N2O Emission and Nitrification/Denitrification Bacterial Communities in Upland Black Soil under Combined Effects of Early and Immediate Moisture
Abstract
:1. Introduction
2. Materials and Methods
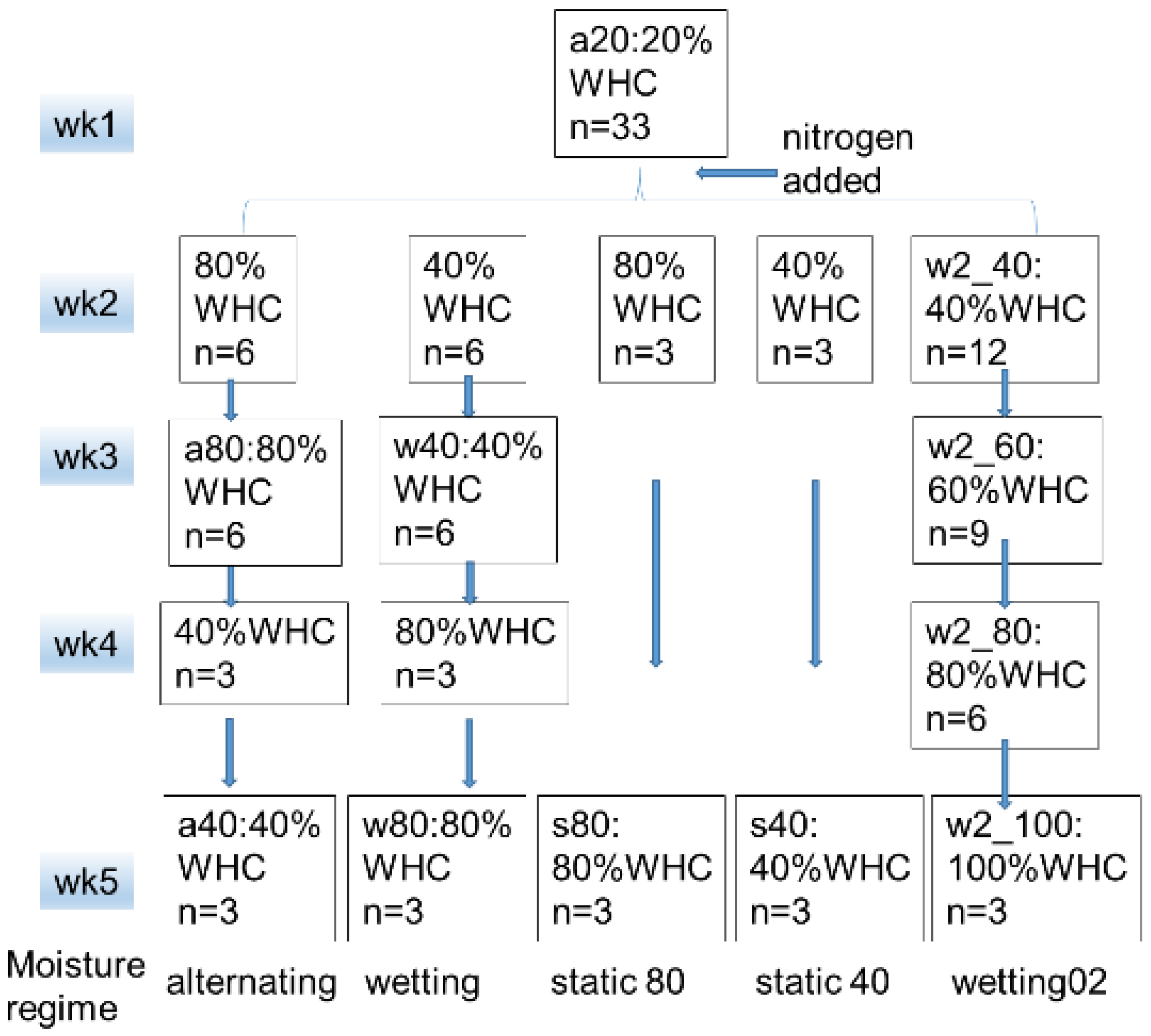
2.1. Soil Sampling and Treatment
2.2. Soil Physicochemical Analyses
2.3. Soil Microbial Community Analyses
2.3.1. Soil DNA Extraction and Quantitative PCR (qPCR)
2.3.2. 16S rRNA Amplicon Sequencing and Bioinformatics
3. Results
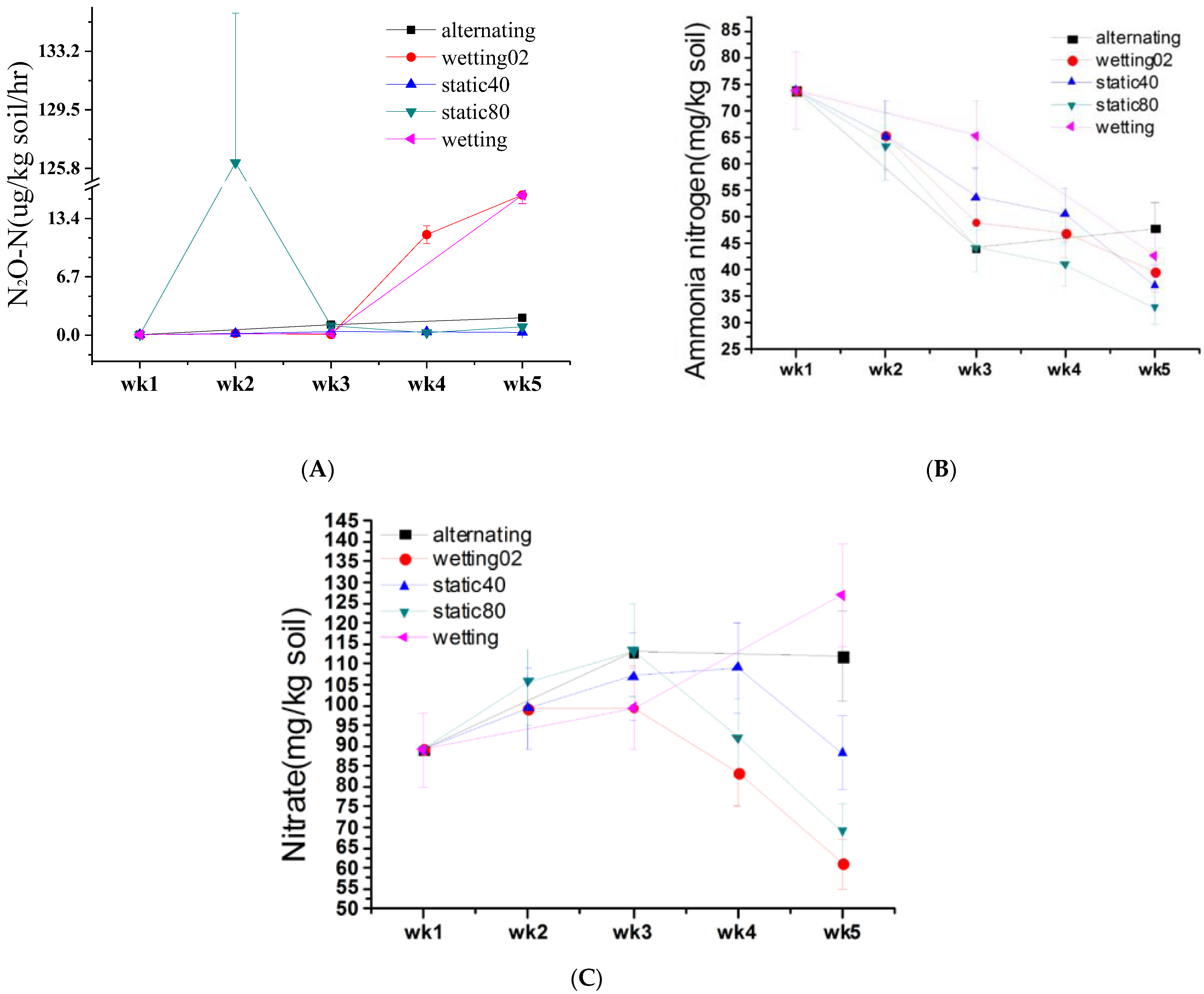
3.1. Impact of Soil Moisture on N2O Emission and Soil Properties
3.1.1. Impact of Soil Moisture on N2O Emission
3.1.2. Impact of Soil Moisture on Soil Properties
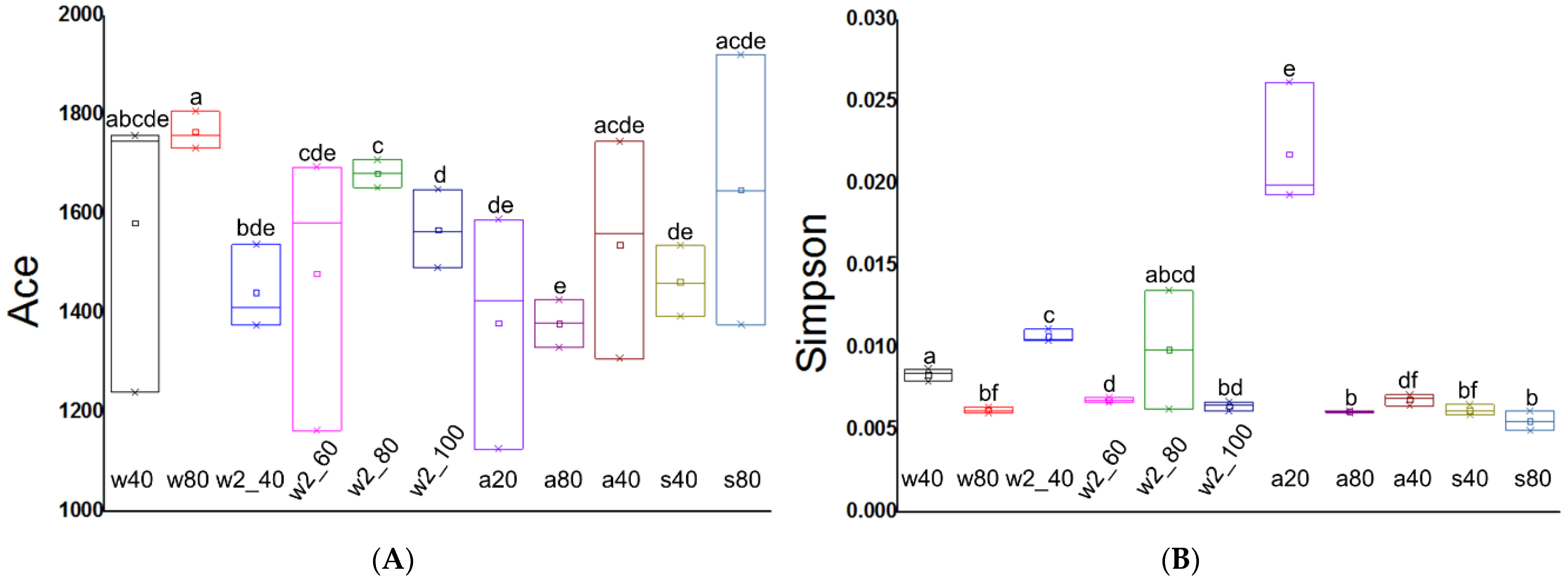
3.2. Alpha Diversity of Soil Microbial Community
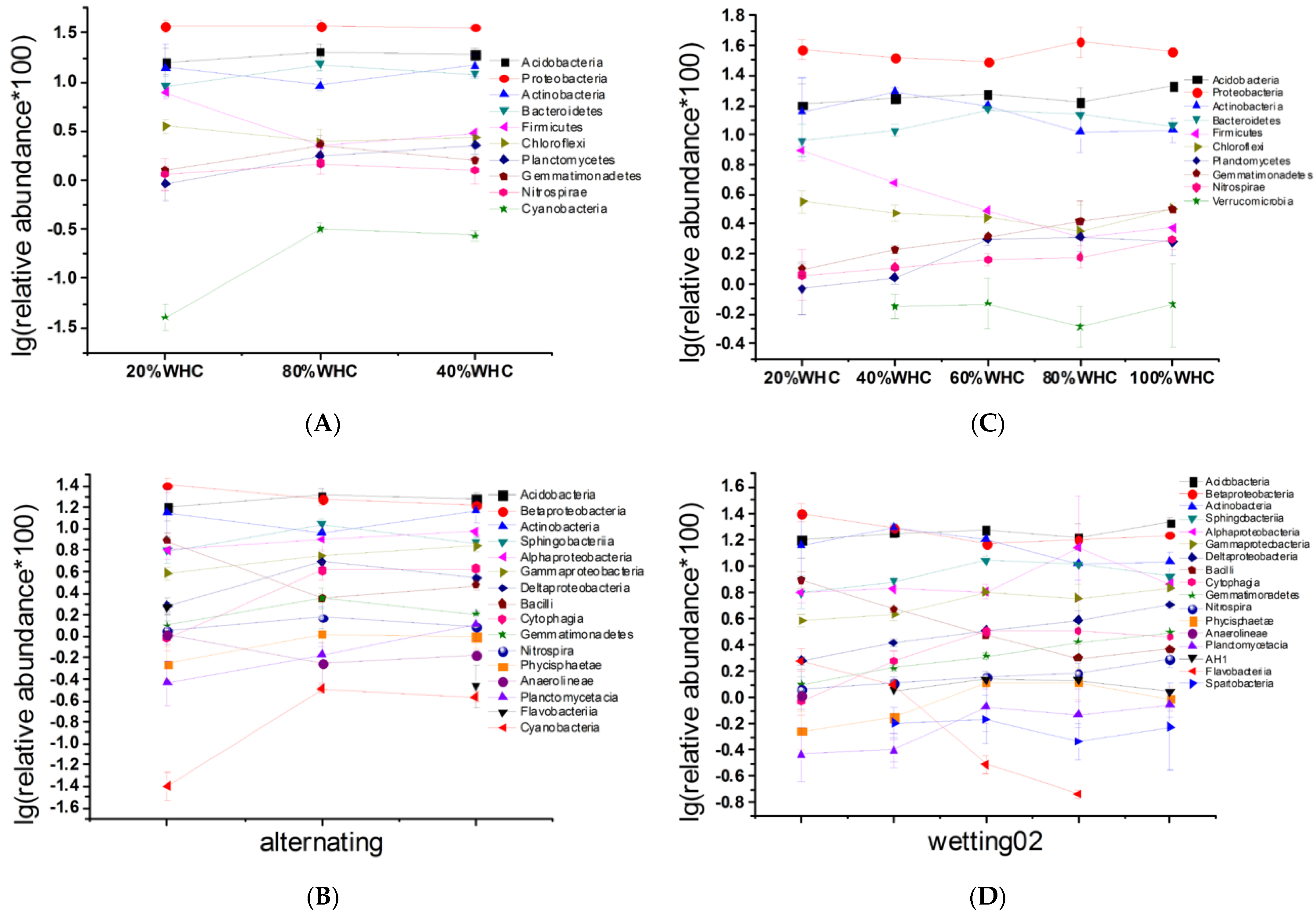
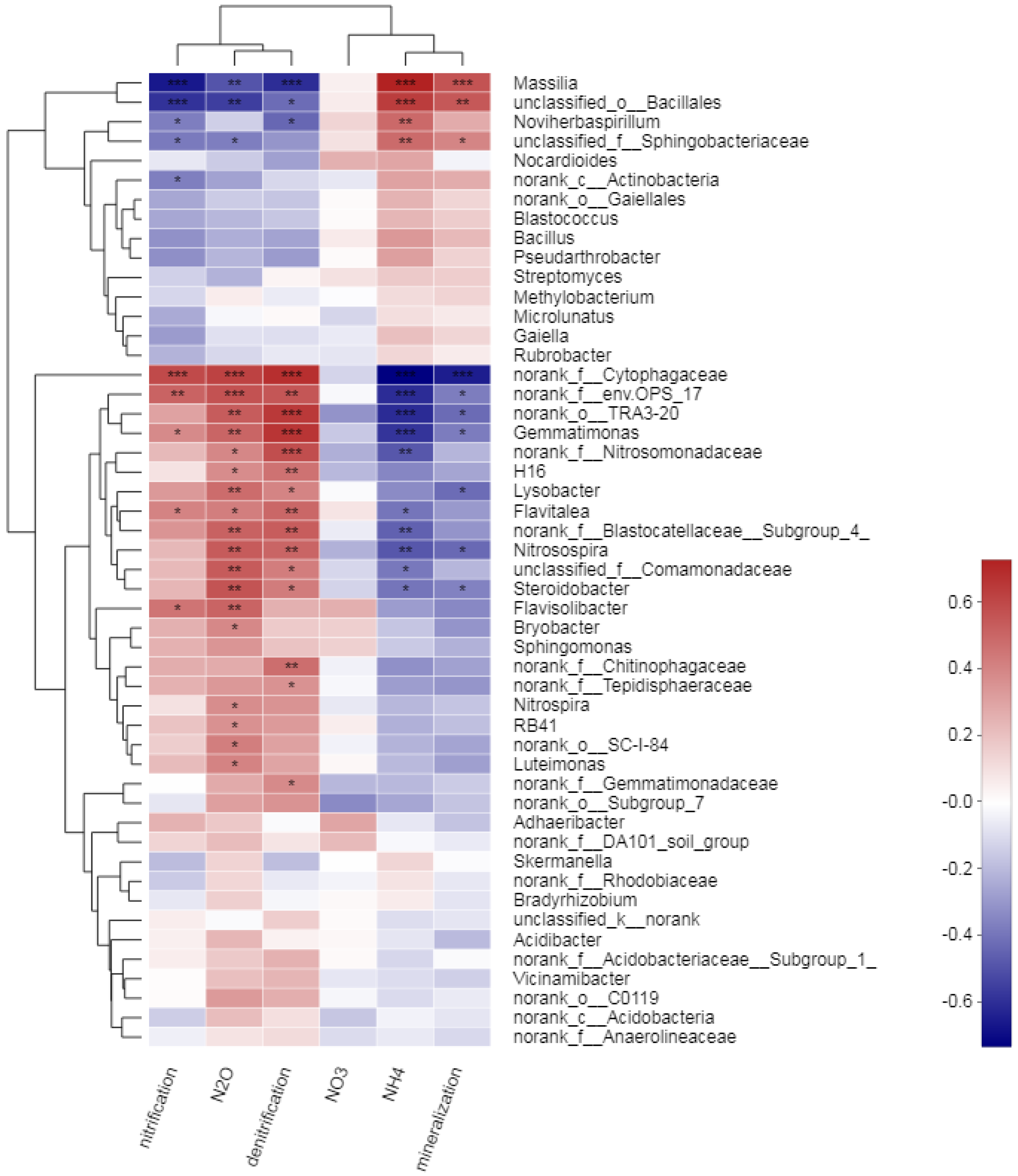
3.3. Microbial Community Composition and Key Responsive Taxonomic Groups
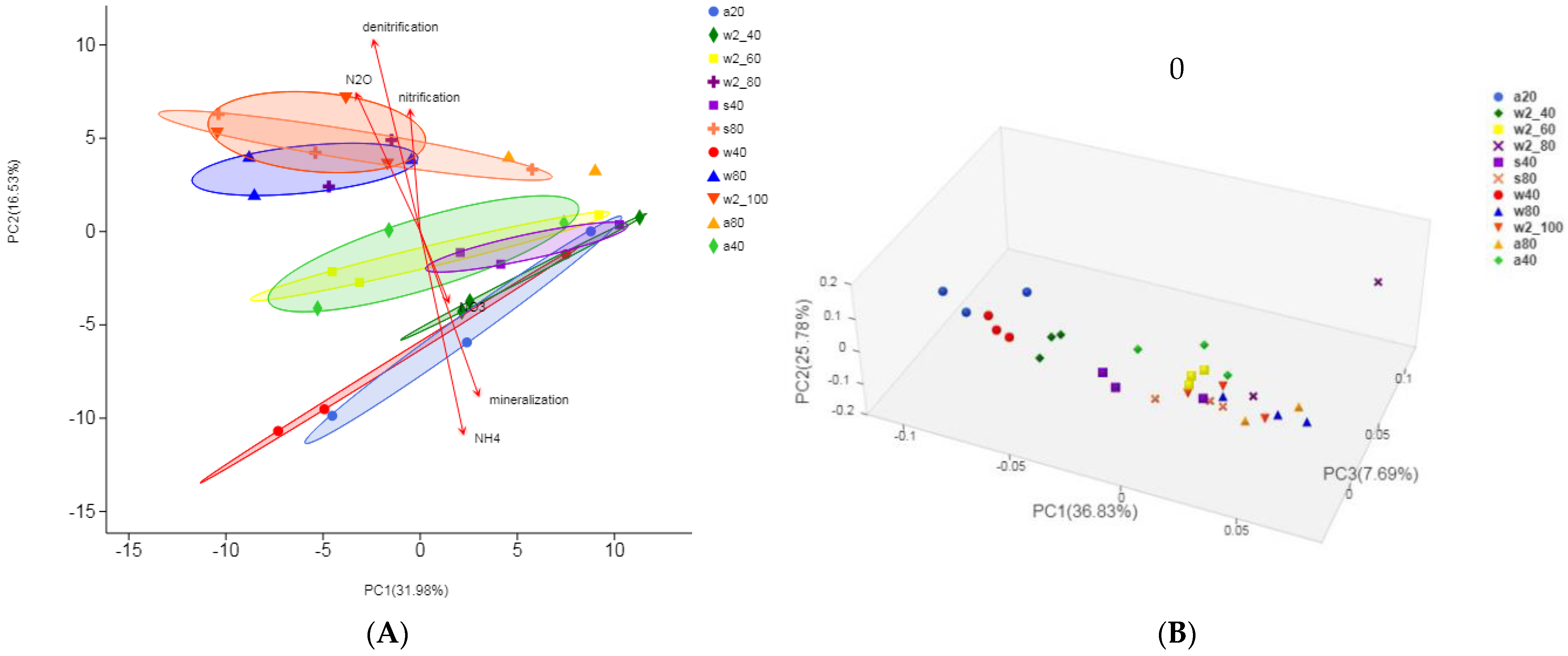
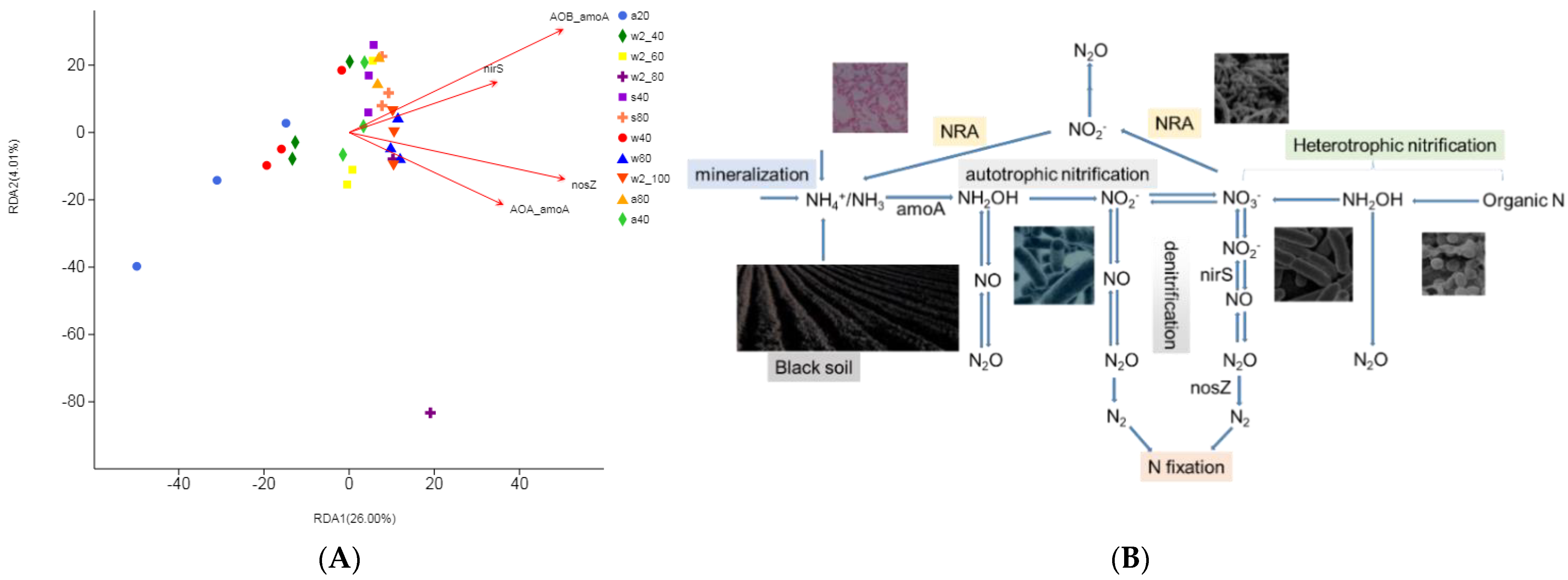
3.4. Beta Diversity of Soil Microbial Community and Taxonomic Difference Analysis
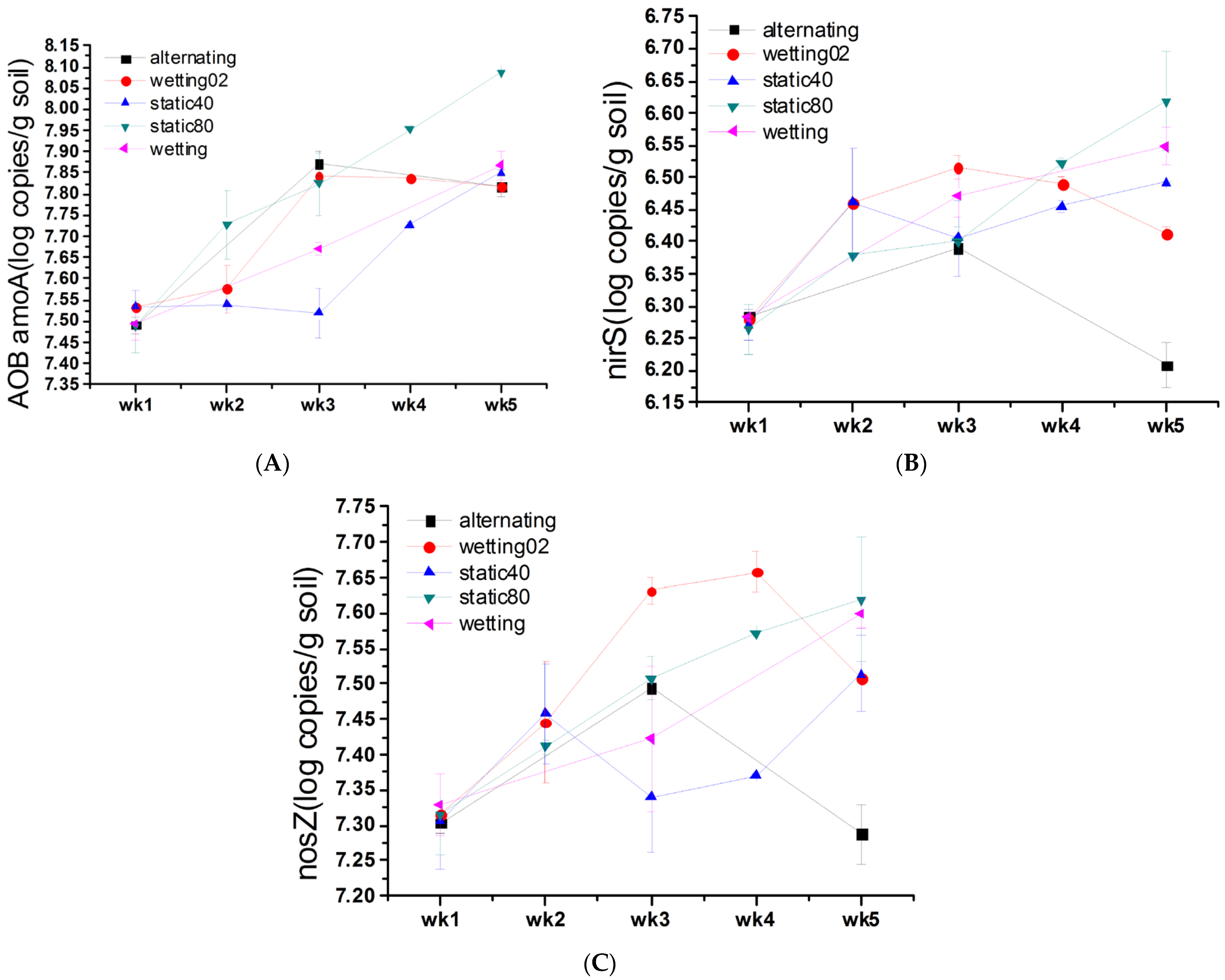
3.5. Impact of Soil Moisture on Gene Abundance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Han, Z.; Deng, M.; Yuan, A.; Wang, J.; Li, H.; Ma, J. Vertical variation of a black soil’s properties in response to freeze-thaw cycles and its links to shift of microbial community structure. Sci. Total Environ. 2018, 625, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.; Li, P.; Han, X.; Qiao, Y.F.; Miao, S.J. Gross nitrogen transformations in black soil under different land uses and management systems. Biol. Fertil. Soils 2016, 52, 233–241. [Google Scholar] [CrossRef]
- Wang, L.F.; Cai, Z.C.; Yang, L.F.; Meng, L. Effect of disturbance and glucose addition on nitrous oxide and carbon dioxide emissions from a paddy soil. Soil Till. Res. 2005, 82, 185–194. [Google Scholar] [CrossRef]
