Abstract
Rice salt tolerance at the germination stage directly affects the germination rate and seedling establishment of rice directly seeded in saline soils, which in turn affects yield. In this study, we determined the relative germination potential (RGP) and relative germination index (RGI) under 200 mM salt stress and control conditions using 295 japonica rice accessions. Statistical analysis showed extensive phenotypic variations under salt stress conditions. Twenty-one varieties with an RGP ≥ 80% and an RGI ≥ 80% were screened. Based on genotypic data including, 788,396 single-nucleotide polymorphisms (SNPs), 40 quantitative trait loci (QTL) were localized on rice chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, and 12, which were shown to be significantly associated with rice salt tolerance at the germination stage, including 20 for RGP and 20 for RGI, using genome-wide association analysis. Six QTL with ≥ 3 consecutive significant SNP loci and localized in the same LD interval were selected for further analysis. Four rice genes (LOC_Os01g04920, LOC_Os10g38350, LOC_Os10g38470, and LOC_Os10g38489) were selected as important candidates for salt tolerance based on haplotype analysis and functional annotation. The findings could facilitate the development of valuable rice varieties for direct seeding in salinized soil and improve japonica rice salt tolerance at the germination stage through molecular breeding.
1. Introduction
Rice is one of the most important staple crops, providing food for more than half of the world’s population [1]. The demand for high quality rice and higher yield has increased in recent years owing to the rapidly improving socioeconomic status and the growing population. However, salt stress is a serious constraint to rice production, which has become a global ecological problem [2]. Direct seeding of rice is a low-cost, highly efficient, and less labor-intensive cultivation method for simplified, large-scale rice production; this method alleviates the water crisis, positively improves soil salinization, and increases rice yield on saline land [3]. With the widespread implementation of direct seeding of rice in saline lands, the selection and breeding of rice varieties suitable for this practice has become more and more in demand. The chosen variety must be suitable for the natural conditions of the production area, and the emergence of its seedlings must also be considered. Therefore, identifying and screening salt tolerant sprouting varieties suitable for direct seeding, studying their genetic mechanisms, and mining salt tolerance candidate genes have become popular research topics.
The genetic basis of salt tolerance in rice is complex; salt tolerance is a quantitative trait controlled by multiple genes and strongly influenced by the environment [4]. Over 900 quantitative trait loci (QTL) for salt tolerance have been identified in rice [5,6,7,8,9,10,11]; key salt tolerance genes include SKC1 [12], HST [13], DST [14], and qSE3 [15]. These genes have been used in rice breeding, and the salt tolerance mechanisms have been clarified. The identification of new QTL for salinity tolerance will help us to further understand the mechanism of salinity tolerance in rice and promote breeding of salt tolerant rice. There is no significant correlation between salinity tolerance during the rice germination period and other plant stages [16]. Breeding salt tolerant varieties of rice at the germination stage can help protect rice from irreversible damage in the early stages of growth and thus, enhance the rice yield. Most studies on salt tolerance in rice have focused on the seedling stage [17], while only few have addressed the germination stage [16,18,19].
With the rapid development of biotechnology approaches, such as high-throughput sequencing and gene chip, in recent years, genome-wide association analysis (GWAS) has been widely used to study complex plant traits [20,21,22,23]. For example, An et al. [24] located 17 QTL significantly associated with dry weight ratio for salt tolerance in rice, using GWAS and a population consisting of 181 varieties. Yuan et al. [25] characterized 664 rice varieties at the seedling stage grown under a 0.9% salt solution and localized 21 salt tolerance-related QTL by GWAS. Rohila et al. [26] identified nine unknown salt tolerance-associated QTL by combining approximately three million SNP loci and eight salt tolerance phenotypic traits. More salt tolerance genes and QTL must be discovered to further understand the mechanism of salt tolerance in rice and breed salt tolerant rice varieties.
