised by Pan, Liu [81]). These authors presented a table describing some key photosynthetic enzymes and structural proteins, including a substantial increase in cytosolic and stromal Na+ concentrations that inhibit many key metabolic processes such as protein synthesis and oxidative phosphorylation. Increased cytosolic Na+ levels also disturb intracellular K+ and Ca+2 homeostasis in leaf mesophylls, thus affecting photosynthesis and light signal transduction [81].
How to improve plant salt stress tolerance has become a focus of plant biology research. Some authors have reported that Cl− uptake and transport in plants is less controlled than those of Na+ [80]. The result is a wide range of physiological and biochemical changes leading to the inhibition of growth and development; reductions in photosynthesis, respiration, and protein synthesis; and the disruption of nucleic acid metabolism [82]. To survive salt stress, plants respond and adapt through sophisticated mechanisms. For this, modifications of membrane transport, signal transduction, redox reaction, and other processes have been shown to be involved in the salt-stress response [36].
Corte-Real, Miranda [21] studied the effect of NaCl on growth and gas exchange in J. curcas, and they reported that these parameters were more seriously affected in salt-treated plants, though this effect were dose- and genotype-dependent. However, even the plants that were the most affected by NaCl could be restored after 6 days in recovery conditions, suggesting that J. curcas is moderately sensitive to salt. This finding was in agreement with that of Cabral, Binneck [74], who first expressed genes after a short salt stimulus and showed that the expression of a protoplast inner protein in J. curcas (JcPIP1 and JcPIP2) was significantly decreased in the roots of J. curcas plants treated with 150 mM of NaCl. In J. curcas, salt tolerance was achieved by silencing of the JcPIP1 and JcPIP2 using tobacco rattle mosaic virus (TRV)-based on virus-induced gene silencing (VIGS). At 3 days after infiltration with TRV:JcPIP1 or TRV:JcPIP2, the leaves of J. curcas plants became yellowish, and 10–18 days later, the leaves of the treated plants displayed a withering phenotype [83]. Plants infiltrated with TRV:JcPIP1 or TRV:JcPIP2 displayed necrosis spots on the leaves under a salt stress condition; Azevedo, Santos-Rosa [84] described upregulated roots, the plasma-membrane-intrinsic protein PIP2;2, and four tons of plastic. These authors also described the putative 1-aminocyclopropane-1-carboxylate oxidase 4 and S-adenosylmethionine synthetase (SAM2) associated with the ethylene biosynthesis pathway.
3.3. Effect of Salt on Cellular Metabolism and Defense
Among all salt-tolerance mechanisms, the exclusion of Na+ and Cl− from the cytosol via compartmentalization into vacuoles has been most frequently observed in glycophyte species [78,79,85]. The overaccumulation of excessive Na+ in the plant cytoplasm can be prevented using three main strategies in plants: (i) inhibiting Na+ influx, (ii) elevating vacuolar Na+ compartmentation, and (iii) enhancing Na+ efflux (Figure 3). The adaptive response to salinity is multigenic in nature; however, a single gene can also increase the salt tolerance of a plant species [86]. When plants suffer from high salinity, Na+ can enter cells through non-selective cation channels (NSCCs), K+ permeases, and other transporters. Na+/H+ antiporters are expressed by functional gene(s), play an important role in plant salt tolerance, and are examples of single genes that can increase salt tolerance [35,87]. Ion homeostasis is carried out either via the sequestration of excess sodium into vacuoles via vacuolar Na+/H+ antiporters (NHX1) energized by the proton gradient generated by vacuolar membrane H+-ATPase and H+-pyrophosphatases [87,88] or via active exclusion through Na+/H+ antiporters in salt overly sensitive (SOS) 1 located on the plasma membrane [86].
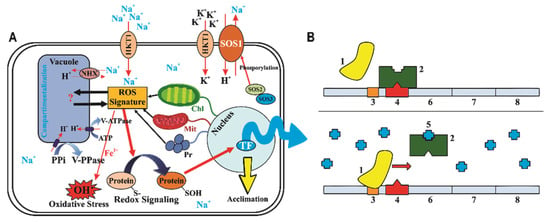
Figure 3.
(A) The interaction between reactive oxygen species (ROS) produced from Na+ signals. The ROS signature, promoted by an unknown pathway, compartmentalizes Na+ into vacuoles. In the tonoplast, H+ pumps equilibrate this compartmentalization. The ROS signature promotes the upregulation of a protein oxidation system that connects to the nucleus, promoting transcription factor (TF) activation and promoting some regulation of mitochondria (Mit), chloroplasts (Chl), and peroxisomes (Pr). In another way, the SOS1 channel in the plasma membrane promotes an antiporter flux of Na+ and H+. Na+ and K+ compete with the same plasma membrane channel (HKT1). (B) Model proposed for the interaction between NaCl and catalase (CAT) gene expression. On non-salt-stressed plants (upper panel), the RNA polymerase (1) does not translate the CAT genes because there is a repressor (2) occupying the operon (4) on the CAT-encoding gene. Therefore, in salt-stressed plants (bottom panel), the H2O2 (5) binds to repressor that remains inactive and then dissociates from the CAT-encoding gene. Afterwards, the RNA polymerase binds to promoter region (3) on the CAT-encoding gene and then promotes the translation of CAT1 (6), CAT2 (7), and CAT3 (8). Modified from the work of Choudhary, Rivero [85] and Muchate, Nikalje [79].
Jha, Mishra [87] expressed the SbNHX1 from Salicornia brachiata in J. curcas and confirmed a salt tolerance of up to 200 mM of NaCl through the estimation of different growth parameters, such as increased sodium and chlorophyll levels, reduced rates of electrolyte leakage, and less malondialdehyde in the leaves, all indicative of reduced oxidative damage in these transgenic plants. H+-ATPase also provides the driving force for potassium retention, which is also driven by sodium exclusion through NHX exchangers. The differential expression/function of SOS1 and HKT (high-affinity potassium transporters) depends on habitat, demonstrating the complexity of salinity tolerance [89]. However, the development of transgenic J. curcas plants via the overexpression of selected salt-responsive gene(s) is a better choice for the genetic improvement of the plant regarding salt tolerance and breeding [87].
Several halophyte genes associated with cation/proton antiporters on the plasma membrane (SOS1), vacuolar membrane (NHX), plasma membrane, vacuolar H+-ATPases, vacuolar H+-pyrophosphatase, and potassium transporters (as well as other genes involved in salt-tolerance mechanisms apart from defined pathways, i.e., crosstalk, osmolytes, antioxidant enzymes, salt glands, and bladders) are the current targets of researchers seeking strategies to transform glycophyte plants so that they can tolerate high salinity. Ubiquitous membrane proteins that catalyze the electroneutral exchange of Na+ or H+ across the membrane, thereby playing important roles in cellular Na+/K+ and pH homeostasis, are the most studied mechanisms to cope with salinity (Figure 2A) [79,85]. Among the five identified SOS genes, only three genes (SOS1, SOS2, and SOS3) are involved in the salt-signaling pathway for salt tolerance in Arabidopsis [38]. It is probable that vacuolar antiporters are more suitable and promising genes for genetic transformation because the entire process takes place in the cell itself, unlike SOS1, which has shown different functions depending on the external saline environment [79,85]. The removal and sequestration of Na+ and the retention and uptake of K+ from the cytoplasm are the mechanisms that plants use to reduce excess Na+ or maintain K+ content, respectively. In addition, the genes involved in the synthesis of compatible solutes or osmoprotectants such as proline, glycine-betaine, and inositol may comprise another interesting tolerance mechanism to protect cells from salinity damage [90].
The regulation of Na+ plasma membrane signaling (HKT1) by calcium [79], phosphorylation, hormones such as nitric oxide (NO) [78] and calcium-dependent protein kinase (CDPKs) [79], or NADPH availability may generate signaling ROS at the apoplast. This signaling ROS then moves into the cytoplasm via regulated aquaporins, and together with the metabolic and signaling ROS produced in the chloroplast, peroxisome, and mitochondria (Figure 2A), they alter the redox status of key regulatory proteins such as transcription factors (TFs) affecting gene expression, thus promoting the expression of specific genes such as SOS2 and SOS3 that activate via phosphorylation with SOS1 (Figure 3A) to exchange Na+/H+ antiporters and CAT in the plasma membrane [79,85]. NO mediates the post-translational modification of target proteins through S-nitrosylation and nitration. Under water-deficit conditions, induced or not by salt stress, ABA induces NO and ROS synthesis. Both NO and ROS form 8-nitro-cGMP-inducing stomata closure that, in turn, reduces CO2 assimilation [91]. S-nitrosylation, the covalent binding of NO to thiol groups of cysteine, is a post-translational modification that can regulate the function of some enzymes involved in respiration, antioxidation, and photorespiration during salinity stress [92]. Gadelha, Miranda [78] reported that NO-pretreated J. curcas seedlings presented a significant mitigation of salt stress due to low toxic ion and ROS accumulation via powerful antioxidant systems, resulting in the elevated growth of NO-treated seedlings under saline conditions (Figure 4). Moreover, salt stress was found to promote significant increases in lipid peroxidation and H2O2 contents at all analyzed time points and plant tissues; nevertheless, the NO pre-treatment effectively alleviated oxidative damage and ROS accumulation in J. curcas seedlings [78] (Figure 3). These findings suggest that the improved K+/Na+ of NO-treated stressed plants could be related to restrictions in Na+ influx across the root plasma membrane or to vacuolar compartmentalization, contributing to enhanced salt tolerance [79]. Gadelha, Miranda [78] suggested that exogenous NO can also partially prevent lipid peroxidation and ROS accumulation, thereby alleviating the oxidative damage normally caused by salinity stress.