- Lin, S.; Hernandez-Ramirez, G. Nitrous oxide emissions from manured soils as a function of various nitrification inhibitor rates and soil moisture contents. Sci. Total Environ. 2020, 738, 139669. [Google Scholar] [CrossRef]
- Thilakarathna, S.K.; Hernandez-Ramirez, G. Primings of soil organic matter and denitrification mediate the effects of moisture on nitrous oxide production. Soil Biol. Biochem. 2021, 155, 108166. [Google Scholar] [CrossRef]
- Wu, D.M.; Dong, W.X.; Oenema, O.; Wang, Y.Y.; Trebs, I.; Hu, C.S. N2O consumption by low-nitrogen soil and its regulation by water and oxygen. Soil Biol. Biochem. 2013, 60, 165–172. [Google Scholar] [CrossRef]
- Wang, L.F.; Cai, Z.C. Nitrous oxide and carbon dioxide emissions from upland acidic soils under flooding and moistening pretreatments. Acta Sci. Circumstantiae 2011, 31, 1736–1744. [Google Scholar]
- Peralta, A.L.; Ludmer, S.; Kent, A.D. Hydrologic history influences microbial community composition and nitrogen cycling under experimental drying/wetting treatments. Soil Biol. Biochem. 2013, 66, 29–37. [Google Scholar] [CrossRef]
- Banerjee, S.; Helgason, B.; Wang, L.F.; Winsley, T.; Ferrari, B.C.; Siciliano, S.D. Legacy effects of soil moisture on microbial community structure and N2O emissions. Soil Biol. Biochem. 2016, 95, 40–50. [Google Scholar] [CrossRef]
- Krichels, A.; DeLucia, E.H.; Sanford, R.; Chee-Sanford, J.; Yang, W.H. Historical soil drainage mediates the response of soil greenhouse gas emissions to intense precipitation events. Biogeochemistry 2019, 142, 425–442. [Google Scholar] [CrossRef]
- Maljanen, M.; Yli-Pirilä, P.; Hytönen, J.; Joutsensaari, J.; Martikainen, P.J. Acidic northern soils as sources of atmospheric nitrous acid (HONO). Soil Biol. Biochem. 2013, 67, 94–97. [Google Scholar] [CrossRef]