This study aimed to understand the genetic basis of salt tolerance at the germination stage and provide a molecular basis for the selection and breeding of rice varieties to improve salt tolerance in japonica rice. We identified 295 japonica rice accessions for salt tolerance and analyzed them by GWAS. Then, haplotype analysis was performed and combined with gene function annotation to mine salt tolerance candidate genes.
2. Materials and Methods
2.1. Plant Materials and Salt Tolerance Identification
The natural population consisted of 295 temperate japonica germplasm resources (Table S1), mainly from the Asian countries China, Japan, Korea, and North Korea. Twenty-two representative rice accessions were selected for a preliminary experiment to determine the appropriate salt stress concentration. To disrupt dormancy, rice seeds were dried for 48 h at 55 °C in an incubator (RXZ-500C, Ningbo Jiangnan Instrument Factory, Ningbo China) and then sterilized with 1% NaClO for 20 min, followed by washing with sterile water. One hundred plump seeds of each variety were placed in a 9-cm Petri dish containing 25 mL of 100-, 200-, 300-, 400-, 500-, and 600-mM sodium chloride solution or distilled water (control group) and placed at 28 °C, 14 h/10 h day/night cycle, and 70% relative humidity. Every day the number of germinated seeds was tallied. Seeds were considered germinated when the radicle broke through the seed coat and reached a length ≥ 1 mm (Figure 1) [27].

Figure 1.
Criteria of seed germination. (a,b) Photographs of rice seeds showing the definition of germination (a) and no germination (b).
Two hundred and ninety-five japonica rice accessions were tested in an incubator based on the results of the preliminary experiment. One hundred full seeds of each rice variety were tested under salt stress (200 mM) and control (distilled water) conditions, with three replicates. The solution was changed daily throughout the test. Until germination halted, the number of seeds that germinated was counted every day for statistical analysis. Salt stress tolerance was assessed using two parameters: relative germination potential (RGP), defined as RGP = N1/N2 × 100%, where N1 is the number of treated seeds that germinated on day 9 and N2 is the number of control seeds that germinated on day 5—the days differed because the seeds in the control group ended up germinating on day 5; and relative germination index (RGI) was defined as ∑(Gt/Dt) where Gt is the germination percentage on each day, and Dt is the number of days it took the seeds to germinate [28]. All rice accessions were classified according to the magnitudes of the RGP and RGI: 80–100%, seeds were considered highly salt tolerant; 60–80%, salt tolerant; 40–60%, moderately salt tolerant; 20–40%, salt sensitive; and 0–20%, highly salt sensitive.
2.2. GWAS for Salt Tolerance
Two hundred and ninety-five japonica rice accessions were genotyped using a total of 788,369 SNPs with a minor allele frequency of ≥ 5% and a missing rate of ≤ 20% for GWAS [29]. For that, we used a mixed linear model (MLM) in TASSEL 5.0 [30], considering kinship (K) and population structure (Q). p < 5.46 × 10−6 was chosen as the cutoff for identifying SNPs strongly linked with traits. The effective number of independent markers was calculated using the genetic type 1 error calculator (GEC, http://statgenpro.psychiatry.hku.hk/gec/ (accessed on 7 September 2020)) [29]. Using the R package “qqman”, Manhattan and Q–Q plots were produced from the GWAS results.
To obtain independent association signals, we performed LD calculations for SNPs in a 5-Mb region above the threshold, and if r2 was ≥0.25, the SNP with the smallest p value in this SNP cluster was taken as the lead SNP [31]. The LD decay distance upstream and downstream of the lead SNP was taken as the candidate interval of QTL, and the calculation was as follows: r2 values of all SNPs in the 2-Mb region upstream and downstream of the lead SNP were calculated; r2 values of the top 10% of the region from 1.5 to 2 Mb from the lead SNP were taken as the background r2 values, and background r2 values plus 0.2 were used as the chaining region. LDBlockshow [32] was used to estimate the local LD block region: white, r2 = 0; yellow, 0 < r2 <1; red, r2 = 1.
