Overexpressing OsPYL/RCAR7 Improves Drought Tolerance of Maize Seedlings by Reducing Stomatal Conductance
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Materials and Genetic Transformation
2.2. Verification of Transgenic Maize
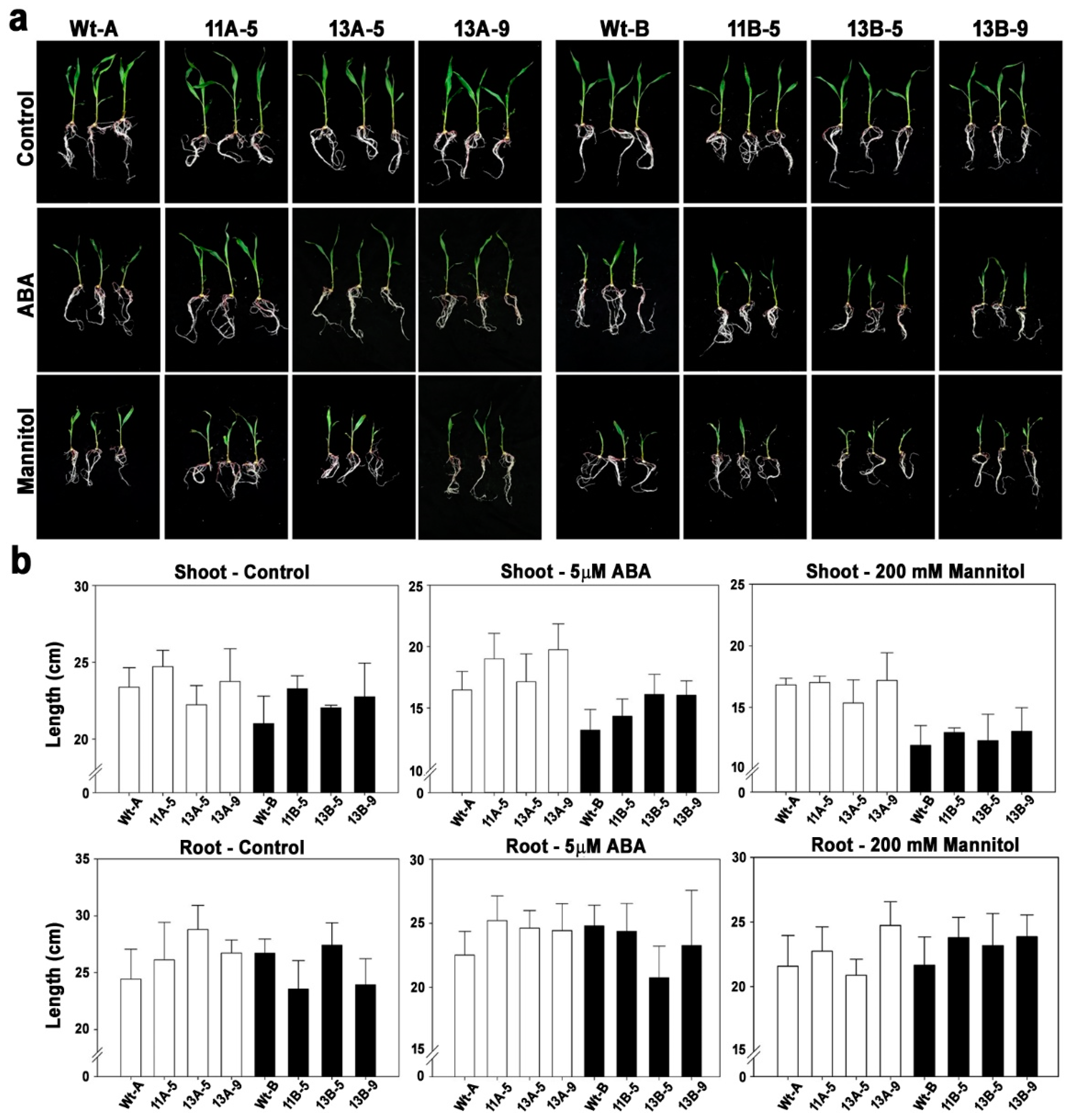
2.3. Postgermination Assay
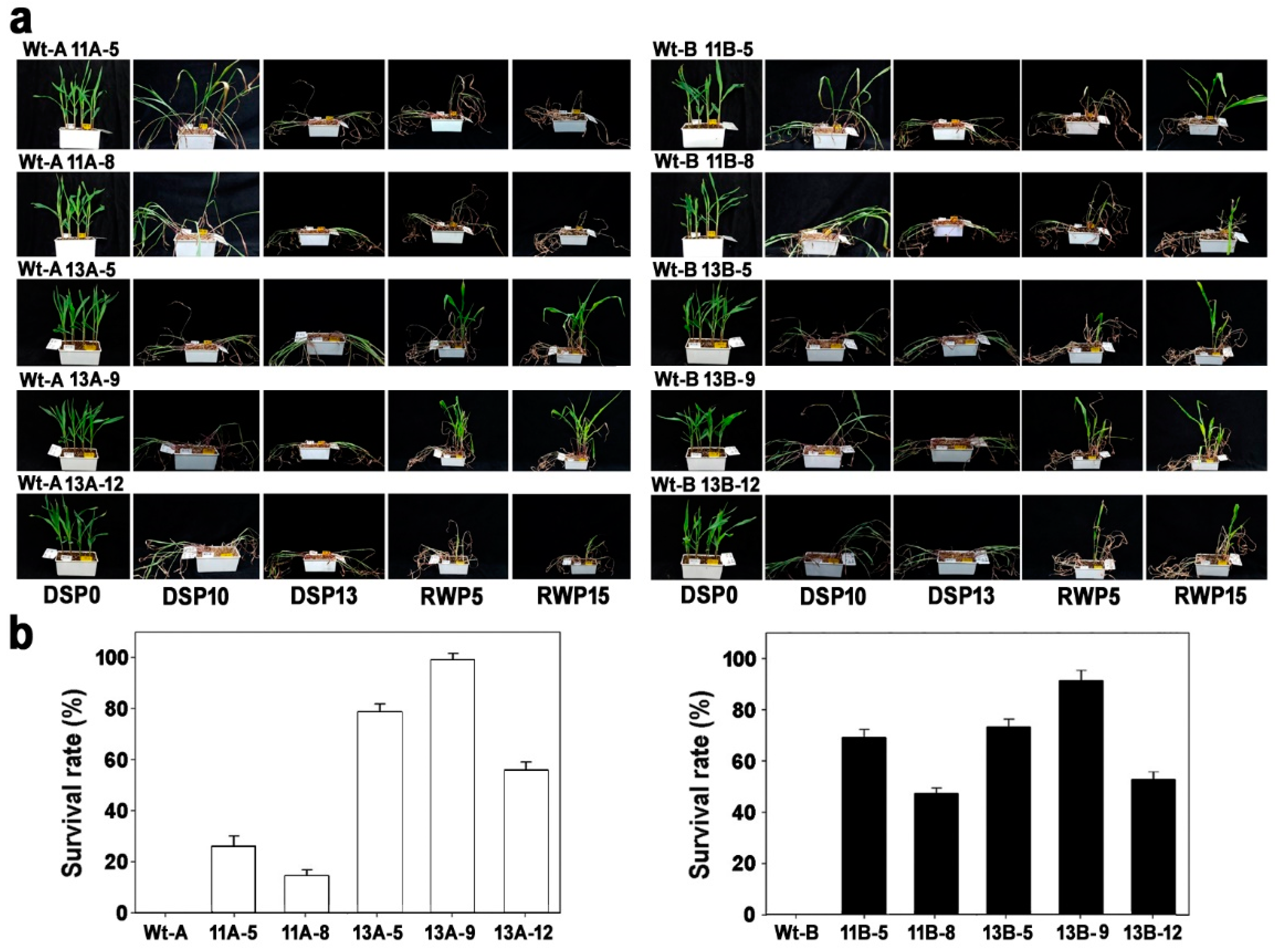
2.4. Phenotypic Analysis under Drought Stress
2.5. Analysis of Water Loss and Stomatal Conductance
2.6. Statistical Analysis
3. Results
3.1. Generation of Transgenic Maize Overexpressing OsPYL/RCAR7
3.2. Phenotypes of OsPYL/RCAR7-OX Transgenic Maize
3.3. Analysis of Water Loss and Stomatal Conductance
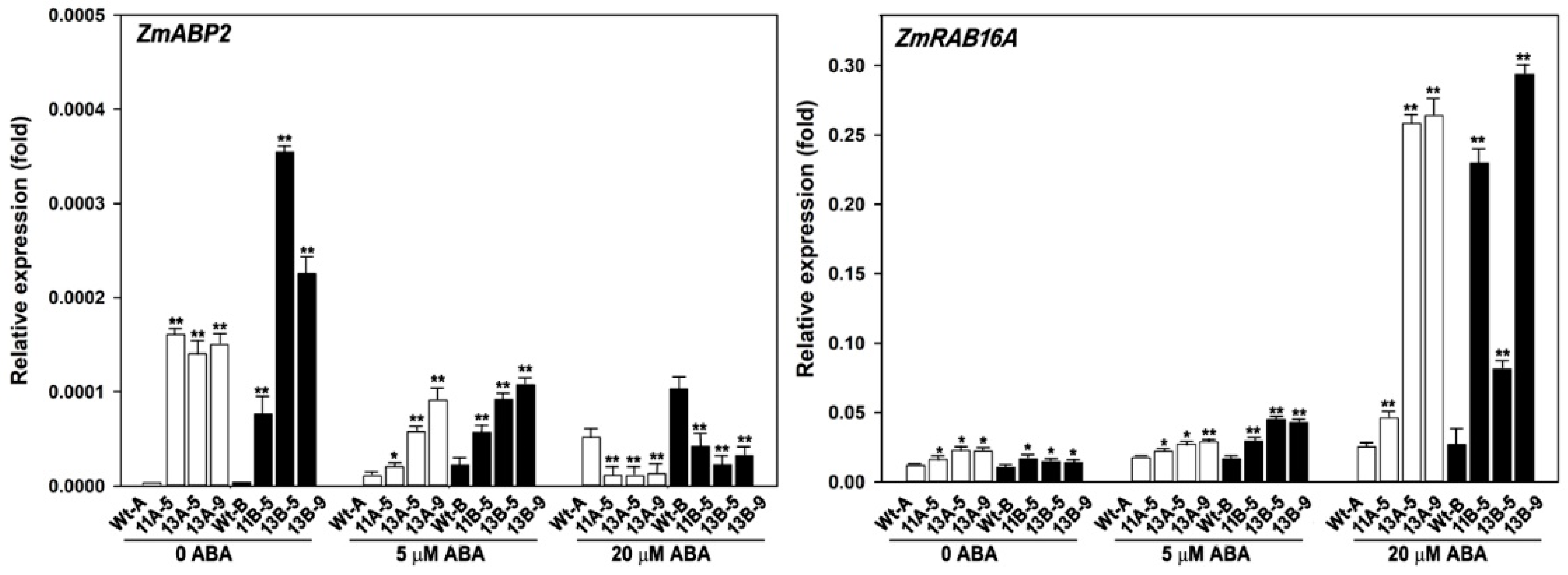
3.4. Expression Patterns of ABA-Related Genes in Transgenic Maize Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, C.L. Maize and grace: Africa’s encounter with a New World crop, 1500–2000—By James, C.; McCann. Dev. Econ. 2007, 45, 242–245. [Google Scholar]
- Mueller, N.D.; Gerber, J.S.; Johnston, M.; Ray, D.K.; Ramankutty, N.; Foley, J.A. Closing yield gaps through nutrient and water management. Nature 2012, 490, 254–257. [Google Scholar] [CrossRef]
- Edgerton, M.D. Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiol. 2009, 149, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Khatoon, T.; Hussain, K.; Majeed, A.; Nawaz, K.; Nisar, M.F. Morphological variations in maize (Zea mays L.) under different levels of NaCl at germination stage. World Appl. Sci. J. 2010, 8, 1294–1297. [Google Scholar]
- Nepolean, T.; Kaul, J.; Mukri, G.; Mittal, S. Genomics enabled next-generation breeding approaches for developing system-specific drought tolerant hybrids in maize. Front. Plant Sci. 2018, 9, 361. [Google Scholar] [CrossRef] [Green Version]
