The Use of Near-Infrared Imaging (NIR) as a Fast Non-Destructive Screening Tool to Identify Drought-Tolerant Wheat Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Imaging Experiments
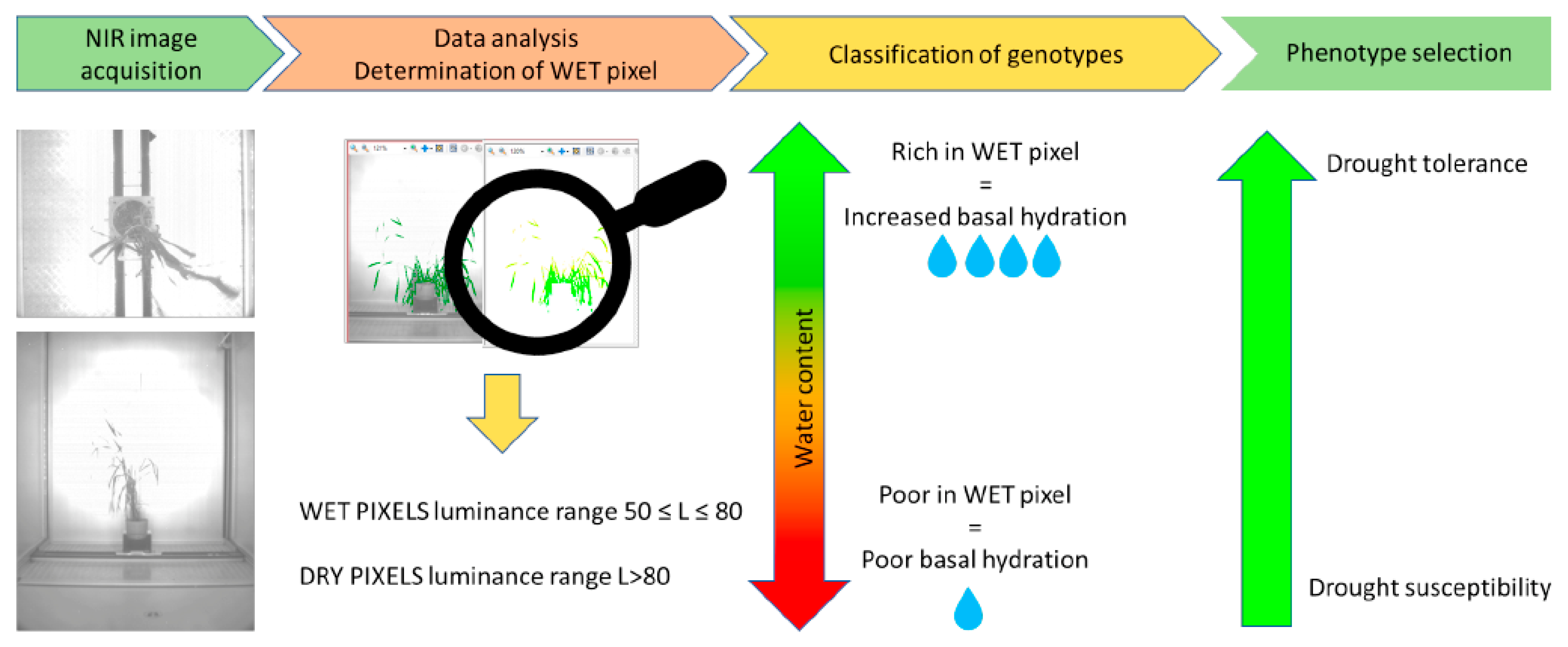
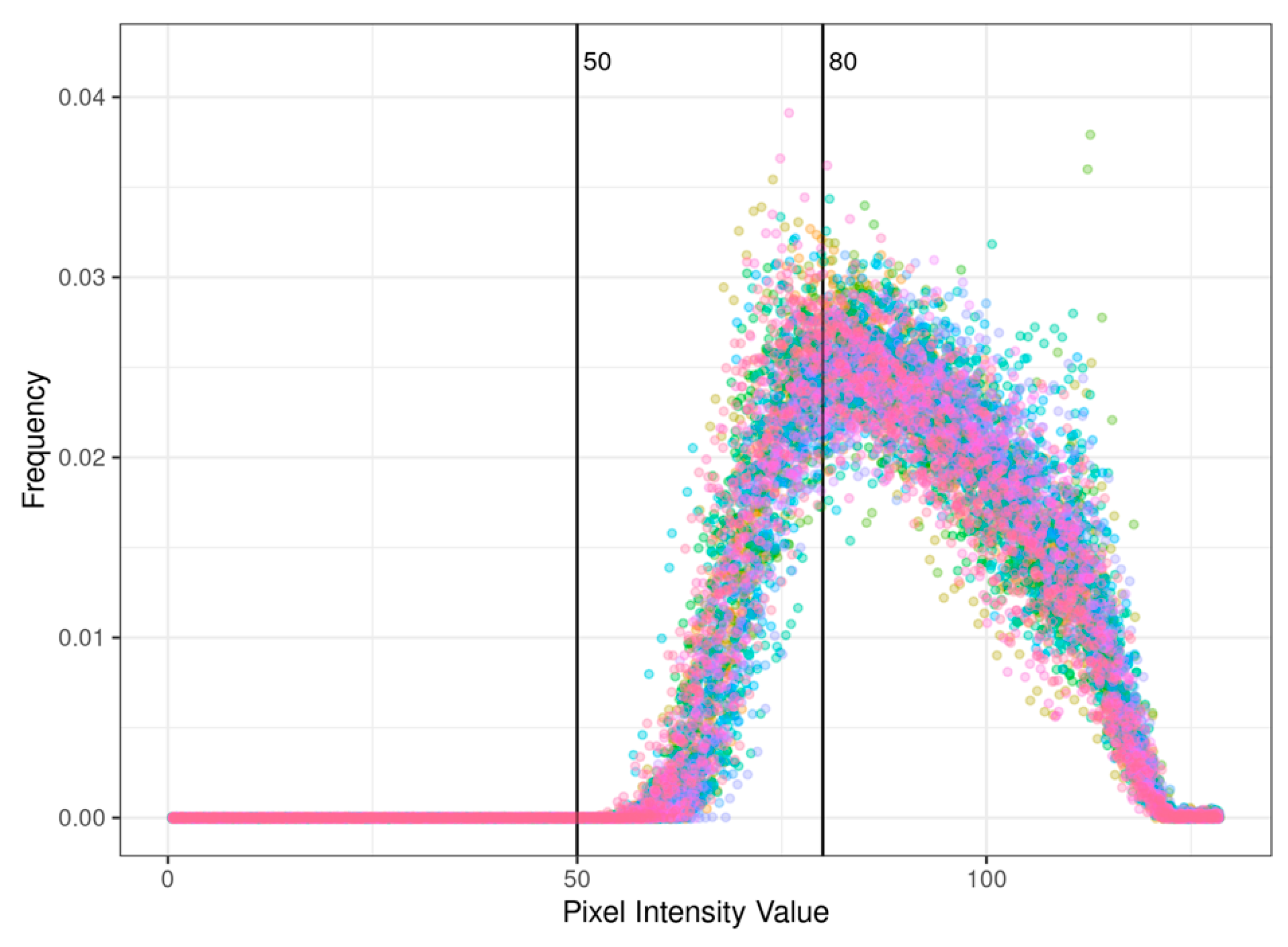
2.3. Data Analysis
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pistorius, R. Scientists, Plants and Politics. Scientists, Plants and Politics: A History of the Plant Genetic Resources Movement; International Plant Genetic Resources Institute: Rome, Italy, 1997; ISBN 978-9290433088. [Google Scholar]
- Adamski, N.M.; Borrill, P.; Brinton, J.; Harrington, S.A.; Marchal, C.; Bentley, A.R.; Bovill, W.D.; Cattivelli, L.; Cockram, J.; Contreras-Moreira, B.; et al. A Roadmap for Gene Functional Characterisation in Crops with Large Genomes: Lessons from Polyploid Wheat. eLife 2020, 9, e55646. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Redondo, J.; Piñero, D.; Eguiarte, L.E. Genomic, Transcriptomic and Epigenomic Tools to Study the Domestication of Plants and Animals: A Field Guide for Beginners. Front. Genet. 2020, 11, 742. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Mujeeb-Kazi, A.; Ogbonnaya, F.C.; He, Z.; Rajaram, S. Wheat Genetic Resources in the Post-Genomics Era: Promise and Challenges. Ann. Bot. 2018, 121, 603–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, M.; Tattaris, M.; Cossani, C.M.; Ellis, M.; Yamaguchi-Shinozaki, K.; Pierre, C.S. Exploring Genetic Resources to Increase Adaptation of Wheat to Climate Change. In Proceedings of the Advances in Wheat Genetics: From Genome to Field; Ogihara, Y., Takumi, S., Handa, H., Eds.; Springer: Tokyo, Japan, 2015; pp. 355–368. [Google Scholar]
- Segelbacher, G.; Bosse, M.; Burger, P.; Galbusera, P.; Godoy, J.A.; Helsen, P.; Hvilsom, C.; Iacolina, L.; Kahric, A.; Manfrin, C.; et al. New Developments in the Field of Genomic Technologies and Their Relevance to Conservation Management. Conserv. Genet. 2022, 23, 217–242. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Ndjiondjop, M.-N.; Henry, R.J. Role of Genomics in Promoting the Utilization of Plant Genetic Resources in Genebanks. Brief. Funct. Genom. 2018, 17, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Schurr, U.; Loreto, F.; Menesatti, P.; Carpentier, S. Plant Phenotyping Research Trends, a Science Mapping Approach. Front. Plant Sci. 2019, 9, 1933. [Google Scholar] [CrossRef] [Green Version]