2.3. Identification of Important QTL and Haplotype Analysis of Candidate Genes
QTL regions meeting the following criteria were considered important: (1) consistently identified in RGP and RGI; and (2) QTL with ≥3 consecutive above-threshold SNPs.
Haplotype analysis was performed for all genes in the candidate region as follows: nonsynonymous mutant SNPs in the exonic region of each gene in the QTL interval were extracted from the Rice SNP-Seek Database (https://snp-seek.irri.org/ (accessed on 3 October 2021)). DnaSP software (www.ub.edu/dnasp/ (accessed on 12 October 2021)) was used for haplotype analysis and multiple comparisons of phenotypic values between different haplotypes (≥10 materials) to screen out genes with significant differences in phenotypic values between haplotypes. For genes with non-significant differences in phenotypic values between haplotypes, SNPs in the promoter region (first 2 kb of ATG) were extracted for haplotype analysis. The screened genes were combined with gene function annotations to predict the most likely candidate genes.
3. Results
3.1. Identification of Salt Tolerant Japonica Rice at the Germination Stage
The RGP and RGI of 22 representative rice accessions were measured under 6 salt treatments (100, 200, 300, 400, 500, and 600 mM NaCl) to obtain the appropriate salt concentration. The results of the preliminary experiments showed that the differences between varieties were best represented at 200 mM (Table 1). Therefore, 200 mM NaCl was used to determine the salt tolerance of all 295 japonica rice accessions at the germination stage. Rice seeds did not germinate under the 500 or 600 mM NaCl treatments.

Table 1.
Statistical analysis of RGP and RGI of 22 representative rice accessions.
The 295 japonica rice accessions showed a continuous distribution and an approximately symmetric distribution, indicating that these indices are quantitative traits under the joint control of multiple genes (Table 2, Figure 2). Finally, 21 highly salt tolerant varieties with both RGP ≥ 80% and RGI ≥ 80% were screened (Table 3).

Table 2.
Statistical analysis of RGP and RGI of 295 japonica rice accessions.
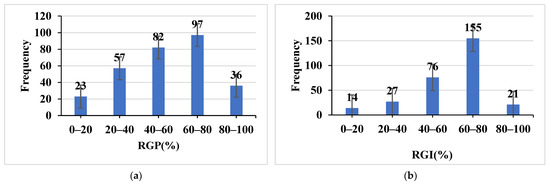
Figure 2.
Frequency distribution of RGP (a) and RGI (b) among the 295 japonica rice accessions.

Table 3.
Specific information of 21 high salt tolerance accessions.
3.2. GWAS of Salt Tolerance at the Germination Stage in Japonica Rice
Combining the 788,369 SNPs obtained from our previous study [29], we used the MLM of TASSEL 5.0, with a significance threshold of p ≤ 5.46 × 10−6, to perform GWAS on the RGP and RGI in the 295 japonica rice accessions. In total, 40 QTL were detected (Table 4), among which 20 QTL associated with RGP were located on chromosomes 1, 3, 5, 6, 7, 9, 10, 11, and 12, with r2 ranging from 10.02% to 14%. The 20 QTL associated with RGI were located on chromosomes 1, 2, 3, 5, 6, 7, 8, 9, 10, and 12, with r2 ranging from 10.10% to 12%. By comparison with the results of previous studies, 10 QTL in the present study were located at intervals similar to those of previously identified salt tolerance QTL, and 10 QTL existed for cloned salt tolerance-related genes.

Table 4.
Chromosomal sites with significant correlation of salt tolerance traits during germination.
Among these QTL, qRGP1.1 and qRGI1.2, qRGP1.3 and qRGI1.5, qRGP3.1 and qRGI3, qRGP6.1 and qRGI6, qRGP7.1 and qRGI7.2, and qRGP10 and qRGI10 were significantly associated with both RGP and RGI, with ≥3 consecutive significant SNP loci and localized in the same LD interval. QTL considered important for salt tolerance were renamed qRG1.1, qRG1.2, qRG3, qRG6, qRG7, and qRG10 (Figure 3).