- Farooq, M.; Hussain, M.; Wakeel, A.; Siddique, K.H. Salt stress in maize: Effects, resistance mechanisms, and management. A review. Agron. Sustain. Dev. 2015, 35, 461–481. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Mu, C.; Zheng, H.; Lu, S.; Zhang, H.; Zhang, X.; Liu, X. Exogenous Pi supplementation improved the salt tolerance of maize (Zea mays L.) by promoting Na+ exclusion. Sci. Rep. 2018, 8, 16203. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, B.; Sa, K.J.; Lee, K. Drought tolerance screening of maize inbred lines at an early growth stage. Plant Breed. Biotechnol. 2019, 7, 326–339. [Google Scholar] [CrossRef]
- Li, Z.; Liu, C.; Zhang, Y.; Wang, B.; Ran, Q.; Zhang, J. The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J. Exp. Bot. 2019, 70, 5471–5486. [Google Scholar] [CrossRef]
- Mehta, R.H.; Ponnuchamy, M.; Kumar, J.; Reddy, N.R. Exploring drought stress-regulated genes in senna (Cassia angustifolia Vahl.): A transcriptomic approach. Funct. Integr. Genom. 2017, 17, 1–25. [Google Scholar] [CrossRef]
- Bray, E.A. Plant responses to water deficit. Trends Plant Sci. 1987, 2, 48–54. [Google Scholar] [CrossRef]
- Dai, A. Increasing drought under global warming in observations and models. Nat. Clim. Chang. 2013, 3, 52–58. [Google Scholar] [CrossRef]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought-from genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, H.; Song, X.; Jin, J.; Zhang, X. The responses of plant leaf CO2/H2O exchange and water use efficiency to drought: A meta-analysis. Sustainability 2018, 10, 551. [Google Scholar] [CrossRef] [Green Version]
- Zenda, T.; Liu, S.; Wang, X.; Liu, G.; Jin, H.; Dong, A.; Yang, Y.; Duan, H. Key maize drought-responsive genes and pathways revealed by comparative transcriptome and physiological analysis of contrasting inbred lines. Int. J. Mol. Sci. 2019, 20, 1268. [Google Scholar] [CrossRef] [Green Version]
- Jewell, M.C.; Campbell, B.C.; Godwin, I.D. Transgenic plants for abiotic stress resistance. In Transgenic Crop Plants; Kole, C., Michler, C.H., Abbott, A.G., Hall, T.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 67–132. [Google Scholar]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [Green Version]