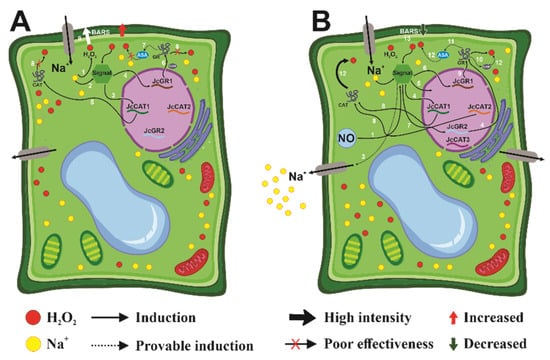
Figure 4.
Model proposed for responses to salt stress. (A) Upon salt stress exposure, (1) the accumulation of sodium (Na+—yellow circles) inside the cells promotes an increase in hydrogen peroxide (H2O2—red circles) content. In parallel, (2) a still-unknown signaling pathway recognizes the H2O2 and Na+ accumulation and triggers an upregulation of (3) JcCAT1 and (4) JcGR1 gene expression, resulting in improved (5) catalase (CAT) and (6) glutathione reductase (GR) activities, respectively. The enhanced GR activity is caused by a greater (7) ascorbate (ASA) content. Nonetheless, these responses are not effective (8) for H2O2 scavenging, resulting in severe damage to (9) membrane lipids. (B) Yet, NO priming (1) quickly activates the signaling pathway, reprogramming the salt responses. First, (2) it recognizes Na+ accumulation and seems to activate (3) the effective extrusion of Na+ back into the medium. Secondly, it induces the upregulation of (4) JcCAT1, (5) JcCAT2, (6) JcGR1, and (7) JcGR2 gene transcripts, which better direct (8) CAT and (9) GR activity. At the same time, the GR increases the contents of (10) glutathione (GSH) and (11) ASA. Finally, (12) the H2O2 is almost scavenged, (13) avoiding damage to membrane lipids in embryo axis cells and enhancing the salt tolerance of J. curcas seedlings. Modified from the work of Gadelha, Miranda [78].
Plants have several strategies to protect the photosystems against photoinhibition and photodamage, such as downregulation in light harvesting, excess energy dissipation by non-photochemical quenching, increases in water–water cycle and cyclic electron flow, and other important biochemical pathways such as the ascorbate–glutathione cycle, photorespiration, and nitrate and ammonia assimilation. The role of the efficiency of these processes in salt tolerance is possibly species-dependent, but the involved mechanisms are not understood [93]. The PSII D1 protein is the specific light-induced protease required to remove damaged D1 proteins, which are replaced with new copies produced by a de novo synthesis process termed reversible photoinhibition. At the molecular level, salt stress affects the repair via inhibiting the expression of the psbA genes for preD1 at both the transcriptional and the translational levels [94]. Salt stress also inhibits the degradation of the D1 protein in the photodamaged PSII. Similar results have also been reported for higher plants (reviewed in [81]). The involvement of the nitrate assimilation pathway is also closely linked to photosynthesis because nitrite and ammonia assimilation may consume electrons from reduced ferredoxin, thus reducing the overproduction of ROS, due to lower levels of NADP+ [95]. The re-assimilation of NH4+, which arises from glycine decarboxylation, consumes reducing equivalents from ferredoxin and/or NAD(P)H, as well as ATP. Similarly, the conversion of glycerate to glycerate-3-phosphate (PGA) consumes ATP and regenerates adenosine diphosphate (ADP). The requirement of NADH for hydroxypyruvate reduction could provide an additional sink for reducing equivalents, which could originate from chloroplasts via the malate valve or the mitochondrial tricarboxylic acid (TCA) cycle. Thus, photorespiration is a significant and important component of the processes involved in minimizing ROS production (by dissipating excess reduced equivalents and energy), even though it is itself a source of H2O2 [96].
According to the work of Corte-Real, Oliveira [75], proteome data of salt-stressed J. curcas have evidenced of major molecular processes regulated by saline conditions that appear to be genotype-dependent (Figure 4). Differential proteomic analysis data revealed that the tolerant-like genotype showed proteins of different pathways related to the salinity response, including the production of antioxidant enzymes, as well as signaling and stress regulation pathways, especially with high sensitivity and responsivity to abscisic acid (ABA). The higher physiological efficiency of the tolerant-like genotype is also due to its ability to produce important enzymes from different energy and metabolic pathways, such as photosynthesis and glycolysis, thus ensuring their development. Though the sensitive-like genotype showed the exclusive accumulation of the glutarredoxin (Grxs) class protein, the tolerant-like genotype accumulated CAT, with both acting on H2O2 dismutation. Unlike CAT, Grxs are proteins of the glutathione cycle, an NADPH-dependent pathway. Thus, the reduction in NADPH production under salt stress may be part of this antioxidative pathway, suggesting that the sensitive-like genotype leads to the greater activation of a less efficient pathway in ROS scavengers compared to CAT activity in the tolerant-like J. curcas genotype [75].
3.4. Metabolite Profile of Salt-Stressed J. curcas Plants
In excess, ROS can result in extensive damage to proteins, DNA, and lipids, thereby impairing normal cellular functions and resulting in perpetual metabolic dysfunction and plant death [97]. It has been well-documented that plant species employing highly efficient antioxidant systems have improved salt stress tolerance, a response that may be signaled by some small molecules, such as H2O2, oligochitosan, NaCl, proline, ABA, and NO (see the work of Gadelha, Miranda [78] and references therein).
Lin, Li [98] reported that eight CO2 assimilation-related proteins—two RuBisCO activases, RuBisCO large subunit-binding protein subunit beta, ribulose 1,5-bisphosphate carboxylase large subunit, transketolase, fructose 1,6-bisphosphatase, sedoheptulose 1,7-bisphosphatase, and ribose-5-phosphate isomerase—showed significant changes in response to salt treatment and were downregulated under salt stress (Figure 5). It was reported that salt stress suppressed the carboxylase activity of RuBisCO while enhancing its oxygenase activity [98]. Moreover, in the energy category, a root mitochondrial ATP synthase beta subunit was found to be downregulated by salt stress, potentially causing low ATP production under high salt stress (Figure 6).
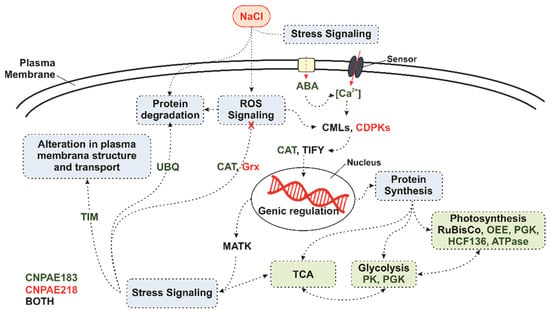
Figure 5.
Model proposed for the gene modulation pathway in response to NaCl in two different genotypes of Jatropha curcas subjected to 48 h of 750 mM of NaCl. ABA, abscisic acid. CMLs, calcium-binding proteins. CDPKs, calcium-dependent protein kinase. MYB, MYB Transcription factor family. TIFY, TIFY transcription factor family. RuBisCO, ribulose 1,5-bisphosphatase carboxylase-oxygenase. OEE, oxygen-evolving protein. PGK, phosphoglycerate kinase. HCF136, photosystem II assembly/stability factor. ATPase, ATP synthase. PK, pyruvate kinase. MATK, maturase K. TIM, translocase of the mitochondrial inner membrane. UBQ, ubiquitin transferase. CAT, catalase. Grx, glutaredoxin. Modified from the work of Corte-Real, Oliveira [75].
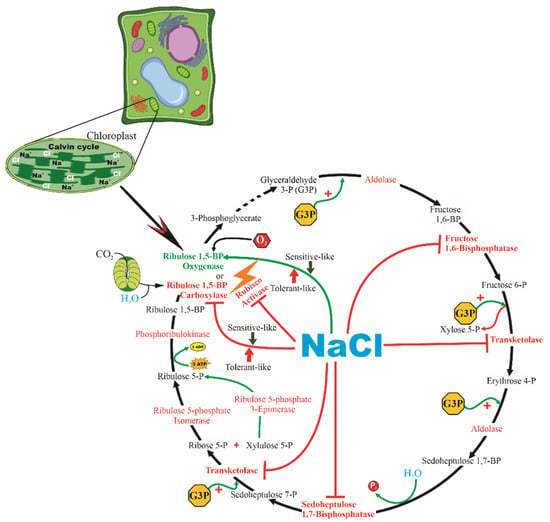
Figure 6.
Model proposing that the Na+ and Cl− ions promote the downregulation of many enzymes, such as RuBisCO activase, fructose 1,6-bisphosphatase, transketolase, sedoheptulose 1,7-bisphosphatase, and ribulose 5-phosphate isomerase, inhibiting the conversion of ribose 5-phosphate to ribulose 5-phosphate. On tolerant-like plants, Na+ and Cl− promote the downregulation of the carboxylase activity and promote the oxygenase activity of ribulose 1,5-bisphosphatase carboxylase-oxygenase. On the other hand, on sensitive-like plants, Na+ and Cl− promote the downregulation of the oxygenase activity of ribulose 1,5-bisphosphatase carboxylase-oxygenase, thus promoting carboxylase activity. Model constructed using data presented by Lin, Li [98] and Sivakumar, Sharmila [48].
Downstream from this transcriptional regulatory network, a number of osmolyte (such as trehalose, raffinose, and proline) synthesis genes are involved in osmotic homeostasis and the stabilization of proteins and cellular structures [99]. Similarly to Zhang, Zhang [99], our team studied J. curcas subjected to 150 mM of NaCl for 3 h and detected 2.4-fold more raffinose in the salt-sensitive-like genotype and 12.7% less raffinose in the salt-tolerant-like genotype [77].