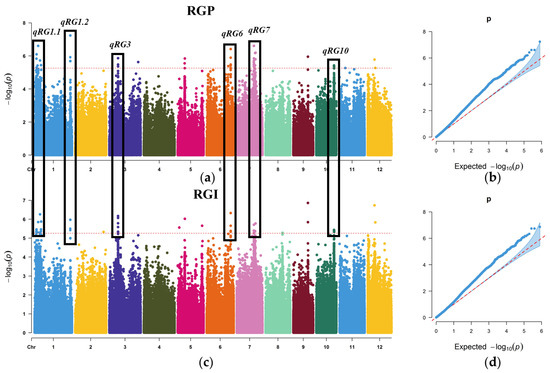
Figure 3.
Manhattan plots and quantile–quantile (Q–Q) plots of GWAS for the salt tolerance. Red horizontal dashed line indicates the genome-wide significance threshold. (a) Manhattan plot for the RGP. (b) Q–Q plot for the RGP. (c) Manhattan plot for the RGI. (d) Q–Q plot for the RGI.
3.3. Haplotype Analyses of the Candidate Genes
Haplotype analysis was conducted on nonsynonymous mutant SNPs of the exon region and SNPs of the promoter for all genes within the six QTL (qRG1.1, qRG1.2, qRG3, qRG6, qRG7, and qRG10) intervals to identify important candidate genes. There were no significant differences in phenotypic values among haplotypes for qRG1.1, qRG3, qRG6, or qRG7 intervals.
The LD block region for qRG1.2 on chromosome 1 was predicted from 2.06 to 2.37 Mb (310 kb) and contained 43 genes, according to the Rice Annotation Project (RAP) [40] (Figure 4 and Table S2). This candidate region includes two salt stress response genes, OsRAV2 [33] and OsPTR7 [34]; however, their phenotypic values did not significantly differ between haplotypes. Four genes (LOC_Os01g04650, LOC_Os01g04720, LOC_Os01g04870, and LOC_Os01g04920) were associated with significant differences in RGP and RGI among the different haplotypes. The nonsynonymous mutant SNPs of LOC_Os01g04650 were divided into two haplotypes, and Hap1 (A) had significantly higher RGP and RGI than did Hap2 (C) (Figure 5a,b). The nonsynonymous mutant SNPs of LOC_Os01g04720 were divided into two haplotypes, and Hap2 (G) had significantly higher RGP and RGI than did Hap1 (C) (Figure 5c,d). The nonsynonymous mutant SNPs of LOC_Os01g04870 were divided into two haplotypes, and Hap1 (A) had significantly higher RGP and RGI values than did Hap2 (G) (Figure 5e,f). The nonsynonymous mutant SNPs of LOC_Os01g04920 were divided into two haplotypes, and Hap2 (T) had significantly higher RGP and RGI than did Hap1 (C) (Figure 5g,h). Gene function annotations showed that LOC_Os01g04920 was the cloned gene OsSQD2.2 (Table S2), encoding a group I glycosyltransferase-containing structural domain. The overexpression of this gene increases flavonoid glycoside levels [41], and high levels of flavonoid glycosides improve stress resistance in rice [42]. This gene may affect salt tolerance in rice by regulating flavonoid glycoside metabolism; thus, LOC_Os01g04920 is hypothesized to be the most likely candidate gene for salt tolerance.
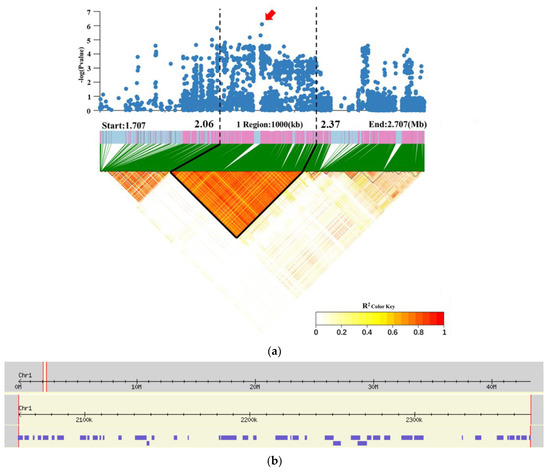
Figure 4.