- Jones, A.M. A new look at stress: Abscisic acid patterns and dynamics at high-resolution. New Phytol. 2015, 210, 38–44. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, H.; Hong, J.W.; Lee, Y.N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.D.; Lee, S.; Lee, S.C.; Kim, B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Lee, K.; Hwang, H.; Bhatnagar, N.; Kim, D.Y.; Yoon, I.S.; Byun, M.O.; Kim, S.T.; Jung, K.H.; Kim, B.G. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2014, 65, 453–456. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Hao, Q.; Li, W.; Yan, C.; Yan, N.; Yin, P. Identification and characterization of ABA receptors in Oryza sativa. PLoS ONE 2014, 9, e95246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Liu, J.; Tischer, S.V.; Christmann, A.; Windisch, W.; Schnyder, H.; Grill, E. Leveraging abscisic acid receptors for efficient water use in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6791–6796. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Yang, L.; Liu, X.; Tang, R.; Wang, Y.; Ge, H.; Wu, M.; Zhang, J.; Zhao, F.; Luan, S.; et al. Overexpression of poplar pyrabactin resistance-like abscisic acid receptors promotes abscisic acid sensitivity and drought resistance in transgenic Arabidopsis. PLoS ONE 2016, 11, e0168040. [Google Scholar] [CrossRef] [Green Version]
- Mega, R.; Tsujimoto, H.; Okamoto, M. Genetic manipulation of abscisic acid receptors enables modulation of water use efficiency. Plant Signal. Behav. 2019, 14, e1642039. [Google Scholar] [CrossRef]
- Mega, R.; Abe, F.; Kim, J.S.; Tsuboi, Y.; Tanaka, K.; Kobayashi, H.; Sakata, Y.; Hanada, K.; Tsujimoto, H.; Kikuchi, J.; et al. Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 2019, 5, 153–159. [Google Scholar] [CrossRef]
- Santiago, J.; Rodrigues, A.; Saez, A.; Rubio, S.; Antoni, R.; Dupeux, F.; Park, S.Y.; Márquez, J.A.; Cutler, S.R.; Rodriguez, P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009, 60, 575–588. [Google Scholar] [CrossRef]
- Park, S.Y.; Peterson, F.C.; Mosquna, A.; Yao, J.; Volkman, B.F.; Cutler, S.R. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature 2015, 520, 545–548. [Google Scholar] [CrossRef]