3.5. Transcriptome Analysis of Water and Salt-Stressed J. curcas Plants
A transcriptomic analysis via RNA sequencing in Illumina HiSeq™ 2000, performed for samples collected under moderate and severe stress followed by rewatering, revealed shoot- and root-specific molecular adaptations under moderate stress [100]. Under severe stress, a dramatic transcriptomic reorganization at the root and shoot level surpassed organ specificity. Roots respond rapidly to drought, with more than 400 reported differentially expressed genes (DEGs) under moderate stress. Souza, Silva [77] described 4646 suppressed DEGs in sensitive-like J. curcas and only 57 DEGS were in salt tolerant-like J. curcas. In leaves, however, impressive changes in expression patterns were only observed under severe stress, indicating that J. curcas leaves do not respond to drought as rapidly as roots, even though the leaf transcriptome returned to control conditions faster than the root transcriptome after stress relief [100].
Based on the existence of conserved Tryp, Arg, Lys, and Tyr motifs in comparison to the Arabidopsis genome, a total of 58 Tryp, Arg, Lys, and Tyr genes were identified in the J. curcas genome [101]. The analysis of transcript accumulation, based on the RNA-seq of these 58 Tryp, Arg, Lys, and Tyr genes, showed 26 transcripts with at least 2-fold increases or decreases in expression in response to drought and 17 transcripts that responded to drought and salinity. These genes are good candidates for improving tolerance to both stresses with a biotechnological approach.
Plant hormone signals, such as JA and ABA, play indispensable roles in plant response to drought stress [102]. Overexpressing downstream transcription factors (NAC, MYB, C2H2, CBF/DREB, WRKY, and bHLH) can also improve plant drought tolerance [103]. The C-repeat-binding factor/dehydration-responsive element-binding factor (CBF/DREB) is widely considered a key regulator of plant abiotic stress response [53]. Transgenic N. benthamiana, overexpressing JcCBF2 of J. curcas, showed improved survival rate under drought stress [104]. Its leaf water retention and active oxygen-scavenging capacity but reduced photosynthesis and transpiration rate suggest that JcCBF2 plays an important role in improving plant drought tolerance. Overexpressing JcCBF2 was shown to lead to a decreased leaf area and increased leaf thickness. These overexpression lines of transgenic plants showed the expression of NbMYB21, NbMYB86, and NbMYB44, and both ABA- and JA-related genes increased under drought stress. These data indicate that JcCBF2 is able to positively regulate the plant drought response by changing the leaf anatomical structure and possibly through JA and ABA signaling pathways.
In Supplementary Table S1, we show the gene(s) studied, the analytical methods applied, the plant conditions, the salinity treatments, the exposure time, and related references. In this set of manuscripts, J. curcas genes showed participation in phytohormone signaling, the synthesis of osmoprotectant compounds, redox metabolism, ionic and osmotic adjustment, and secondary metabolism. A general analysis of Supplementary Table S1 demonstrates that in addition to roots (the tissue in direct contact with NaCl), leaves were also targeted. Additionally, most studies on J. curcas genes have concerned transgenic events caused by Agrobacterium in Arabidopsis or other model plants, followed by the validation of the transgene expression, usually by qPCR assay. In the studied set of manuscripts, the majority of J. curcas plants developed up to two months after germination when they received a salt treatment (which ranged from 50 to 300 mM of NaCl), with samples being collected at variable periods of hours (up to 48 h), days (from 2 to 25 days), or weeks (up to three weeks). These studies have contributed to J. curcas breeding programs by increasing salt tolerance in this species and other economically important and equally salt-sensitive ones.
After saline stress (100 mM of NaCl), with exposure times of 2 h, 2 days, and 7 days, Zhang, Zhang [99] observed induction two hours after the salt treatment of genes related to osmotic regulation (as expected) and TF proteins from several families, including DREB, HD-ZIP, NAC, WRKY, ERF, MYB, bHLH, and AUX/IAA (Supplementary Table S1). These TFs are crucial in regulating gene expression by recognizing cis-regulatory elements of the promoters of the genes to be transcribed. The P5CS1 gene (D-1-pyrroline-5-carboxylate synthetase; Supplementary Table S1), related to proline biosynthesis, was identified and was overexpressed after 3 h with 150 mM of NaCl [77].
Zhang, Zhang [99] observed that the APX4 gene was negatively expressed after seven days of 100 mM of NaCl exposure, which may have reflected the inhibition of ROS accumulation. Despite some differences in methodology (tags versus RNA-Seq), origin of the accessions (Chinese versus Brazilian), exposure times (2 h, 2 days, and 7 days versus 3 h), and stress conditions (NaCl: 100 versus 150 mM), the transcriptomic analyses performed by Zhang, Zhang [99] and Souza, Silva [77] enabled the identification of important convergent genes involved in J. curcas responding to the salt stimulus. The data also reinforce the importance of transcriptomic analysis identifying those genes.
The regulation of gene transcription is central to both tissue specific-gene expression and the regulation of gene activity in response to specific stimuli. Thus, whilst some cases of regulation after transcription have been observed, regulation occurs in most cases at the level of transcription via the decision regarding which genes will be transcribed into the primary RNA transcript [74]. In order to produce their effects, transcription factors generally require the ability to bind to DNA and then positively or negatively influence transcription. Our research group recently described the four most outstanding families of transcription factors (ARF, NAC, MYB, and AP2/ERF) that have important roles in responses to the biotic and abiotic stress of plants, especially J. curcas [74]. Wu, Xu [104] identified a total of 100 NAC genes (JcNAC) in Jatropha; based on phylogenetic analysis and gene structures, 83 JcNAC genes were classified as members of or separated from 39 previously predicted orthologous groups (OGs) of NAC sequences. Jatropha has a single intron-containing NAC gene subfamily that has been lost in many plants. The JcNAC genes are non-randomly distributed through the 11 linker groups of the genome of this plant and appear to be preferentially retained duplicates that arose from duplication events. The digital analysis of gene expression has indicated that some of the JcNAC genes have tissue-specific expression profiles (for example, in leaves, roots, stem bark, and seeds), and 29 genes respond differentially to abiotic stress.
The MYB family, which is involved in plant development and abiotic stress tolerance, was identified the entire genome of Jatropha [105], including in reference to family composition, phylogenetic evolution, and functional prediction analysis. Additionally, they specified the function of the JcMYB2 gene, which allowed them to identify 128 MYB genes, including 123 R2R3-MYB, 4 R1R2R3-MYB, and 1 4R-MYB.
The AP2/ERF transcription factors comprise the largest family involved in plant hormones such as ABA and ethylene, which help activate ABA and ethylene-dependent and -independent stress-responsive genes. Tang, Liu [106] found that JcDREB2 was repressed by salinity stress in Jatropha leaves. The overexpression of the JcDREB2 gene in rice reduced tolerance to salt stress, which led to the more pronounced aging and rolling of the leaves; in this study, they reported the low accumulation of proline in the transgenic JcDREB2 rice plants compared to the wild-type plants under salinity stress. Likewise, the authors reported higher activities of SOD and CAT in non-transformed rice plants compared to transgenic rice plants under salinity stress, suggesting that they were at a disadvantage in high salinity conditions. Finally, the overexpression of JcDREB2 was found to improve sensitivity to salt stress in transgenic rice plants. These results have increased our understanding of the roles of this Jatropha transcription factor AP2/ERF in plant growth and responses to abiotic stress.
3.6. Transgenic J. curcas Plants for Cultivation on Saline Conditions
Improving the salinity and drought tolerance of crop plants by genetic means has been an important but largely unfulfilled aim of modern agricultural development. Though acclaimed widely for its oil, many authors have not considered J. curcas economically important enough for domestication, and its seed and oil productivity is hugely variable [107,108]. In terms of viability, a recent study showed that the majority of biofuel projects collapse within the first 5 years of operation [71]. In the case of Ghana, out of the nine verified abandoned J. curcas projects for which we could establish a date of collapse, five failed in the first 3 years of operation and the rest failed within the first 5 years. The reasons for these failures were diverse and varied according to projects and location. Ahmed, Campion [71] reported numerous reasons for the failure of J. curcas plantations around the world, especially in Ghana, and they found that the main reason for the collapse of J. curcas plantations has been the lack of a stable variety, cultivar, or genotype that can guarantee the farmer credibility and stable seed yield of investing in J. curcas plantations. However, the affected farmers did not know about this transaction until Biofuel Africa started land clearing to plant J. curcas. As a result, there was deep skepticism about the leadership and benevolence of the chieftaincy institutions in the Kusawgu Traditional Are.
J. curcas continues to remain semi-wild, and ideal commercial varieties with profitable yields are still lacking. Varieties that produce high and stable yields need to be bred to develop J. curcas into a successful biofuel crop [109]. Accordingly, the genetic transformation of J. curcas has been pursued for the last 10 years, and many transformation methods have been established [110]. Liu, Wang [111] postulated that overexpressing genes of jasmonate pathway (JcAOC) would be valuable in attaining plants with higher tolerance to stresses, including salt stress. These authors reported that the expression of JcAOC mRNA was ~2.7-fold higher in the leaves of WT-J. curcas subjected to 2 h of 300 mM of NaCl compared to control ones. To confirm whether salt stress and low temperature affected the expression of JcAOC, these authors tested the growth performance of Escherichia coli cells transformed with recombinant plasmid pET-JcAOC under stress conditions and showed that the growth of transformed E. coli cells with recombinant plasmid pET-JcAOC was 2.5-fold higher than pET-30a empty vectors. Since E. coli does not contain the JA biosynthetic pathway, these results revealed a potential property of JcAOC in addition to catalyzing JA biosynthesis, as well as that it is a valuable gene that deserves special attention because the genetic engineering of JcAOC could be a potential alternative to enhance J. curcas tolerance against salt stress.