Candidate gene of qRG1.2 on chromosome 1. (a) Local Manhattan plot (top) and LD heatmap (bottom) surrounding the lead SNP. (b) Based on the Rice Annotation Project (RAP) [40], the 310 kb region contained 43 genes.


Figure 5.
Gene structure and haplotype analysis. (a,b) represent the gene structure and haplotype analysis of LOC_Os01g04650. (c,d) represent the gene structure and haplotype analysis of LOC_Os01g04720. (e,f) represent the gene structure and haplotype analysis of LOC_Os01g04870. (g,h) represent the gene structure and haplotype analysis of LOC_Os01g04920. The * and ** suggest significance of ANOVA at p < 0.05 and p < 0.01, respectively.
The LD block region for qRG10 on chromosome 10 was predicted from 20.44 to 20.65 Mb (210 kb). According to the RAP [40], this interval contains 54 genes and does not include any known salt tolerance genes (Figure 6 and Table S3). Four genes (LOC_Os10g38350, LOC_Os10g38450, LOC_Os10g3847, and LOC_Os10g38489) were associated with significant differences in RGP and RGI among the different haplotypes. The nonsynonymous mutant SNPs of LOC_Os10g38350 were divided into two haplotypes, and Hap2 (CAC) had significantly higher RGP and RGI values than did Hap1 (GGT) (Figure 7a,b). The nonsynonymous mutant SNPs of LOC_Os10g38450 were divided into two haplotypes, and Hap2 (AG) had significantly higher RGP and RGI values than did Hap1 (GT) (Figure 7c,d). The nonsynonymous mutant SNPs of LOC_Os10g38470 were divided into two haplotypes, and Hap2 (T) had significantly higher RGP and RGI than did Hap1 (G) (Figure 7e,f). The nonsynonymous mutant SNPs of LOC_Os10g38489 were divided into two haplotypes, and Hap2 (C) had significantly higher RGP and RGI values than did Hap1 (G) (Figure 7g,h). Gene function annotation showed that LOC_Os10g38350, LOC_Os10g38470, and LOC_Os10g38489 encoded glutathione S-transferase (Table S3). This enzyme plays a role in primary metabolism, secondary metabolism, and plant protection from oxidative damage [43]. It is suggested that these three genes may further affect plant salt tolerance by influencing the accumulation of reactive oxygen species in plants [44]. Therefore, LOC_Os10g38350, LOC_Os10g38470, and LOC_Os10g38489 are hypothesized to be the most likely candidates for salt tolerance in rice.
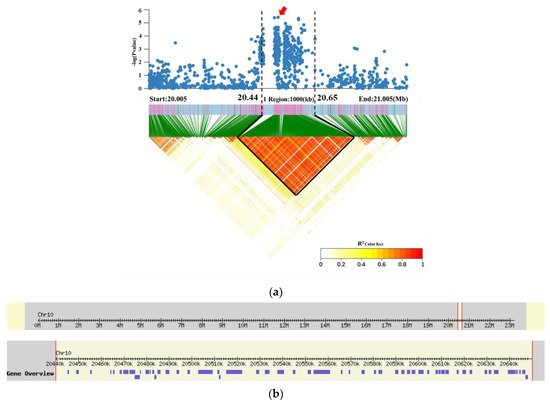
Figure 6.
Candidate gene of qRG10 on chromosome 10. (a) Local Manhattan plot (top) and LD heatmap (bottom) surrounding the lead SNP. (b) Based on the Rice Annotation Project (RAP) [40], the 210 kb region contained 54 genes.