- Morisse, M.; Wells, D.M.; Millet, E.J.; Lillemo, M.; Fahrner, S.; Cellini, F.; Lootens, P.; Muller, O.; Herrera, J.M.; Bentley, A.R.; et al. A European Perspective on Opportunities and Demands for Field-Based Crop Phenotyping. Field Crops Res. 2022, 276, 108371. [Google Scholar] [CrossRef]
- Resende, R.T.; Piepho, H.-P.; Rosa, G.J.M.; Silva-Junior, O.B.; E Silva, F.F.; de Resende, M.D.V.; Grattapaglia, D. Enviromics in Breeding: Applications and Perspectives on Envirotypic-Assisted Selection. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2021, 134, 95–112. [Google Scholar] [CrossRef]
- Würschum, T. Modern Field Phenotyping Opens New Avenues for Selection. In Applications of Genetic and Genomic Research in Cereals; Miedaner, T., Korzun, V., Eds.; Woodhead Publishing: Sawston, UK, 2019; pp. 233–250. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Jimenez-Berni, J.; Fischer, R.A.; Deery, D.M.; Smith, D.J. Review: High-Throughput Phenotyping to Enhance the Use of Crop Genetic Resources. Plant Sci. 2019, 282, 40–48. [Google Scholar] [CrossRef]
- Reynolds, M.; Chapman, S.; Crespo-Herrera, L.; Molero, G.; Mondal, S.; Pequeno, D.N.L.; Pinto, F.; Pinera-Chavez, F.J.; Poland, J.; Rivera-Amado, C.; et al. Breeder Friendly Phenotyping. Plant Sci. 2020, 295, 110396. [Google Scholar] [CrossRef]
- Watt, M.; Fiorani, F.; Usadel, B.; Rascher, U.; Muller, O.; Schurr, U. Phenotyping: New Windows into the Plant for Breeders. Annu. Rev. Plant Biol. 2020, 71, 689–712. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Ferrio, J.P.; Buxó, R.; Voltas, J. The Historical Perspective of Dryland Agriculture: Lessons Learned from 10,000 Years of Wheat Cultivation. J. Exp. Bot. 2007, 58, 131–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- del Pozo, A.; Brunel-Saldias, N.; Engler, A.; Ortega-Farias, S.; Acevedo-Opazo, C.; Lobos, G.A.; Jara-Rojas, R.; Molina-Montenegro, M.A. Climate Change Impacts and Adaptation Strategies of Agriculture in Mediterranean-Climate Regions (MCRs). Sustainability 2019, 11, 2769. [Google Scholar] [CrossRef] [Green Version]