Tsuchimoto, Cartagena [112] successfully transformed J. curcas into a type that overexpressed the Synechococcus GSMT and DMT genes (which encode glycine sarcosine methyltransferase and sarcosine dimethylglycine methyltransferase, respectively, and which catalyze glycine betaine production), and the transgenic plants were noticeably more tolerant to drought than wild-type plants. These findings indicate that these plants can adopt the strategy used by halophytes to survive in drought conditions via the protective nature of glycine betaine [112]. The glycine content was found to be significantly increased in transgenic J. curcas plants that overexpressed GSMT and DMT. Jha, Mishra [87] successfully transformed J. curcas with the salt-responsive SbNHX1 gene to enhance its salt tolerance and improve Na+ inside vacuoles, demonstrating the use of this strategy to promote survival under salt stress. An active vacuolar antiporter utilizes the proton motive force generated by vacuolar ATPases and pyrophosphatases to sequester excess Na+ into the vacuole. Moreover, these authors observed higher Na+ content and less K+ content in transgenic leaf tissues under salt stress, which they attributed to ion homeostasis due to the activity of the vacuolar Na+/H+ antiporter SbNHX1. In case, the transgenic lines showed enhanced tolerance at 200 mM of NaCl, enhancing Na+ sequestration into the vacuole.
Salt-tolerant transgenic J. curcas plants can be used for cultivation in salt-affected marginal areas for their sustainable management with biofuel production. The diffusion of such transgenic plants may help J. curcas during salt cultivation without affecting food production. Recently, Chacuttayapong, Enoki [113] described an efficient method to produce a Jatropha curcas transgenic line that expresses Os03, Os04, Os08, Os10, and Os10L, suggesting that the genes associated with larger seed sizes in Arabidopsis thaliana (which were found using the rice FOX-hunting system) produce larger seeds in Jatropha curcas.
4. Toxicity and Medical Use of Jatropha curcas Seeds
The fact that the oil extracted from J. curcas is excellent-quality biofuel is indisputable. However, there are currently no agronomically improved varieties available for J. curcas, and the seed meal obtained after the extraction of the oil cannot be used as an animal feed due to its toxicity [114]. Nevertheless, in the process of extracting the oil from the seeds of J. curcas, a physic nut cake/meal is produced and can be considered a great source of protein and fiber for animal consumption [114,115]. The ability to use J. curcas meal as animal feed improves the economics of J. curcas production because it means that the crop could produce both fuel and feed [19]. However, the use of this physic nut cake/meal is limited due to the high concentration of two toxic constituents: phorbol esters (PMA) and curcin, as well as other anti-nutritional factors such as lectins, ribosome-inactivating proteins (RIP), and saponins [116,117,118,119]. PMA is a diterpene and can act in two ways, acute (intense inflammatory response) and chronic (inducing tumor formation) [120]. Arroyo, Vanegas [121] described a simple, inexpensive method to reduce the PMA to a level of 0.03% using 92% ethanol, a treatment for which no positive test was obtained for tannins and saponins. The ethanol treatment reserves 64% of proteins, 42.7% of soluble carbohydrates, 36.3% of nitrogen sources, and 41.3% of fiber, leaving this methodology as the best option for PMA, saponin, and tannin detoxification. Accordingly, physic nut cake/meal should be approved for addition into livestock feed according to the Food and Agriculture Organization (FAO) criteria [121].
Das, Uppal [122] reported on PMA detoxification through composting using four different animal dung types, and they found that PMA was reduced to 12%, which means that their method was technically less effective than that described by Arroyo, Vanegas [121] using only ethanol.
Some studies have shown the potential of physic nut cake/meal in fish diets. Saha and Ghosh [122] fed rohu (Labeo rohita) fingerlings with de-oiled J. curcas seed meal (DJSM) fermented for 15 days by an exo-enzyme-producing bacterium, Bacillus cereus, isolated from the hindgut of rohu. This procedure caused a decrease in the contents of anti-nutritional factors but increases in the levels of free amino acids and free fatty acids. Hassaan, Goda [123] obtained similar results using DJSM fermented with Bacillus licheniformis (LFJSM) and Bacillus pumilus (PFJSM). Fermented DJSM was mixed with concentrations of LFJSM and PFJSM, and it was found that PFJSM-25 and PFJSM-50 showed non-significant differences compared to the control and that 50% of fish meal can be replaced by PFJSM in Nile tilapia diets. However, Nile tilapia growth is retarded if they are fed with 0.16% of trypsin inhibitor, as 0.06% is the maximum tolerated level in Nile tilapia growth. A Brazilian study [124] showed that the apparent digestibility coefficients of crude protein for nontoxic and detoxified physic nut cake/meal were 77.51% and 81.11%, respectively—values similar to those reported with another seed meal crude extract. Nontoxic physic and detoxified physic nuts presented apparent digestibility coefficients of 90.48% and 52.10%, respectively [124]. Another Brazilian study [125] demonstrated that 0.025 g of crude extract per kilogram of animal led to 100% animal death, reinforcing the need to detoxify physic nut cake/meal. The acute oral LD50 of the oil was found to be 6 mL kg−1 body weight in Haffkine Wistar strain rats [126]. Elsewhere, the PMA fraction from seed oil was reported as a promising candidate for use as a plant-derived protectant for a variety of crops from a range of pre- and post-harvest insect pests [127]. The results reported so far demonstrate that there is potential for the use of physic nut cake/meal in animal supplementation, mainly in regions not supplied with soybean and its co-products.
Of the toxic compounds present in physic nut cake/meal, curcin is by far the most toxic molecule. Curcin is often classified as a lectin and described as having a similar toxicity to ricin from castor bean. Both curcin and ricin are RIPs that depurinate rRNA, thus arresting protein synthesis. Curcin is a type-I RIP, and ricin is a type-II RIP. Type II RIPs contain a catalytic A-chain (RIP), whereas type I RIPs such as curcin lack this lectin domain. Due to the lack of the lectin domain, the LD50 values of type I RIPs are typically over 1000-fold higher than those observed for type-II RIPs in whole rats [115]. The presence of curcin in the seeds of J. curcas is therefore unlikely to present a significant barrier to the processing of J. curcas seed meal into animal feed. Additionally, curcin has been investigated for its anti-tumor potential. Some scholars studying curcin have shown that like other RIPs, both purified and recombinant curcin protein can inhibit cell-free translation and protein synthesis in a reticulocyte lysate system [118,128,129]. Consequently, RIPs can arrest DNA and protein metastasis before sequentially inhibiting the division and proliferation of cancer cells [119]. This could be one of the reasons for curcin’s anti-tumor activity.Huang, Hou [130] reported that transgenic lines of tobacco plants, expressing cur2p fragment (coding premature curcin 2 protein), led to an increased tolerance to tobacco mosaic virus (TMV) and the fungal pathogen Rhizoctonia solani by delaying the development of systemic symptoms of TMV and reducing the damage caused by the fungal disease. Zhao, Wang [120] showed the effect of purified curcin from seeds of J. curcas on the growth inhibition rate of mouse sarcoma—180 cells; 20% on 3rd day and 40% on 7th day under 100 mg mL−1. The same author also reported that curcin could inhibit S-180 cell growth at 10 mg mL−1. The physic nut cake/meal produced from the non-toxic J. curcas variety was used in the preparation of livestock feed at 20, 40, and 60%, replacing soybean meal (which was used as the reference concentrated protein source) [114]. The results of this experiment were encouraging. The animals showed gains in weight and carcass quality similar to those obtained with the soy concentrate, proving the acceptance of the animals. Moreover, Wu, Goh [131] isolated three curcin genes and the endosperm-specific C1 promoter, thus providing useful information and research materials for further functional studies of curcin proteins and genetic engineering of J. curcas.
5. Concluding Remarks
Salt stress affects various aspects of the biochemistry and physiology of J. curcas, with negative impacts on growth and yield, thus supporting the conclusion that the species is sensitive to salt stress. Important genes and proteins involved in a wide variety of processes, including stress signaling, gene expression, redox regulation, defense systems, stress regulation, and secondary metabolism have been studied. Transcriptomic analyses have provided a global overview of genes expressed by the plants after the stress stimulus. Based on transcriptomic studies, differentially expressed genes could be valuable transgenes in transgenic events. Additionally, the development of functional molecular markers based on these differentially expressed genes could be optimized, and a selected panel of these genes could help breeders to select accessions based on gene expression performance. Genetic engineering using halophyte genes might offer an excellent platform for developing glycophytic crops with improved salinity tolerance. It is now possible to comprehend a mechanistic framework of the responses to water deficit and salinity in this still-undomesticated oil seed species. Nevertheless, much work is still required for J. curcas to become a real biodiesel feedstock. In summary, the data presented here represent a further step to understand salinity and water-deficit stresses responses in J. curcas plants, information that could be used to improve tolerance of this industrial crop. To be environmentally sustainable and non-competitive with existing food sources, the productivity of J. curcas needs to increase to allow for commercial cultivation on marginal land, which would be cost-effective and sustainable only after rational marker-assisted breeding or trait introduction though genetic modification. Taken together, these expression data suggest that the careful selection of candidate genes is a critical step prior to the initiation of a specific genetic engineering project.
Moreover, we have described the beneficial and harmful effects of phorbol ester and curcin from a more global perspective instead of just stating that physic nut cake/meal is a toxic by-product to the environment and animals. This paradigm was overturned with a series of articles describing the effects of both phorbol ester and curcin in J. curcas seeds. Furthermore, Embrapa Agroenergia (Brasília, DF, Brazil) recently reported the identification of a non-toxic variety of J. curcas. The results achieved so far demonstrate that there is potential for the use of physic nut cake/meal in animal food supplementation, especially in regions not supplied with soybean and its by-products. As a result of the research generated by Embrapa, it is expected in the mid-term to establish viable industrial processes capable of detoxifying Jatropha cake/meal and obtaining high-productivity commercial cultivars without the presence of toxic components [132]. Furthermore, Embrapa Semiárido (Petrolina, PE, Brazil) has developed a highly productive variety of J. curcas with a yield of 3800 kg of seed per hectare, yielding about 1000 L of oil ha and thus opening the door to a new market in terms of Embrapa biofuels and carbon sequestration, since J. curcas is a semi-hardwood plant that can develop good harvests for up to 50 years.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture12050594/s1, Table S1. Saline stress-associated genes and metabolic pathways in physic nut [133,134,135,136,137,138].