- Donaldson, M.A.; Bish, D.L.; Raff, J.D. Soil surface acidity plays a determining role in the atmospheric-terrestrial exchange of nitrous acid. Proc. Natl. Acad. Sci. USA 2014, 111, 18472–18477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.F.; Cai, Z.C. Nitrous oxide production at different soil moisture contents in an arable soil in China. Soil Sci. Plant Nutr. 2008, 54, 786–793. [Google Scholar] [CrossRef]
- Qin, H.L.; Xing, X.; Tang, Y.; Zhu, B.L.; Wei, X.M.; Chen, X.B.; Liu, Y. Soil moisture and activity of nitrite- and nitrous oxide-reducing microbes enhanced nitrous oxide emissions in fallow paddy soils. Biol. Fert. Soil. 2020, 56, 53–67. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Osanai, Y.; Lai, K.; Anderson, I.C.; Bange, M.P.; Tissue, D.T.; Singh, B.K. Flooding and prolonged drought have differential legacy impacts on soil nitrogen cycling, microbial communities and plant productivity. Plant Soil 2018, 431, 371–387. [Google Scholar] [CrossRef]
- Shi, X.; Hu, H.W.; Zhu-Barker, X.; Hayden, H.; Wang, J.; Suter, H.; Chen, D.; He, J.Z. Nitrifier-induced denitrification is an important source of soil nitrous oxide and can be inhibited by a nitrification inhibitor 3,4-dimethylpyrazole phosphate. Environ. Microbiol. 2017, 19, 4851–4865. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Burger, M.; Doane, T.A.; Horwath, W.R. Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc. Natl. Acad. Sci. USA 2013, 110, 6328–6333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Changing precipitation pattern alters soil microbial community response to wet-up under a Mediterranean-type climate. ISME J. 2015, 9, 946–957. [Google Scholar] [CrossRef] [Green Version]
- Hao, D.C.; Song, S.M.; Mu, J.; Hu, W.L.; Xiao, P.G. Unearthing microbial diversity of Taxus rhizosphere via MiSeq high-throughput amplicon sequencing and isolate characterization. Sci. Rep. 2016, 6, 22006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obayomi, O.; Edelstein, M.; Safi, J.; Mihiret, M.; Ghazaryan, L.; Vonshak, A.; Bernstein, N.; Gillor, O. The combined effects of treated wastewater irrigation and plastic mulch cover on soil and crop microbial communities. Biol. Fert. Soils 2020, 56, 729–742. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Y.R.; Zhang, C.J.; Zhang, L.M.; Han, L.L.; Shen, J.P.; He, J.Z. Responses of soil nitrous oxide production and abundances and composition of associated microbial communities to nitrogen and water amendment. Biol. Fertil. Soils 2017, 53, 601–611. [Google Scholar] [CrossRef]
- Guo, L.; Wang, X.; Diao, T.; Ju, X.; Niu, X.G.; Zheng, L.; Zhang, X.; Han, X. N2O emission contributions by different pathways and associated microbial community dynamics in a typical calcareous vegetable soil. Environ. Pollut. 2018, 242, 2005–2013. [Google Scholar] [CrossRef] [PubMed]
- Beare, M.H.; Gregorich, E.G.; St-Georges, P. Compaction effects on CO2 and N2O production during drying and rewetting of soil. Soil Biol. Biochem. 2009, 41, 611–621. [Google Scholar] [CrossRef]
- Guo, X.B.; Drury, C.F.; Yang, X.M.; Zhang, R.D. Influence of constant and fluctuating water contents on nitrous oxide emissions from soils under varying crop rotations. Soil Sci. Soc. Am. J. 2010, 74, 2077–2085. [Google Scholar] [CrossRef]
- Barnard, R.L.; Blazewicz, S.J.; Firestone, M.K. Rewetting of soil: Revisiting the origin of soil CO2 emissions. Soil Biol. Biochem. 2020, 147, 107819. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Osanai, Y.; Lai, K.; Anderson, I.C.; Bange, M.P.; Tissue, D.T.; Singh, B.K. Responses of the soil microbial community to nitrogen fertilizer regimes and historical exposure to extreme weather events: Flooding or prolonged-drought. Soil Biol. Biochem. 2018, 118, 227–236. [Google Scholar] [CrossRef]
- Szukics, U.; Abell, G.C.; Hödl, V.; Mitter, B.; Sessitsch, A.; Hackl, E.; Zechmeister-Boltenstern, S. Nitrifiers and denitrifiers respond rapidly to changed moisture and increasing temperature in a pristine forest soil. FEMS Microbiol. Ecol. 2010, 72, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Bowen, H.; Maul, J.E.; Cavigelli, M.A.; Yarwood, S. Denitrifier abundance and community composition linked to denitrification activity in an agricultural and wetland soil. Appl. Soil Ecol. 2020, 151, 103521. [Google Scholar] [CrossRef]