- Dorosh, L.; Kharenko, O.A.; Rajagopalan, N.; Loewen, M.C.; Stepanova, M. Molecular mechanisms in the activation of abscisic acid receptor PYR1. PLoS Comput. Biol. 2013, 9, e1003114. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, M.; Kagiyama, M.; Shibata, N.; Hirano, Y.; Hakoshima, T. Mechanism of high-affinity abscisic acid binding to PYL9/RCAR1. Genes Cells 2014, 19, 386–404. [Google Scholar] [CrossRef]
- Bhatnagar, V.; Kim, R.; Han, S.; Song, J.; Lee, G.S.; Lee, S.; Min, M.K.; Kim, B.-G. Ectopic Expression of OsPYL/RCAR7, an ABA receptor having low signaling activity, improves drought tolerance without growth defects in rice. Int. J. Mol. Sci. 2020, 21, 4163. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.K.; Lee, G.-S.; Park, K.J.; Kim, J.-K.; Jang, H.J.; Suh, E.J.; Kim, K.-H.; Lee, Y.H. Production of transgenic maize plants with herbicide resistance through Agrobacterium-mediated transformation. Korean J. Breed. Sci. 2019, 51, 290–297. [Google Scholar] [CrossRef]
- Lee, H.; Zhang, Z.J. Maize (Zea mays) Hi-II transformation via Agrobacterium-mediated T-DNA transfer. Curr. Protoc. Plant Biol. 2016, 1, 121–137. [Google Scholar] [CrossRef]
- Cho, M.J.; Banh, J.; Yu, M.; Kwan, J.; Jones, T.J. Improvement of Agrobacterium-mediated transformation frequency in multiple modern elite commercial maize (Zea mays L.) inbreds by media modifications. Plant Cell Tissue Organ Cult. 2015, 121, 519–529. [Google Scholar] [CrossRef]
- Hong, J.K.; Park, S.-R.; Suh, E.J.; Park, J.; Lee, H.-H. Effects of overexpression of Brassica rapa SHORT VEGETATIVE PHASE gene on flowering time. Korean J. Breed. Sci. 2020, 52, 244–251. [Google Scholar] [CrossRef]
- Zong, N.; Li, X.-J.; Wang, L.; Wang, Y.; Wen, H.-T.; Li, L.; Zhang, X.; Fan, Y.-L.; Zhao, J. Maize ABP2 enhances tolerance to drought and salt stress in transgenic Arabidopsis. J. Integr. Agric. 2018, 17, 2379–2393. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wu, M.; Zhang, M.; Jiang, W.; Ren, X.; Liang, E.; Zhang, D.; Zhang, C.; Xiao, N.; Li, Y.; et al. A maize phytochrome-interacting factors protein ZmPIF1 enhances drought tolerance by inducing stomatal closure and improves grain yield in Oryza sativa. Plant Biotechnol. J. 2018, 16, 1375–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.K.; Suh, E.J.; Park, S.R.; Park, J.; Lee, Y.-H. Multiplex CRISPR/Cas9 mutagenesis of BrVRN1 delays flowering time in Chinese cabbage (Brassica rapa L. ssp. Pekinensis). Agriculture 2021, 11, 1286. [Google Scholar] [CrossRef]
- Kim, S.L.; Kim, N.; Lee, H.; Lee, E.; Cheon, K.-S.; Kim, M.; Baek, J.; Choi, I.; Ji, H.; Yoon, I.S.; et al. High-throughput phenotyping platform for analyzing drought tolerance in rice. Planta 2020, 252, 38. [Google Scholar] [CrossRef]
- Ravia, S.; Young, T.; Macinnis-Ngc, C.; Nyugena, T.V.; Duxburya, M.; Alfaroa, A.C.; Leuzingera, S. Untargeted metabolomics in halophytes: The role of different metabolites in New Zealand mangroves under multi-factorial abiotic stress conditions. Environ. Exp. Bot. 2020, 173, 103993. [Google Scholar] [CrossRef]
- Carvalhoa, H.D.R.; Heilmana, J.L.; McInnesa, K.J.; Rooneya, W.L.; Lewis, K.L. Epicuticular wax and its effect on canopy temperature and water use of Sorghum. Agric. For. Meteorol. 2020, 284, 107893. [Google Scholar] [CrossRef]
- Spalinskas, R.; Bulcke, M.; Eede, G.; Milcamps, A. LT-RADE: An efficient user-friendly genome walking method applied to the molecular characterization of the insertion site of genetically modified maize MON810 and rice LLRICE62. Food Anal. Methods 2013, 6, 705–713. [Google Scholar] [CrossRef]
- Park, S.-I.; Kwon, H.J.; Cho, M.H.; Song, J.S.; Kim, B.-G.; Baek, J.H.; Kim, S.L.; Ji, H.S.; Kwon, T.-R.; Kim, K.-H.; et al. The OsERF115/AP2EREBP110 transcription factor is involved in the multiple stress tolerance to heat and drought in rice plants. Int. J. Mol. Sci. 2021, 22, 7181. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhong, J.; Sun, X.; Wang, B.; Terzaghi, W.; Dai, M. The maize ABA receptors ZmPYl8, 9, and 12 facilitate plant drought resistance. Front. Plant Sci. 2018, 9, 422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tischer, S.V.; Wunschel, C.; Papacek, M.; Kleigrewe, K.; Hofmsnn, T.; Christmann, A.; Grill, E. Combinatorial interaction network of abscisic acid receptors and coreceptors from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 10280–10285. [Google Scholar] [CrossRef] [Green Version]
- Hong, C.Y.; Chao, Y.Y.; Yang, M.Y.; Cheng, S.Y.; Cho, S.C.; Kao, C.H. NaCl-induced expression of glutathione reductase in roots of rice (Oryza sativa L.) seedlings is mediated through hydrogen peroxide but not abscisic acid. Plant Soil 2009, 320, 103–115. [Google Scholar] [CrossRef]
- Wang, X.S.; Zhu, H.B.; Jin, G.L.; Liu, H.L.; Wu, W.R.; Zhu, J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci. 2007, 172, 414–420. [Google Scholar] [CrossRef]
- Jin, X.; Shi, C.; Yu, C.Y.; Yamada, T.; Sacks, E.J. Determination of leaf water content by visible and near-infrared spectrometry and multivariate calibration in Miscanthus. Front. Plant Sci. 2017, 8, 721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef] [Green Version]
- Murata, Y.; Mori, I.C.; Munemasa, S. Diverse stomatal signaling and the signal integration mechanism. Annu. Rev. Plant Biol. 2015, 66, 369–392. [Google Scholar] [CrossRef]
- Singh, R.; Parihar, P.; Singh, S.; Mishra, R.K.; Singh, V.P.; Prasad, S.M. Reactive oxygen species signaling and stomatal movement: Current updates and future perspectives. Redox Biol. 2017, 11, 213–218. [Google Scholar] [CrossRef] [PubMed]


| F1—Type-A, B73 (♀) × T1 (♂) | F1—Type-B, T1 (♀) × B73 (♂) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Line (F1) | Transgene Ratio (%) | Genotype | No. of Seeds | Line (F1) | Transgene Ratio (%) | Genotype | No. of Seeds | ||
| #11–2- (named 11A-) | 1 | 60 | Ab, ab | 191 | #11–2- (named 11B-) | 1 | 40 | aB, ab | 240 |
| 2 | 0 | ab | 142 | 2 | 0 | ab | 237 | ||
| 3 | 60 | Ab, ab | 187 | 3 | 50 | aB, ab | 262 | ||
| 4 | 40 | Ab, ab | 158 | 4 | 40 | aB, ab | 267 | ||