Author Contributions
Conceptualization, M.F.P., A.J.-O., and L.A.R.-P.; data curation, M.F.P., A.J.-O., and L.A.R.-P.; Investigation, M.F.P., A.J.-O., and L.A.R.-P.; methodology, M.F.P., A.J.-O., and L.A.R.-P.; project administration, M.F.P., A.J.-O., and L.A.R.-P.; writing—original draft, M.F.P., A.J.-O., and L.A.R.-P.; writing—review and editing, M.F.P., A.J.-O., and L.A.R.-P. resources, M.F.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Council for Scientific and Technological Development (CNPq) (Grants 163524/2017-3).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mitra, S.; Ghose, A.; Gujre, N.; Senthilkumar, S.; Borah, P.; Paul, A.; Rangan, L. A review on environmental and socioeconomic perspectives of three promising biofuel plants Jatropha curcas, Pongamia pinnata and Mesua ferrea. Biomass Bioenergy 2021, 151, 106173. [Google Scholar] [CrossRef]
- Satterthwaite, D. Cities’ contribution to global warming: Notes on the allocation of greenhouse gas emissions. Environ. Urban 2008, 20, 539–549. [Google Scholar] [CrossRef]
- IPCC. Deforestation Accounts for about 20% of CO2 Emissions Globally; Climate Central: Princeton, NJ, USA, 2021; Available online: https://www.climatecentral.org/what-we-do#wwd (accessed on 23 September 2021).
- Rodrigues, A.S.; Rocha, A.C.P.; Da Costa, A.S.V.; Lopes, I.A.P. Prospects of raw materials for the production of biodiesel in Brazil. Int. J. Res.-Granthaalayah 2020, 8, 133–143. [Google Scholar] [CrossRef]
- Alherbawi, M.; AlNouss, A.; McKay, G.; Al-Ansari, T. Optimum sustainable utilisation of the whole fruit of Jatropha curcas: An energy, water and food nexus approach. Renew. Sustain. Energy Rev. 2021, 137, 110605. [Google Scholar] [CrossRef]
- Alherbawi, M.; McKay, G.; Mackey, H.R.; Al-Ansari, T. A novel integrated pathway for Jet Biofuel production from whole energy crops: A Jatropha curcas case study. Energy Convers. Manag. 2021, 229, 113662. [Google Scholar] [CrossRef]
- Achten, W.; Verchot, L.; Franken, Y.; Mathijs, E.; Singh, V.; Aerts, R.; Muys, B. Jatropha bio-diesel production and use. Biomass Bioenergy 2008, 32, 1063–1084. [Google Scholar] [CrossRef]
- Becker, K.; Makkar, H.P.S. Jatropha curcas: A potential source for tomorrow’s oil and biodiesel. Lipid Technol. 2008, 20, 104–107. [Google Scholar] [CrossRef]
- Klein-Marcuschamer, D.; Turner, C.; Allen, M.; Gray, P.; Dietzgen, R.G.; Gresshoff, P.M.; Hankamer, B.; Heimann, K.; Scott, P.T.; Stephens, E.; et al. Technoeconomic analysis of renewable aviation fuel from microalgae, Pongamia pinnata, and sugarcane. Biofuels Bioprod. Biorefin. 2013, 7, 416–428. [Google Scholar] [CrossRef]
- Wang, W.-C. Techno-economic analysis for evaluating the potential feedstocks for producing hydro-processed renewable jet fuel in Taiwan. Energy 2019, 179, 771–783. [Google Scholar] [CrossRef]
- Tao, L.; Milbrandt, A.; Zhang, Y.; Wang, W.-C. Techno-economic and resource analysis of hydroprocessed renewable jet fuel. Biotechnol. Biofuels 2017, 10, 261. [Google Scholar] [CrossRef]
- IATA. Jet Fuel Price Monitor; International Air Transport Association: Montreal, QC, Canada, 2021; Available online: https://www.iata.org/en/publications/economics/fuel-monitor/ (accessed on 22 September 2021).
- BRASIL. Lei No. 11.097, 13 January 2005. In Diário Oficial da União; Civil, C., Ed.; Imprensa Nacional: Rio de Janeiro, Brazil, 2005. [Google Scholar]
- Chiappini, G.; Gaudarde, G. Ministro Bento Albuquerque Garante Mistura Obrigatória de Biodiesel. Agência EPBR. 2021. Available online: https://epbr.com.br/ministro-bento-albuquerque-garante-mistura-obrigatorio-de-biodiesel/ (accessed on 3 April 2021).
- ANP. Agência Nacional de Petróleo, Anuário Estatístico 2020; ANP: Brasília, Brazil, 2020. [Google Scholar]
- Maes, W.; Trabucco, A.; Achten, W.; Muys, B. Climatic growing conditions of Jatropha curcas L. Biomass Bioenergy 2009, 33, 1481–1485. [Google Scholar] [CrossRef]
- Divakara, B.N.; Upadhyaya, H.D.; Wani, S.P.; Gowda, C.L. Biology and genetic improvement of Jatropha curcas L.: A review. Appl. Energy 2010, 87, 732–742. [Google Scholar] [CrossRef]
- Pompelli, M.F.; Barata-Luís, R.; Vitorino, H.S.; Gonçalves, E.R.; Rolim, E.V.; Santos, M.G.; Almeida-Cortez, J.S.; Ferreira, V.M.; Lemos, E.E.; Endres, L. Photosynthesis, photoprotection and antioxidant activity of purging nut under drought deficit and recovery. Biomass Bioenergy 2010, 34, 1207–1215. [Google Scholar] [CrossRef]
- Reubens, B.; Achten, W.; Maes, W.; Danjon, F.; Aerts, R.; Poesen, J.; Muys, B. More than biofuel? Jatropha curcas root system symmetry and potential for soil erosion control. J. Arid Environ. 2011, 75, 201–205. [Google Scholar] [CrossRef]
- Bekalu, Y.; Fekad, M. Review on economic importance of Jatropha apart from its use as a biofuel. Int. J. Res. Dev. 2020, 5, 24–33. [Google Scholar] [CrossRef]
- Corte-Real, N.; da Cunha de Miranda, P.V.V.; Endres, L.; de Souza, E.R.; Pompelli, M.F. Tolerance to salinity in Jatropha curcas are genotype-dependent. Braz. J. Dev. 2019, 5, 22169–22199. [Google Scholar] [CrossRef][Green Version]
- Tedla, A.; Minale, M.; Eshetu, R. Determination of the upper limit age of Jatropha curcas plantation for optimum yield production. Asian Bus. Rev. 2020, 10, 37–42. [Google Scholar] [CrossRef]
- Zavala-Hernández, J.T.; Córdova-Téllez, L.; Martínez-Herrera, J.; Molina-Moreno, J.C. Fruit and seed development of Jatropha curcas L. and physiological indicators of seed maturity. Rev. Fitotec. Mex. 2015, 38, 275–284. [Google Scholar]
- Jonas, M.; Ketlogetswe, C.; Gandure, J. Variation of Jatropha curcas seed oil content and fatty acid composition with fruit maturity stage. Heliyon 2020, 6, e03285. [Google Scholar] [CrossRef]
- Jongschaap, R.E.E.; Corré, W.J.; Bindraban, P.S.; Brandenburg, W.A. Claims and Facts on Jatropha curcas L; Wageningen UR, Plant Research International: Wageningen, The Netherlands, 2007; 66p. [Google Scholar]
- Lane, J. Jatropha Biofuel Around the World: A 13-Country Tour of Development Activity. Available online: https://www.renewableenergyworld.com/articles/2014/09/jatropha-biofuel-around-the-world-a-13-country-tour-of-development-activity.html (accessed on 29 July 2019).