- Wang, J.Y.; Jia, J.X.; Xiong, Z.Q.; Khalil, M.A.; Xing, G.X. Water regime-nitrogen fertilizer-straw incorporation interaction: Field study on nitrous oxide emissions from a rice agroecosystem in Nanjing, China. Agri. Ecosys. Environ. 2011, 141, 437–446. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Pajares, S.; Campo, J.; Bohannan, B.J.M.; Etchevers, J.D. Environmental controls on soil microbial communities in a seasonally dry tropical forest. Appl. Environ. Microbiol. 2018, 84, e00342-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azarbad, H.; Constant, P.; Giard-Laliberté, C.; Bainard, L.D.; Yergeau, E. Water stress history and wheat genotype modulate rhizosphere microbial response to drought. Soil Biol. Biochem. 2018, 126, 228–236. [Google Scholar] [CrossRef]
- Shen, Q.L.; Zhang, K.L.; Song, J.W.; Shen, J.X.; Xu, J.M.; Inubushi, K.; Brookes, P.C. Contrasting biomass, dynamics and diversity of microbial community following the air-drying and rewetting of an upland and a paddy soil of the same type. Biol. Fert. Soil. 2018, 54, 871–875. [Google Scholar] [CrossRef]
- Preece, C.; Verbruggen, E.; Liu, L.; Weedon, J.T.; Peñuelas, J. Effects of past and current drought on the composition and diversity of soil microbial communities. Soil Biol. Biochem. 2019, 131, 28–39. [Google Scholar] [CrossRef]
- Calderón, K.; Spor, A.; Breuil, M.C.; Bru, D.; Bizouard, F.; Violle, C.; Barnard, R.L.; Philippot, L. Effectiveness of ecological rescue for altered soil microbial communities and functions. ISME J. 2017, 11, 272–283. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.F.; Xia, Z.W.; Sang, C.P.; Wang, X.; Peng, B.; Wang, C.; Zhang, J.B.; Müller, C.; Bai, E. Soil resource status affects the responses of nitrogen processes to changes in temperature and moisture. Biol. Fert. Soils 2019, 55, 629–641. [Google Scholar] [CrossRef]