- Branlard, G.; Giraldo, P.; He, Z.; Igrejas, G.; Ikeda, T.M.; Janni, M.; Labuschagne, M.T.; Wang, D.; Wentzel, B.; Zhang, K. Contribution of Genetic Resources to Grain Storage Protein Composition and Wheat Quality. In Wheat Quality For Improving Processing And Human Health; Igrejas, G., Ikeda, T., Guzmán, C., Eds.; Springer: Cham, Switzerland, 2020; pp. 39–72. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum Wheat Genome Highlights Past Domestication Signatures and Future Improvement Targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzucotelli, E.; Sciara, G.; Mastrangelo, A.M.; Desiderio, F.; Xu, S.S.; Faris, J.; Hayden, M.J.; Tricker, P.J.; Ozkan, H.; Echenique, V.; et al. The Global Durum Wheat Panel (GDP): An International Platform to Identify and Exchange Beneficial Alleles. Front. Plant Sci. 2020, 11, 2036. [Google Scholar] [CrossRef]
- Pignone, D.; De Paola, D.; Rapanà, N.; Janni, M. Single Seed Descent: A Tool to Exploit Durum Wheat (Triticum Durum Desf.) Genetic Resources. Genet. Resour. Crop Evol. 2015, 62, 1029–1035. [Google Scholar] [CrossRef]
- Wang, C.; Hu, S.; Gardner, C.; Lübberstedt, T. Emerging Avenues for Utilization of Exotic Germplasm. Trends Plant Sci. 2017, 22, 624–637. [Google Scholar] [CrossRef] [Green Version]
- Danzi, D.; Briglia, N.; Petrozza, A.; Summerer, S.; Povero, G.; Stivaletta, A.; Cellini, F.; Pignone, D.; De Paola, D.; Janni, M. Can High Throughput Phenotyping Help Food Security in the Mediterranean Area? Front. Plant Sci. 2019, 10, 15. [Google Scholar] [CrossRef]
- Pignone, D.; De Paola, D.; Rapanà, N.; Janni, M. Capturing Wild Relative and Landrace Diversity for Crop Improvement Using a New Selection Tool to Exploit Genetic Resources in Durum Wheat. In Enhancing Crop Genepool Use: Capturing Wild Relative and Landrace Diversity for Crop Improvement; CAB Internationa: Wallingford, UK, 2016; pp. 47–53. [Google Scholar]