- Rodríguez, O.A.V.; Vázquez, A.P.; Gamboa, C.M. Drivers and Consequences of the First Jatropha curcas Plantations in Mexico. Sustainability 2014, 6, 3732–3746. [Google Scholar] [CrossRef]
- Soto, I.; Ellison, C.; Kenis, M.; Diaz, B.; Muys, B.; Mathijs, E. Why do farmers abandon jatropha cultivation? The case of Chiapas, Mexico. Energy Sustain. Dev. 2018, 42, 77–86. [Google Scholar] [CrossRef]
- Hsie, B.S.; Mendes, K.R.; Antunes, W.C.; Endres, L.; Campos, M.L.; Souza, F.C.; Santos, N.D.; Singh, B.; Arruda, E.C.; Pompelli, M.F. Jatropha curcas L. (Euphorbiaceae) modulates stomatal traits in response to leaf-to-air vapor pressure deficit. Biomass Bioenergy 2015, 81, 273–281. [Google Scholar] [CrossRef]
- Pompelli, M.F.; Ferreira, P.P.; Chaves, A.R.; Figueiredo, R.C.; Martins, A.O.; Jarma-Orozco, A.; Bhatt, A.; Batista-Silva, W.; Endres, L.; Araújo, W.L. Physiological, metabolic, and stomatal adjustments in response to salt stress in Jatropha curcas. Plant Physiol. Biochem. 2021, 168, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Pompelli, M.F.; França, S.C.S.; Tigre, R.C.; Oliveira, M.T.; Sacilot, M.; Pereira, E.C.G. Spectrophotometric determinations of chloroplastidic pigments in acetone, ethanol and dimethylsulphoxide. Braz. J. Biosc. 2013, 11, 52–58. [Google Scholar]
- Campos, M.L.O.; Hsie, B.S.; Granja, J.A.A.; Correia, R.M.; Silva, S.R.S.; Almeida-Cortez, J.S.; Pompelli, M.F. Photosynthesis and antioxidant activity mechanisms in Jatropha curcas L. under salt stress. Braz. J. Plant Physiol. 2012, 24, 55–67. [Google Scholar] [CrossRef]
- Silva, E.N.; Ferreira-Silva, S.L.; Fontenele, A.D.V.; Ribeiro, R.V.; Viégas, R.A.; Silveira, J.A.G. Photosynthetic changes and protective mechanisms against oxidative damage subjected to isolated and combined drought and heat stresses in Jatropha curcas plants. J. Plant Physiol. 2010, 167, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.C. Turgor maintenance by osmotic adjustment: 40 years of progress. J. Exp. Bot. 2018, 69, 3223–3233. [Google Scholar] [CrossRef]
- Quintal, E.B.; Velarde-Buerdía, A.; Ku-González, A.; Carillo-Pech, M.; Ortega, D.; Echevarría-Machado, I.; Epottosin, I.; Martínez-Estévez, M. Mechanisms of salt tolerance in habanero pepper plants (Capsicum chinense Jacq.): Proline accumulation, ions dynamics and sodium root-shoot partition and compartmentation. Front. Plant Sci. 2014, 5, 605. [Google Scholar] [CrossRef]
- Yang, L.; Ma, C.; Wang, L.; Chen, S.; Li, H. Salt stress induced proteome and transcriptome changes in sugar beet monosomic addition line M14. J. Plant Physiol. 2012, 169, 839–850. [Google Scholar] [CrossRef]
- Silva, E.N.; Ferreira-Silva, S.L.; Viégas, R.A.; Silveira, J.A.G. The role of organic and inorganic solutes in the osmotic adjustment of drought-stressed Jatropha curcas plants. Environ. Exp. Bot. 2010, 69, 279–285. [Google Scholar] [CrossRef]
- Wang, H.; Ao, P.; Yang, S.; Zou, Z.; Wang, S.; Gong, M. Molecular Cloning and Expression Analysis of the Gene Encoding Proline Dehydrogenase from Jatropha curcas L. Appl. Biochem. Biotechnol. 2015, 175, 2413–2426. [Google Scholar] [CrossRef] [PubMed]
- Naliwajski, M.; Skłodowska, M. The Relationship between the Antioxidant System and Proline Metabolism in the Leaves of Cucumber Plants Acclimated to Salt Stress. Cells 2021, 10, 609. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Xiao, X.; Zhang, W.; Lang, D.; Li, Z.; Zhang, X. Exogenous silicon relieve drought stress and salt stress of Glycyrrhiza uralensis seedlings by regulating proline metabolism and nitrogen assimilation. J. Hortic. Sci. Biotechnol. 2021, 96, 728–737. [Google Scholar] [CrossRef]
- Masoudniaragh, A.; Oraei, M.; Gohari, G.; Akbari, A.; Faramarzi, A. Using halloysite nanotubes as carrier for proline to alleviate salt stress effects in sweet basil (Ocimum basilicum L.). Sci. Hortic. 2021, 285, 110202. [Google Scholar] [CrossRef]
- Hare, P.D.; Cress, W.A.; van Staden, J. Proline synthesis and degradation: A model system for elucidating stress-related signal transduction. J. Exp. Bot. 1999, 50, 413–434. [Google Scholar] [CrossRef]
- Drake, P.L.; Froend, R.H.; Franks, P.J. Smaller, faster stomata: Scaling of stomatal size, rate of response, and stomatal conductance. J. Exp. Bot. 2013, 64, 495–505. [Google Scholar] [CrossRef]
- Ehleringer, J.R.; Sandquist, D. Photosynthesis: Physiological and ecological considerations. In Plant Physiology, 5th ed.; Taiz, L., Zeiger, E., Eds.; Sinauer Associates Inc.: Sunderland, MA, USA, 2010; pp. 244–371. [Google Scholar]
- Buchanan, B.B.; Wolosiuk, R.A. Photosynthesis: The carbon reactions. In Plant Physiology, 5th ed.; Taiz, L., Zeiger, E., Eds.; Sinauer Associates Inc.: Sunderland, MA, USA, 2010; pp. 199–242. [Google Scholar]
- Blankenship, R.E. Photosynthesis: The light reactions. In Plant Physiology and Development, 6th ed.; Taiz, L., Zeiger, E., Møller, I.M., Murphy, A., Eds.; Sinauer Associates, Inc.: Sunderland, MA, USA, 2014; pp. 163–197. [Google Scholar]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Sivakumar, P.; Sharmila, P.; Saradhi, P.P. Proline Alleviates Salt-Stress-Induced Enhancement in Ribulose-1,5-Bisphosphate Oxygenase Activity. Biochem. Biophys. Res. Commun. 2000, 279, 512–515. [Google Scholar] [CrossRef]
- Shabala, S.; Cuin, T.A. Potassium transport and plant salt tolerance. Physiol. Plant. 2008, 133, 651–669. [Google Scholar] [CrossRef]
- Babita, M.; Maheswari, M.; Rao, L.; Shanker, A.; Rao, D.G. Osmotic adjustment, drought tolerance and yield in castor (Ricinus communis L.) hybrids. Environ. Exp. Bot. 2010, 69, 243–249. [Google Scholar] [CrossRef]
- Leandro, D.D.S.; Tessio, A.D.S.; Priscila, S.D.O.; Bruno, G.L.; Marcio, G.C.D.C.; Alan, F.D.A.A.; Fabio, P.G. Abscisic acid-mediated stomatal closure and antioxidant defenses in Jatropha curcas L. seedlings submitted to moderate water deficit. Afr. J. Agric. Res. 2016, 11, 2806–2816. [Google Scholar] [CrossRef]
- Tominaga, J.; Inafuku, S.; Coetzee, T.; Kawamitsu, Y. Diurnal regulation of photosynthesis in Jatropha curcas under drought during summer in a semi-arid region. Biomass Bioenergy 2014, 67, 279–287. [Google Scholar] [CrossRef]
- Yang, S.L.; Chen, K.; Wang, S.S.; Gong, M. Osmoregulation as a key factor in drought hardening-induced drought tolerance in Jatropha curcas. Biol. Plant. 2015, 59, 529–536. [Google Scholar] [CrossRef]
- Silva, E.N.; Ribeiro, R.V.; Ferreira-Silva, S.L.; Vieira, S.A.; Ponte, L.F.A.; Silveira, J.A.G. Coordinate changes in photosynthesis, sugar accumulationand antioxidative enzymes improve the performance of Jatropha curcas plants under drought stress. Biomass Bioenergy 2012, 45, 270–279. [Google Scholar] [CrossRef]
- Silva, E.N.; Silveira, J.A.G.; Aragão, R.M.; Vieira, C.F.; Carvalho, F. Photosynthesis impairment and oxidative stress in Jatropha curcas exposed to drought are partially dependent on decreased catalase activity. Acta Physiol. Plant. 2019, 41, 4. [Google Scholar] [CrossRef]
- Silva, E.N.; Silveira, J.A.G.; Ribeiro, R.V.; Oliveira, J.; Cardoso, R.A. Photosynthetic and Antioxidant Responses of Jatropha curcas Plants to Heat Stress: On the Relative Sensitivity of Shoots and Roots. J. Plant Growth Regul. 2018, 37, 255–265. [Google Scholar] [CrossRef]
- Imlay, J.A. Pathways of Oxidative Damage. Annu. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Beis, A.; Patakas, A. Relative contribution of photoprotection and anti-oxidative mechanisms to differential drought adaptation ability in grapevines. Environ. Exp. Bot. 2012, 78, 173–183. [Google Scholar] [CrossRef]
- Kramer, P.J.; Koslowski, T. Physiology of Wood Plants; Academic Press: New York, NY, USA, 1989; 811p. [Google Scholar]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Rajaona, A.; Brueck, H.; Seckinger, C.; Asch, F. Effect of salinity on canopy water vapor conductance of young and 3-year old Jatropha curcas L. J. Arid Environ. 2012, 87, 35–41. [Google Scholar] [CrossRef]
- Pompelli, M.F. Benefits and Harms of Wastewater in Agriculture. Acta Sci. Agric. 2021, 5, 112–113. [Google Scholar] [CrossRef]
- Da Fonseca, A.F.; Melfi, A.J.; Montes, C.R. Maize Growth and Changes in Soil Fertility After Irrigation with Treated Sewage Effluent. I. Plant Dry Matter Yield and Soil Nitrogen and Phosphorus Availability. Commun. Soil Sci. Plant Anal. 2005, 36, 1965–1981. [Google Scholar] [CrossRef]