- Palmer, K.; Horn, M.A. Actinobacterial nitrate reducers and proteobacterial denitrifiers are abundant in N2O-metabolizing palsa peat. Appl. Environ. Microbiol. 2012, 78, 5584–5596. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Horn, M.A.; Behrendt, T.; Müller, S.; Li, J.; Cole, J.A.; Xie, B.; Ju, X.; Li, G.; Ermel, M.; et al. Soil HONO emissions at high moisture content are driven by microbial nitrate reduction to nitrite: Tackling the HONO puzzle. ISME J. 2019, 13, 1688–1699. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, B.; Zhang, W.; Zhang, Y.; Zhang, Y.; Zhang, T. Nitrous oxide emission from stormwater biofilters in alternating dry and wet weather. Environ. Res. 2020, 191, 110137. [Google Scholar] [CrossRef]
- Zhang, J.B.; Müller, C.; Cai, Z.C. Heterotrophic nitrification of organic N and its contribution to nitrous oxide emissions in soils. Soil Biol. Biochem. 2015, 84, 199–209. [Google Scholar] [CrossRef]
- Shang, Q.Y.; Yang, X.X.; Gao, C.M.; Wu, P.P.; Liu, J.J.; Xu, Y.C.; Shen, Q.R.; Zou, J.W.; Guo, S.W. Net annual global warming potential and greenhouse gas intensity in Chinese double rice-cropping systems: A 3-year field measurement in long-term fertilizer experiments. Glob. Change Biol. 2011, 17, 2196–2210. [Google Scholar] [CrossRef]
- Yue, J.; Shi, Y.; Liang, W.; Wu, J.; Wang, C.R.; Huang, G.H. Methane and nitrous oxide emissions from rice field and related microorganism in black soil, northeastern China. Nutr. Cycl. Agroecosys. 2005, 73, 293–301. [Google Scholar] [CrossRef]
- Senbayram, M.; Chen, R.; Mühling, K.H.; Dittert, K. Contribution of nitrification and denitrification to nitrous oxide emissions from soils after application of biogas waste and other fertilizers. Rapid Comm. Mass Spectro. 2009, 23, 2489–2498. [Google Scholar] [CrossRef] [PubMed]