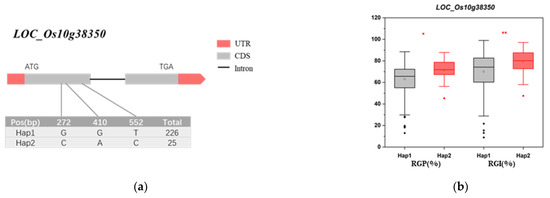
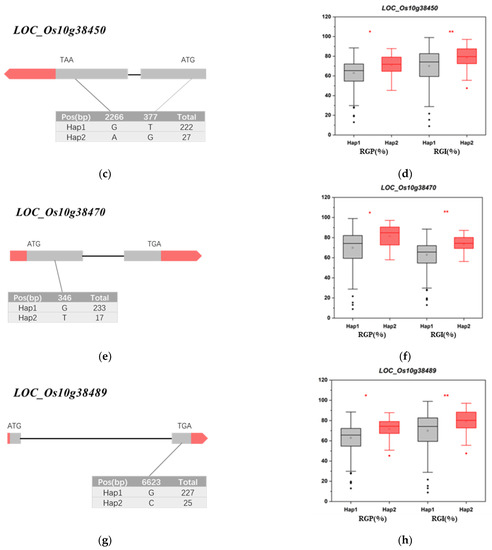
Figure 7.
Gene structure and haplotype analysis. (a,b) represent the gene structure and haplotype analysis of LOC_Os10g38350. (c,d) represent the gene structure and haplotype analysis of LOC_Os10g38450. (e,f) represent the gene structure and haplotype analysis of LOC_Os10g38470. (g,h) represent the gene structure and haplotype analysis of LOC_Os10g38489. The * and ** suggest significance of ANOVA at p < 0.05 and p < 0.01, respectively.
4. Discussion
Identifying salinity tolerance in rice is important, and different methods of identification may lead to different salinity tolerance quantification in rice [45]. In this study, RGP and RGI were selected as indicators for the identification of salt tolerance at the germination stage of rice, which have also been used in previous studies [18,19]. The concentration of salt in seed treatment is an important factor for the accurate determination of salt tolerance during rice germination. Shi et al. [46] measured SSI, VI, and MGT at three different concentrations of NaCl (60, 80, and 100 mM) in 35 randomly selected rice varieties (from 478 varieties) to determine the most appropriate salt concentration. The results showed the most genetic variation was observed in the 60 mM NaCl treatment among all treatments. Yu et al. [19] randomly selected 12 rice varieties for screening experiments using GP with 0, 200, and 300 mM NaCl. According to the screening results, 200 mM NaCl adequately exposed their phenotypic variation and facilitated the differentiation of rice varieties with different levels of salt tolerance. In this study, we analyzed and compared phenotypic values under six different concentrations of NaCl (100, 200, 300, 400, 500, and 600 mM NaCl) (Table 1) and found that 200 mM NaCl best reflected the differences among varieties. Combining RGP and RGI, we finally screened 21 highly salt tolerant varieties among 295 japonica rice accessions, which provided material for the direct seeding of rice. We found that salt stress did both: exerted a significant inhibitory effect and delay seed germination.
The identification of genes related to salinity tolerance in different rice varieties is important to improve rice productivity in saline soils. GWAS technology is based on millions of SNP loci in the genome. Phenotypic traits are directly associated with genotypes through mathematical models to identify loci or master effect genes associated with the target traits [47]. This study used 295 japonica rice accessions to investigate candidate genes and 40 QTL on chromosomes 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, and 12 that regulate important phenotypes under salt stress in rice at the germination stage. Some QTL detected in this study were located at the same or similar intervals as previously localized QTL or cloned genes, based on the information published in the National Rice Center gene database (http://www.ricedata.cn/gene/ (accessed on 14 October 2021)). For example, the salt tolerance-related QTL qSHL1.7b [35] was in the same interval as qRGP1.5 and qRGI1.3 in our study; the salt sensitive gene OsDREB2A [36] was located within the qRGP1.6 and qRGI1.6 intervals. Similarly, salt tolerance-related QTL, qSTL1-1 [25], qSTL8-1 [25], and qRNC-9 [12], were located near qRGP1.2, qRGI1.4, qRGP9, qRGI9, and qRGI8. Moreover, salt tolerance-related genes, OsPP2A-2 [37], OsHAK13 [38], and SRWD4 [39], were located within qRGP3.2, qRGP6.1, qRGI6, and qRGI8. The results indicates that GWAS accurately detected and localized QTL intervals.