- Ribeiro, G.A.S. Acompanhamento da Safra Brasileira de Grãos; Companhia Nacional de Abastecimento: Brasília, Brazil, 2022. [Google Scholar]
- Asano, T.; Maeda, M.; Takaki, M. Wastewater reclamation and reuse in Japan: Overview and implementation examples. Water Sci. Technol. 1996, 34, 219–226. [Google Scholar] [CrossRef]
- Dorta-Santos, M.; Tejedor, M.; Jiménez, C.; Hernández-Moreno, J.M.; Palacios-Díaz, M.P.; Díaz, F.J. Recycled Urban Wastewater for Irrigation of Jatropha curcas L. in Abandoned Agricultural Arid Land. Sustainability 2014, 6, 6902–6924. [Google Scholar] [CrossRef]
- Dorta-Santos, M.; Tejedor, M.; Jiménez, C.; Hernández-Moreno, J.M.; Díaz, F.J. Using marginal quality water for an energy crop in arid regions: Effect of salinity and boron distribution patterns. Agric. Water Manag. 2016, 171, 142–152. [Google Scholar] [CrossRef]
- Valdes-Rodriguez, O.A.; Sánchez-Sánchez, O.; Pérez-Vázquez, A.; Ruiz-Bello, R. Soil texture effects on the development of Jatropha seedlings—Mexican variety ‘piñón manso’. Biomass Bioenergy 2011, 35, 3529–3536. [Google Scholar] [CrossRef]
- Ahmed, A.; Campion, B.B.; Gasparatos, A. Biofuel development in Ghana: Policies of expansion and drivers of failure in the jatropha sector. Renew. Sustain. Energy Rev. 2017, 70, 133–149. [Google Scholar] [CrossRef]
- Silva, E.N.; Silveira, J.A.; Ribeiro, R.V.; Vieira, S.A. Photoprotective function of energy dissipation by thermal processes and photorespiratory mechanisms in Jatropha curcas plants during different intensities of drought and after recovery. Environ. Exp. Bot. 2015, 110, 36–45. [Google Scholar] [CrossRef]
- Isla, F.L.; Campos, M.L.; Endres, L.; Bezerra-Neto, E.; Pompelli, M.F. Effects of seed storage time and salt stress on the germination of Jatropha curcas L. Ind. Crop. Prod. 2018, 118, 214–224. [Google Scholar] [CrossRef]
- Cabral, G.; Binneck, E.; De Souza, M.C.P.; Da Silva, M.D.; Neto, J.R.C.F.; Pompelli, M.F.; Endres, L.; Kido, É.A. First Expressed TFome of Physic Nut (Jatropha curcas L.) After Salt Stimulus. Plant Mol. Biol. Rep. 2020, 38, 189–208. [Google Scholar] [CrossRef]
- Corte-Real, N.; Oliveira, M.S.; Jarma-Orozco, A.; Fernandes, D.; dos Santos, M.A.; Endres, L.; Junior, T.C.; Pompelli, M.F. Comparative analysis of salt-induced changes in the leaves proteome of two contrasting Jatropha curcas genotypes. Braz. J. Dev. 2020, 6, 39845–39872. [Google Scholar] [CrossRef]
- Liang, J.; Zhou, M.; Zhou, X.; Jin, Y.; Xu, M.; Lin, J. JcLEA, a Novel LEA-Like Protein from Jatropha curcas, Confers a High Level of Tolerance to Dehydration and Salinity in Arabidopsis thaliana. PLoS ONE 2013, 8, e83056. [Google Scholar] [CrossRef]
- de Souza, M.C.P.; da Silva, M.D.; Binneck, E.; Cabral, G.; Iseppon, A.M.B.; Pompelli, M.F.; Endres, L.; Kido, É.A. RNA-Seq transcriptome analysis of Jatropha curcas L. accessions after salt stimulus and unigene-derived microsatellite mining. Ind. Crop. Prod. 2020, 147, 112168. [Google Scholar] [CrossRef]
- Gadelha, C.G.; Miranda, R.D.S.; Alencar, N.L.M.; Costa, J.H.; Prisco, J.T.; Gomes-Filho, E. Exogenous nitric oxide improves salt tolerance during establishment of Jatropha curcas seedlings by ameliorating oxidative damage and toxic ion accumulation. J. Plant Physiol. 2017, 212, 69–79. [Google Scholar] [CrossRef]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant Salt Stress: Adaptive Responses, Tolerance Mechanism and Bioengineering for Salt Tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Díaz-López, L.; Gimeno, V.; Lidon, V.; Simón, I.; Martinez, V.; Garcia-Sanchez, F. The tolerance of Jatropha curcas seedlings to NaCl: An ecophysiological analysis. Plant Physiol. Biochem. 2012, 54, 34–42. [Google Scholar] [CrossRef]
- Pan, T.; Liu, M.; Kreslavski, V.D.; Zharmukhamedov, S.K.; Nie, C.; Yu, M.; Kuznetsov, V.V.; Allakhverdiev, S.I.; Shabala, S. Non-stomatal limitation of photosynthesis by soil salinity. Crit. Rev. Environ. Sci. Technol. 2020, 51, 791–825. [Google Scholar] [CrossRef]
- Sairam, R.K.; Rao, K.; Srivastava, G. Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci. 2002, 163, 1037–1046. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Jiang, L.; Xu, Y.; Wang, Y.; Lu, D.; Chen, F. Aquaporin JcPIP2 is Involved in Drought Responses in Jatropha curcas. Acta Biochim. Biophys. Sin. 2007, 39, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Santos-Rosa, M.J.; Shirasu, K. The U-box protein family in plants. Trends Plant Sci. 2001, 6, 354–358. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ni, Z.; Chen, Q.; Qu, Y. The wheat salinity-induced R2R3-MYB transcription factor TaSIM confers salt stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2017, 491, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Jha, B.; Mishra, A.; Jha, A.; Joshi, M. Developing Transgenic Jatropha Using the SbNHX1 Gene from an Extreme Halophyte for Cultivation in Saline Wasteland. PLoS ONE 2013, 8, e71136. [Google Scholar] [CrossRef]
- Apse, M.P.; Aharon, G.S.; Snedden, W.A.; Blumwald, E. Salt Tolerance Conferred by Overexpression of a Vacuolar Na+/H+ Antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar] [CrossRef]
- Katschnig, D.; Bliek, T.; Rozema, J.; Schat, H. Constitutive high-level SOS1 expression and absence of HKT1;1 expression in the salt-accumulating halophyte Salicornia dolichostachya. Plant Sci. 2015, 234, 144–154. [Google Scholar] [CrossRef]
- Zheng, L.; Dang, Z.; Li, H.; Zhang, H.; Wu, S.; Wang, Y. Isolation and characterization of a Δ1-pyrroline-5-carboxylate synthetase (NtP5CS) from Nitraria tangutorum Bobr. and functional comparison with its Arabidopsis homologue. Mol. Biol. Rep. 2014, 41, 563–572. [Google Scholar] [CrossRef]
- Niu, L.; Liao, W. Hydrogen Peroxide Signaling in Plant Development and Abiotic Responses: Crosstalk with Nitric Oxide and Calcium. Front. Plant Sci. 2016, 7, 230. [Google Scholar] [CrossRef]
- Camejo, D.; Romero-Puertas, M.C.; Rodríguez-Serrano, M.; Sandalio, L.M.; Lázaro, J.J.; Jiménez, A.; Sevilla, F. Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J. Proteom. 2013, 79, 87–99. [Google Scholar] [CrossRef]
- Takahashi, S.; Badger, M.R. Photoprotection in plants: A new light on photosystem II damage. Trends Plant Sci. 2011, 16, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Allakhverdiev, S.; Murata, N. Environmental stress inhibits the synthesis de novo of proteins involved in the photodamage–repair cycle of Photosystem II in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 2004, 1657, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Aragão, R.M.; Silva, E.N.; Vieira, C.F.; Silveira, J.A.G. High supply of NO3 − mitigates salinity effects through an enhancement in the efficiency of photosystem II and CO2 assimilation in Jatropha curcas plants. Acta Physiol. Plant. 2012, 34, 2135–2143. [Google Scholar] [CrossRef]
- Voss, I.; Sunil, B.; Scheibe, R.; Raghavendra, A.S. Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol. 2013, 15, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Singh, S.; Parihar, P.; Mishra, R.K.; Tripathi, D.K.; Singh, V.P.; Chauhan, D.K.; Prasad, S.M. Reactive Oxygen Species (ROS): Beneficial Companions of Plants’ Developmental Processes. Front. Plant Sci. 2016, 7, 1299. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, J.P.; Yuan, F.; Yang, Z.; Wang, B.S.; Chen, M. Transcriptome profiling of genes involved in photosynthesis in Elaeagnus angustifolia L. under salt stress. Photosynthetica 2018, 56, 998–1009. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, C.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Global Analysis of Gene Expression Profiles in Physic Nut (Jatropha curcas L.) Seedlings Exposed to Salt Stress. PLoS ONE 2014, 9, e97878. [Google Scholar] [CrossRef]
- Sapeta, H.; Lourenco, T.; Lorenz, S.; Grumaz, C.; Kirstahler, P.; Barros, P.M.; Costa, J.M.; Sohn, K.; Oliveira, M.M. Transcriptomics and physiological analyses reveal co-ordinated alteration of metabolic pathways in Jatropha curcas drought tolerance. J. Exp. Bot. 2016, 67, 845–860. [Google Scholar] [CrossRef]
- Xiong, W.; Xu, X.; Zhang, L.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas L.). Gene 2013, 524, 124–132. [Google Scholar] [CrossRef]
- Kaur, R.; Bhardwaj, R.; Sharma, R.; Kapoor, D.; Kohli, S.; Kumar, V.; Kaur, P. Hormonal regulation of drought stress in plants. In Water Stress and Crop Plants: A Sustainable Approach; Ahmad, P., Ed.; John Wiley & Sons, Ltd.: West Sussex, UK, 2016; Volume I, pp. 582–599. [Google Scholar]