- Bergstermann, A.; Cardenas, L.; Bol, R.; Gilljam, L.; Gounlding, K.; Meijide, A.; Scholefield, D.; Vallejo, A.; Well, R. Effect of antecedent soil moisture conditions on emissions and isotoplogue distribution of N2O during denitrification. Soil Biol. Biochem. 2011, 43, 240–250. [Google Scholar] [CrossRef]
- Cai, X.; Wen, P.; Yuan, Y.; Tang, J.; Yu, Z.; Zhou, S. Identification of nitrogen-incorporating bacteria in a sequencing batch reactor: A combining cultivation-dependent and cultivation-independent method. Bioreso. Technol. 2020, 316, 123964. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Wang, L.; Gao, Y.; Zhang, W.; Zhang, H.; Niu, L. Grain size tunes microbial community assembly and nitrogen transformation activity under frequent hyporheic exchange: A column experiment. Water Res. 2020, 182, 116040. [Google Scholar] [CrossRef] [PubMed]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. 2013. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Parinussa, R.M.; Dorigo, W.A.; De Jeu, R.A.M.; Wagner, W.; van Dijk, A.I.J.M.; McCabe, M.F.; Evans, J.P. Developing an improved soil moisture dataset by blending passive and active microwave satellite-based retrievals. Hydrol. Earth Syst. Sci. 2011, 15, 425–436. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Wen, J.; Dente, L.; van der Velde, R.; Wang, L.; Ma, Y.; Yang, K.; Hu, Z. The Tibetan Plateau observatory of plateau scale soil moisture and soil temperature (Tibet-Obs) for quantifying uncertainties in coarse resolution satellite and model products. Hydrol. Earth Syst. Sci. 2011, 15, 2303–2316. [Google Scholar] [CrossRef] [Green Version]