- Buffagni, V.; Vurro, F.; Janni, M.; Gullì, M.; Keller, A.A.; Marmiroli, N. Shaping Durum Wheat for the Future: Gene Expression Analyses and Metabolites Profiling Support the Contribution of BCAT Genes to Drought Stress Response. Front. Plant Sci. 2020, 11, 891. [Google Scholar] [CrossRef]
- Beverly, D.P.; Guadagno, C.R.; Ewers, B.E. Biophysically Informed Imaging Acquisition of Plant Water Status. Front. For. Glob. Change 2020, 3, 125. [Google Scholar] [CrossRef]
- Petrozza, A.; Santaniello, A.; Summerer, S.; Di Tommaso, G.; Di Tommaso, D.; Paparelli, E.; Piaggesi, A.; Perata, P.; Cellini, F. Physiological Responses to Megafol® Treatments in Tomato Plants under Drought Stress: A Phenomic and Molecular Approach. Sci. Hortic. 2014, 174, 185–192. [Google Scholar] [CrossRef]
- Briglia, N.; Nuzzo, V.; Petrozza, A.; Summerer, S.; Cellini, F.; Montanaro, G. Preliminary High-Throughput Phenotyping Analysis in Grapevines under Drought. BIO Web Conf. 2019, 13, 02003. [Google Scholar] [CrossRef]
- Berger, B.; Parent, B.; Tester, M. High-Throughput Shoot Imaging to Study Drought Responses. J. Exp. Bot. 2010, 61, 3519–3528. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| DAS | 60 | 67 | 74 | 81 | 88 |
|---|---|---|---|---|---|
| Zadoks stage | 31 | 32 | 33 | 35 | 36 |
| Read number | 1 | 2 | 3 | 4 | 5 |
| SSD Genotype | Mean | se | SSD Genotype | Mean | se | SSD Genotype | Mean | se |
|---|---|---|---|---|---|---|---|---|
| SSD393 | 0.1234 | 0.0199 | SSD182 | 0.2577 | 0.0355 | SSD255 | 0.311918 | 0.044896 |
| SSD240 | 0.1285 | 0.0232 | SSD336 | 0.2580 | 0.0794 | SSD125 | 0.317003 | 0.031154 |
| SSD266 | 0.1521 | 0.0309 | SSD52 | 0.2583 | 0.0426 | SSD412 | 0.31701 | 0.03785 |
| SSD511 | 0.1550 | 0.0115 | SSD59 | 0.2601 | 0.0176 | SSD246 | 0.318829 | 0.013851 |
| SSD322 | 0.1691 | 0.0292 | SSD315 | 0.2627 | 0.0055 | SSD447 | 0.320201 | 0.043648 |
| SSD338 | 0.1709 | 0.0127 | SSD220 | 0.2665 | 0.0359 | SSD407 | 0.321094 | 0.037618 |
| SSD416 | 0.1985 | 0.0141 | SSD345 | 0.2672 | 0.0512 | SSD467 | 0.323395 | 0.061267 |
| SSD330 | 0.2016 | 0.0765 | SSD99 | 0.2683 | 0.0549 | SSD79 | 0.323652 | 0.046922 |
| SSD397 | 0.2037 | 0.0345 | SSD86 | 0.2706 | 0.0708 | SSD499 | 0.323756 | 0.018017 |
| SSD343 | 0.2038 | 0.0233 | SSD96 | 0.2729 | 0.0280 | SSD262 | 0.324868 | 0.062367 |