| 5 | 100 | Ab | 159 | 5 | 100 | aB | 239 | ||
| 6 | 70 | Ab, ab | 177 | 6 | 40 | aB, ab | 365 | ||
| 7 | 60 | Ab, ab | 142 | 7 | 50 | aB, ab | 270 | ||
| 8 | 100 | Ab | 160 | 8 | 100 | aB | 238 | ||
| #13–2- (named 13A-) | 1 | 60 | Ab, ab | 202 | #13–2- (named 13B-) | 1 | 50 | aB, ab | 330 |
| 2 | 0 | ab | 196 | 2 | 0 | ab | 289 | ||
| 3 | 0 | ab | 191 | 3 | 0 | ab | 274 | ||
| 4 | 40 | Ab, ab | 192 | 4 | 40 | aB, ab | 287 | ||
| 5 | 100 | Ab | 199 | 5 | 100 | aB | 304 | ||
| 6 | 50 | Ab, ab | 268 | 6 | 50 | aB, ab | 309 | ||
| 7 | 40 | Ab, ab | 191 | 7 | 40 | aB, ab | 283 | ||
| 8 | 70 | Ab, ab | 195 | 8 | 40 | aB, ab | 352 | ||
| 9 | 100 | Ab | 197 | 9 | 100 | aB | 318 | ||
| 10 | 60 | Ab, ab | 272 | 10 | 40 | aB, ab | 305 | ||
| 11 | 50 | Ab, ab | 442 | 11 | 40 | aB, ab | 354 | ||
| 12 | 100 | Ab | 239 | 12 | 100 | aB | 345 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, J.K.; Lee, Y.-H.; Kim, B.-G.; Lee, G.S.; Jang, H.J.; Song, G.; Suh, E.J.; Park, S.R. Overexpressing OsPYL/RCAR7 Improves Drought Tolerance of Maize Seedlings by Reducing Stomatal Conductance. Agriculture 2022, 12, 2140. https://doi.org/10.3390/agriculture12122140
Hong JK, Lee Y-H, Kim B-G, Lee GS, Jang HJ, Song G, Suh EJ, Park SR. Overexpressing OsPYL/RCAR7 Improves Drought Tolerance of Maize Seedlings by Reducing Stomatal Conductance. Agriculture. 2022; 12(12):2140. https://doi.org/10.3390/agriculture12122140
Chicago/Turabian StyleHong, Joon Ki, Yeon-Hee Lee, Beom-Gi Kim, Gang Seob Lee, Hee Jeung Jang, Giha Song, Eun Jung Suh, and Sang Ryeol Park. 2022. "Overexpressing OsPYL/RCAR7 Improves Drought Tolerance of Maize Seedlings by Reducing Stomatal Conductance" Agriculture 12, no. 12: 2140. https://doi.org/10.3390/agriculture12122140
APA StyleHong, J. K., Lee, Y.-H., Kim, B.-G., Lee, G. S., Jang, H. J., Song, G., Suh, E. J., & Park, S. R. (2022). Overexpressing OsPYL/RCAR7 Improves Drought Tolerance of Maize Seedlings by Reducing Stomatal Conductance. Agriculture, 12(12), 2140. https://doi.org/10.3390/agriculture12122140