- Wani, S.H.; Tripathi, P.; Zaid, A.; Challa, G.S.; Kumar, A.; Kumar, V.; Upadhyay, J.; Joshi, R.; Bhatt, M. Transcriptional regulation of osmotic stress tolerance in wheat (Triticum aestivum L.). Plant Mol. Biol. 2018, 97, 469–487. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Y.; Tian, Y.; Dai, T.; Xie, G.; Xu, Y.; Chen, F. Overexpressing Jatropha curcas CBF2 in Nicotiana benthamiana improved plant tolerance to drought stress. Gene 2020, 742, 144588. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xu, X.; Xiong, W.; Wu, P.; Chen, Y.; Li, M.; Wu, G.; Jiang, H. Genome-Wide Analysis of the NAC Gene Family in Physic Nut (Jatropha curcas L.). PLoS ONE 2015, 10, e0131890. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peng, X.; Liu, H.; Wang, D.; Shen, S. Genome-wide identification of the Jatropha curcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2. BMC Genom. 2016, 17, 251. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, K.; Zhang, J.; Li, X.; Xu, K.; Zhang, Y.; Qi, J.; Yu, D.; Wang, J.; Li, C. JcDREB2, a Physic Nut AP2/ERF Gene, Alters Plant Growth and Salinity Stress Responses in Transgenic Rice. Front. Plant Sci. 2017, 8, 306. [Google Scholar] [CrossRef] [PubMed]
- Reijnders, L.; Huijbregts, M.A.J. Biofuels for Road Transport: A Seed to Wheel Perspective; Springer: Dordrecht, The Netherlands, 2009. [Google Scholar]
- Moniruzzaman, M.; Yaakob, Z.; Shahinuzzaman, M.; Khatun, R.; Aminul Islam, A.K.M. Jatropha Biofuel Industry: The Challenges. In Frontiers in Bioenergy and Biofuels, 1st ed.; Jacob-Lopes, E., Queiroz, L.Q., Eds.; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef]
- Montes, J.M.; Melchinger, A.E. Domestication and Breeding of Jatropha curcas L. Trends Plant Sci. 2016, 21, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Tao, Y.-B.; Xu, Z.-F. Genetic transformation and transgenics of Jatropha curcas, a biofuel plant. In Jatropha, Challenges for a New Energy Crop; Mulpuri, S., Carels, N., Bahadur, B., Eds.; Springer: Singapore, 2019; pp. 79–92. [Google Scholar]
- Liu, B.; Wang, W.; Gao, J.; Chen, F.; Wang, S.; Xu, Y.; Tang, L.; Jia, Y. Molecular cloning and characterization of a jasmonate biosynthetic pathway gene for allene oxide cyclase from Jatropha curcas. Acta Physiol. Plant. 2010, 32, 531–539. [Google Scholar] [CrossRef]
- Tsuchimoto, S.; Cartagena, J.; Khemkladngoen, N.; Singkaravanit, S.; Kohinata, T.; Wada, N.; Sakai, H.; Morishita, Y.; Suzuki, H.; Shibata, D.; et al. Development of transgenic plants in jatropha with drought tolerance. Plant Biotechnol. 2012, 29, 137–143. [Google Scholar] [CrossRef]
- Chacuttayapong, W.; Enoki, H.; Nabetani, Y.; Matsui, M.; Oguchi, T.; Motohashi, R. Transformation of Jatropha curcas L. for production of larger seeds and increased amount of biodiesel. Plant Biotechnol. 2021, 38, 247–256. [Google Scholar] [CrossRef]
- King, A.; He, W.; Cuevas, J.A.; Freudenberger, M.; Ramiaramanana, D.; Graham, I.A. Potential of Jatropha curcas as a source of renewable oil and animal feed. J. Exp. Bot. 2009, 60, 2897–2905. [Google Scholar] [CrossRef]
- Tosin, O.V.; Ojonogecha, M.S.; Oloyede, T.L.; Gabriel, S.S.; Chidiebere, A.C.; Bolong, A.-M.A. Dietary implications of toasted Jatropha curcas seed: Insight on zootechnical and hematological parameters of Clarias gariepinus. Comp. Clin. Pathol. 2022, 31, 81–90. [Google Scholar] [CrossRef]
- Devappa, R.K.; Makkar, H.P.S.; Becker, K. Jatropha Toxicity—A Review. J. Toxicol. Environ. Health Part B 2010, 13, 476–507. [Google Scholar] [CrossRef] [PubMed]
- Rakshit, K.; Darukeshwara, J.; Raj, K.R.; Narasimhamurthy, K.; Saibaba, P.; Bhagya, S. Toxicity studies of detoxified Jatropha meal (Jatropha curcas) in rats. Food Chem. Toxicol. 2008, 46, 3621–3625. [Google Scholar] [CrossRef] [PubMed]
- Francis, G.; Makkar, H.P.S.; Becker, K. Antinutritional factors present in plant-derived alternate fish feed ingredients and their effects in fish. Aquaculture 2001, 199, 197–227. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, W.; Wang, Y.; Xu, Y.; Chen, F. The effect of curcin from Jatropha curcas on apoptosis of mouse sarcoma-180 cells. Fitoterapia 2012, 83, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, B.J.; Vanegas, Y.O.; Pompelli, M.F.; Garrido, C.G.; Bezerra Neto, E.; Orozco, A.J. Detoxification of Jatropha curcas L. seed meal as possible alternative livestock feed in the Colombian Caribbean. Rev. Actual. Divulg. Científica 2014, 17, 171–178. [Google Scholar]
- Das, M.; Uppal, H.; Singh, R.; Beri, S.; Mohan, K.; Gupta, V.C.; Adholeya, A. Co-composting of physic nut (Jatropha curcas) deoiled cake with rice straw and different animal dung. Bioresour. Technol. 2011, 102, 6541–6546. [Google Scholar] [CrossRef]
- Saha, S.; Ghosh, K. Evaluation of Nutritive Value of Raw and Fermented De-oiled Physic Nut, Jatropha curcas Seed Meal in the Formulated Diets for Rohu, Labeo rohita (Hamilton) Fingerlings. Proc. Zool. Soc. 2013, 66, 41–50. [Google Scholar] [CrossRef]
- Hassaan, M.; Goda, A.-S.; Kumar, V. Evaluation of nutritive value of fermented de-oiled physic nut, Jatropha curcas, seed meal for Nile tilapia Oreochromis niloticus fingerlings. Aquac. Nutr. 2017, 23, 571–584. [Google Scholar] [CrossRef]
- Hisano, H.; Della Flora, M.A.L.; Pilecco, J.L.; Mendonça, S. Apparent digestibility of nutrients, energy, and amino acid of nontoxic and detoxified physic nut cakes for Nile tilapia. Pesquisa Agropecuária Brasileira 2015, 50, 849–853. [Google Scholar] [CrossRef][Green Version]
- Mendonça, S.; Laviola, B.G. Uso potencial e toxidez da torta de pinhão-manso; Embrapa: Brasília, Brazil, 2009. [Google Scholar]
- Gandhi, V.; Cherian, K.; Mulky, M. Toxicological studies on ratanjyot oil. Food Chem. Toxicol. 1995, 33, 39–42. [Google Scholar] [CrossRef]
- Ratnadass, A.; Wink, M. The Phorbol Ester Fraction from Jatropha curcas Seed Oil: Potential and Limits for Crop Protection against Insect Pests. Int. J. Mol. Sci. 2012, 13, 16157–16171. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Yan, F.; Tang, L.; Chen, F. Antitumor effects of curcin from seeds of Jatropha curcas. Acta Pharmacol. Sin. 2003, 24, 241–246. [Google Scholar] [PubMed]
- Huang, M.-X.; Hou, P.; Wei, Q.; Xu, Y.; Chen, F. A ribosome-inactivating protein (curcin 2) induced from Jatropha curcas can reduce viral and fungal infection in transgenic tobacco. Plant Growth Regul. 2008, 54, 115–123. [Google Scholar] [CrossRef]
- Wu, L.; Goh, M.L.; Tian, D.; Gu, K.; Hong, Y.; Yin, Z. Isolation and characterization of curcin genes with distinct expression patterns in leaves and seeds of Jatropha curcas L. Plant Gene 2017, 9, 34–44. [Google Scholar] [CrossRef]
- Laviola, B.G.; Rodrigues, C.M.; Mendonça, S. Pinhão-Manso Pode ser usado Como Alimentação Animal; Infobibos: Campinas, Brazil, 2012; Available online: http://www.infobibos.com/ (accessed on 17 April 2022).
- Eswaran, N.; Parameswaran, S.; Anantharaman, B.; Raja Krishna Kumar, G.; Sathram, R.K.; Johnson, T.S. Generation of an expressed sequence tag (EST) library from salt stressed roots of Jatropha curcas for identification of abiotic stress-responsive genes. Plant Biol. 2012, 14, 428–437. [Google Scholar] [CrossRef]
- Liu, D.; Wang, L.; Zhai, H.; Song, X.; He, S.; Liu, Q. A novel α/β-hydrolase gene IbMas enhances salt tolerance in transgenic sweet potato. PLoS ONE 2014, 9, e115128. [Google Scholar] [CrossRef]
- Liu, D.; He, S.; Song, X.; Zhai, H.; Liu, N.; Zhang, D.; Liu, Q. IbSIMT1, a novel salt induced methyltransferase gene from Ipomoea batatas, is involved in salt tolerance. Plant Cell Tissue Organ. Cult. 2015, 120, 701–715. [Google Scholar] [CrossRef]
- Chen, M.; Chen, J.; Fang, J.; Guo, Z.; Lu, S. Down-regulation of S-adenosylmethionine decarboxylase genes results in reduced plant length, pollen viability, and abiotic stress tolerance. Plant Cell Tissue Organ. Cult. 2014, 116, 311–322. [Google Scholar] [CrossRef]
- Tang, Y.F.; Zhou, Q.; Zhao, Y.M.; Peng, Y.Z. Efficient removal of methyl violet from aqueous solution by a low-cost adsor-bent—C. camphora fallen leaves powder. J. Disper. Sci. Technol. 2017, 38, 1135–1141. [Google Scholar] [CrossRef]
- Wang, H.; Zou, Z.; Wang, S.; Gong, M. Deep sequencing-based transcriptome analysis of the oil-bearing plant Physic Nut (Jatropha curcas L.) under cold stress. Plant Omics 2014, 7, 178–187. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).