| SSD278 | 0.2052 | 0.0355 | SSD83 | 0.2740 | 0.0146 | SSD281 | 0.326212 | 0.048101 |
| SSD432 | 0.2052 | 0.0226 | SSD54 | 0.2744 | 0.0211 | SSD178 | 0.327289 | 0.059534 |
| SSD128 | 0.2056 | 0.0435 | SSD195 | 0.2746 | 0.0847 | SSD65 | 0.328755 | 0.03194 |
| SSD239 | 0.2063 | 0.0427 | SSD415 | 0.2747 | 0.0299 | SSD64 | 0.334353 | 0.056684 |
| SSD271 | 0.2064 | 0.0676 | SSD6 | 0.2773 | 0.0608 | SSD244 | 0.335239 | 0.008056 |
| SSD253 | 0.2083 | 0.0288 | SSD91 | 0.2810 | 0.0395 | SSD477 | 0.335793 | 0.030472 |
| SSD269 | 0.2087 | 0.0336 | SSD431 | 0.2817 | 0.0738 | SSD335 | 0.337223 | 0.080187 |
| SSD180 | 0.2151 | 0.0251 | SSD424 | 0.2828 | 0.0787 | SSD36 | 0.337427 | 0.026768 |
| SSD326 | 0.2216 | 0.0304 | SSD480 | 0.2833 | 0.0371 | SSD70 | 0.338503 | 0.040525 |
| SSD453 | 0.2237 | 0.0254 | SSD24 | 0.2849 | 0.0507 | SSD470 | 0.347294 | 0.042785 |
| SSD155 | 0.2261 | 0.0369 | SSD280 | 0.2858 | 0.0421 | SSD509 | 0.348098 | 0.018469 |
| SSD409 | 0.2276 | 0.0335 | SSD288 | 0.2859 | 0.0234 | SSD15 | 0.348783 | 0.050455 |
| SSD146 | 0.2281 | 0.1248 | SSD142 | 0.2889 | 0.0958 | SSD231 | 0.351929 | 0.033402 |
| SSD399 | 0.2306 | 0.0190 | SSD414 | 0.2895 | 0.0456 | SSD219 | 0.352659 | 0.055812 |
| SSD112 | 0.2310 | 0.0647 | SSD400 | 0.2896 | 0.0093 | SSD494 | 0.35297 | 0.028618 |
| SSD328 | 0.2320 | 0.0289 | SSD348 | 0.2906 | 0.0453 | SSD122 | 0.367246 | 0.00449 |
| SSD123 | 0.2325 | 0.0651 | SSD2 | 0.2941 | 0.0400 | SSD256 | 0.36811 | 0.030159 |
| SSD157 | 0.2329 | 0.0286 | SSD173 | 0.2966 | 0.0542 | SSD451 | 0.371517 | 0.058267 |
| SSD43 | 0.2352 | 0.0368 | SSD298 | 0.2977 | 0.0149 | SSD526 | 0.374167 | 0.043784 |
| SSD290 | 0.2358 | 0.0458 | SSD7 | 0.2984 | 0.0337 | SSD111 | 0.376019 | 0.011487 |
| SSD421 | 0.2363 | 0.0210 | SSD308 | 0.2985 | 0.0212 | SSD227 | 0.378514 | 0.023269 |
| SSD350 | 0.2370 | 0.0048 | SSD44 | 0.2987 | 0.0525 | SSD325 | 0.385445 | 0.024594 |
| SSD487 | 0.2390 | 0.0071 | SSD69 | 0.3000 | 0.0637 | SSD158 | 0.391514 | 0.066688 |
| SSD422 | 0.2396 | 0.0070 | SSD426 | 0.3001 | 0.0170 | SSD245 | 0.397842 | 0.064106 |
| SSD411 | 0.2425 | 0.0340 | SSD507 | 0.3004 | 0.0561 | SSD162 | 0.400995 | 0.057963 |