Cloning QTL affecting complex traits is a major challenge for plant geneticists and molecular biologists because of the labor-intensive and time-consuming nature of the traditional gene map cloning. The GWAS method for QTL and haplotype analyses for all genes within the QTL interval targets candidate genes with significant differences between different haplotype phenotype values. It also combines gene annotation and previous results, which may improve the efficiency and accuracy of candidate gene screening [48]. This study performed a GWAS for salinity tolerance in rice using high-density SNP markers, and the upstream and downstream LD decay distances of lead SNPs were used as candidate intervals to locate QTL regions significantly associated with salinity tolerance. Haplotype analysis of candidate genes was conducted for QTL co-localized with ≥ 3 consecutive significant SNPs for RGP and RGI; four genes in the qRG1.2 interval had significant differences in RGP and RGI between haplotypes. This included LOC_Os01g04920 (OsSQD2.2), which encodes a group I glycosyltransferase-containing structural domain. The overexpression of the this gene increases flavonoid glycoside levels [41], whereas the overexpression of SQD2.1 (SQD2.2 homolog) enhances tolerance to salinity and drought [49]. The QTL GSA1 regulates grain size and stress tolerance in rice, and its overexpression positively regulates flavonoid glycoside levels, which increases salt stress tolerance in rice. In contrast, the knockdown of GSA1 flavonoid glycoside synthesis under salt stress reduces resistance to salt stress in rice [42]. Therefore, it is hypothesized that OsSQD2.2 is most likely a candidate gene for salt tolerance in qRG1.2 and that it may regulate flavonoid metabolism.
Similarly, four genes in the qRG10 interval showed significant differences among haplotypes, including LOC_Os10g38350, LOC_Os10g38470, and LOC_Os10g38489 encoding glutathione S-transferase, which play important roles in primary and secondary metabolism and protection against oxidative damage in plants. The three candidate genes are located on chromosome 10 in a gene cluster containing 28 Tau class GST genes that are present in tandem at a single locus [50]. Members of the Tau class appear to respond more frequently to various stimuli; the expression of two GST genes of the Tau class (OsGSTU3 and OsGSTU4) is induced by hypoxia and salt stress [51]. Transgenic tobacco seedlings overexpressing cDNA encoding glutathione S-transferase grow significantly faster than do control seedlings under salt stress [52]. Glutathione S-transferase catalyzes the scavenging of free radicals in plants, and the overexpression of glutathione S-transferase confers salt stress tolerance [43]. Therefore, we hypothesized that LOC_Os10g38350, LOC_Os10g38470, and LOC_Os10g38489 are the most likely candidate genes for salt tolerance in rice. The next steps of our research will focus on the functional validation of transgenes and utilization of LOC_Os01g04920, LOC_Os10g38350, LOC_Os10g38470, and LOC_Os10g38489 for molecular breeding to provide a theoretical basis for improving salt tolerance in japonica rice.