- Francis, C.A.; Roberts, K.J.; Beman, J.M.; Santoro, A.E.; Oakley, B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 2005, 102, 14683–14688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szukics, U.; Hackl, E.; Zechmeister-Boltenstern, S.; Sessitsch, A. Rapid and dissimilar response of ammonia oxidizing archaea and bacteria to nitrogen and water amendment in two temperate forest soils. Microbiol. Res. 2012, 167, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Throbäck, I.N.; Enwall, K.; Jarvis, A.; Hallin, S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 2004, 49, 401–417. [Google Scholar] [CrossRef] [PubMed]


| Water Regime | No. of Phyla with Statistically Significant (p < 0.05) Change in Relative Abundance | No. of Classes with Statistically Significant (p < 0.05) Change in Relative Abundance |
|---|---|---|
| Alternating | 3/30 | 7/30 |
| Wetting | 9/30 | 18/30 |
| Wetting02 | 8/30 | 16/30 |
| Static/constant | 8/30 | 19/30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Hao, D.-C.; Fan, S.; Xie, H.; Bao, X.; Jia, Z.; Wang, L. N2O Emission and Nitrification/Denitrification Bacterial Communities in Upland Black Soil under Combined Effects of Early and Immediate Moisture. Agriculture 2022, 12, 330. https://doi.org/10.3390/agriculture12030330
Wang L, Hao D-C, Fan S, Xie H, Bao X, Jia Z, Wang L. N2O Emission and Nitrification/Denitrification Bacterial Communities in Upland Black Soil under Combined Effects of Early and Immediate Moisture. Agriculture. 2022; 12(3):330. https://doi.org/10.3390/agriculture12030330
Chicago/Turabian StyleWang, Lei, Da-Cheng Hao, Sisi Fan, Hongtu Xie, Xuelian Bao, Zhongjun Jia, and Lianfeng Wang. 2022. "N2O Emission and Nitrification/Denitrification Bacterial Communities in Upland Black Soil under Combined Effects of Early and Immediate Moisture" Agriculture 12, no. 3: 330. https://doi.org/10.3390/agriculture12030330
APA StyleWang, L., Hao, D.-C., Fan, S., Xie, H., Bao, X., Jia, Z., & Wang, L. (2022). N2O Emission and Nitrification/Denitrification Bacterial Communities in Upland Black Soil under Combined Effects of Early and Immediate Moisture. Agriculture, 12(3), 330. https://doi.org/10.3390/agriculture12030330