| SSD303 | 0.2450 | 0.0061 | SSD292 | 0.3009 | 0.0239 | SSD168 | 0.404066 | 0.059416 |
| SSD237 | 0.2451 | 0.0324 | SSD66 | 0.3010 | 0.0080 | SSD459 | 0.404357 | 0.007627 |
| SSD107 | 0.2457 | 0.0629 | SSD137 | 0.3034 | 0.0306 | SSD500 | 0.413246 | 0.078537 |
| SSD243 | 0.2458 | 0.0623 | SSD113 | 0.3035 | 0.0221 | SSD171 | 0.415962 | 0.010175 |
| SSD116 | 0.2484 | 0.0245 | SSD294 | 0.3036 | 0.0441 | SSD483 | 0.428288 | 0.050466 |
| SSD441 | 0.2509 | 0.0627 | SSD120 | 0.3037 | 0.0184 | SSD283 | 0.433356 | 0.064512 |
| SSD135 | 0.2514 | 0.0221 | SSD423 | 0.3047 | 0.0519 | SSD525 | 0.442475 | 0.011191 |
| SSD92 | 0.2517 | 0.0157 | SSD274 | 0.3066 | 0.0707 | SSD505 | 0.450073 | 0.031612 |
| SSD302 | 0.2545 | 0.0346 | SSD147 | 0.3080 | 0.0631 | SSD457 | 0.450712 | 0.046915 |
| SSD443 | 0.2546 | 0.0348 | SSD427 | 0.308078 | 0.060848 | SSD109 | 0.508032 | 0.057536 |
| SSD35 | 0.2562 | 0.0656 | SSD513 | 0.311494 | 0.045601 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danzi, D.; De Paola, D.; Petrozza, A.; Summerer, S.; Cellini, F.; Pignone, D.; Janni, M. The Use of Near-Infrared Imaging (NIR) as a Fast Non-Destructive Screening Tool to Identify Drought-Tolerant Wheat Genotypes. Agriculture 2022, 12, 537. https://doi.org/10.3390/agriculture12040537
Danzi D, De Paola D, Petrozza A, Summerer S, Cellini F, Pignone D, Janni M. The Use of Near-Infrared Imaging (NIR) as a Fast Non-Destructive Screening Tool to Identify Drought-Tolerant Wheat Genotypes. Agriculture. 2022; 12(4):537. https://doi.org/10.3390/agriculture12040537
Chicago/Turabian StyleDanzi, Donatella, Domenico De Paola, Angelo Petrozza, Stephan Summerer, Francesco Cellini, Domenico Pignone, and Michela Janni. 2022. "The Use of Near-Infrared Imaging (NIR) as a Fast Non-Destructive Screening Tool to Identify Drought-Tolerant Wheat Genotypes" Agriculture 12, no. 4: 537. https://doi.org/10.3390/agriculture12040537
APA StyleDanzi, D., De Paola, D., Petrozza, A., Summerer, S., Cellini, F., Pignone, D., & Janni, M. (2022). The Use of Near-Infrared Imaging (NIR) as a Fast Non-Destructive Screening Tool to Identify Drought-Tolerant Wheat Genotypes. Agriculture, 12(4), 537. https://doi.org/10.3390/agriculture12040537