5. Conclusions
Overall, the present study identified 295 japonica rice accessions with salt tolerance at the germination stage, screened 21 highly salt tolerant rice varieties to provide material support for direct seeding of rice. Two-hundred and ninety five japonica rice varieties exhibited considerable differences in salt stress tolerance, strongly suggesting a wide range of genetic variation in the natural population that could be exploited. Therefore, we further performed GWAS. We localized 40 QTL for salt tolerance at the germination stage based on a high density of SNPs. The 23 QTL detected in the present study have not been reported in previous studies and may be new QTL affecting salt tolerance in japonica rice. In addition, based on haplotype analysis and functional annotation of genes, we screened four genes (LOC_Os01g04920, LOC_Os10g38350, LOC_Os10g38470, and LOC_ Os10g38489) that may contribute to salt tolerance at the germination stage. These new salt tolerance candidate genes may be useful for the molecular breeding of japonica rice during germination to increase salt tolerance and improve quality.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture12101588/s1, Table S1: Geographical distribution and phenotypic value of 295 japonica rice accessions; Table S2: Summary of functional annotation results for genes in the qRG1.2 on chromosome 1; Table S3: Summary of functional annotation results for genes in the qRG10 on chromosome 10.
Author Contributions
Conceptualization, Y.D. and H.Z.; methodology, Y.D., H.Z. and H.W.; software, Y.D. and H.W.; validation, Y.D., H.Z. and J.C.; formal analysis, Y.D. and H.W.; investigation, Y.D., H.Z., H.W., D.Q. and J.C.; resources, D.Z.; data curation, Y.D., H.Z., H.W., D.Q., J.C., J.W., H.L., L.Y., Y.J., W.X., S.L., D.Z. and C.L.; writing—original draft preparation, Y.D. and H.Z.; writing—review and editing, Y.D., H.Z. and H.W.; visualization, Y.D., H.Z. and H.W.; supervision, D.Z.; project administration, D.Z.; funding acquisition, D.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant No. U20A2025, 31872884), and the “Academic Backbone” Project of Northeast Agricultural University (grant No. 20XG24).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Deng, N.; Grassini, P.; Yang, H.; Huang, J.; Cassman, K.G.; Peng, S. Closing yield gaps for rice self-sufficiency in China. Nat. Commun. 2019, 10, 1725. [Google Scholar] [CrossRef] [PubMed]
- Kamran, M.; Parveen, A.; Ahmar, S.; Malik, Z.; Hussain, S.; Chattha, M.S.; Saleem, M.H.; Adil, M.; Heidari, P.; Chen, J.T. An overview of hazardous impacts of soil salinity in crops, tolerance mechanisms, and amelioration through selenium supplementation. Int. J. Mol. Sci. 2019, 21, 148. [Google Scholar] [CrossRef]
- Ishfaq, M.; Akbar, N.; Anjum, S.A.; Anwar-Ul-Haq, M. Growth, yield and water productivity of dry direct seeded rice and transplanted aromatic rice under different irrigation management regimes. J. Integr. Agric. 2020, 19, 2656–2673. [Google Scholar] [CrossRef]
- Liang, J.L.; Qu, Y.P.; Yang, C.G.; Ma, X.D.; Cao, G.L.; Zhao, Z.W.; Zhang, S.Y.; Zhang, T.; Han, L.Z. Identification of QTLs associated with salt or alkaline tolerance at the seedling stage in rice under salt or alkaline stress. Euphytica 2015, 201, 441–452. [Google Scholar] [CrossRef]
- Lei, L.; Han, Z.; Cui, B.; Yang, L.; Liu, H.; Wang, J.; Zhao, H.; Xin, W.; Li, X.; Li, J.; et al. Mapping of a major QTL for salinity tolerance at the bud burst stage in rice (Oryza sativa L.) using a high-density genetic map. Euphytica 2021, 217, 167. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, J.; Chen, Z.; Huang, J.; Bao, Y.; Wang, J.; Zhang, H. Identification of QTLs with main, epistatic and QTL × environment interaction effects for salt tolerance in rice seedlings under different salinity conditions. Theor. Appl. Genet. 2012, 125, 807–815. [Google Scholar] [CrossRef]
- Hossain, H.; Rahman, M.; Alam, M.; Singh, R. Mapping of quantitative trait loci associated with reproductive-stage salt tolerance in rice. J. Agron. Crop Sci. 2015, 201, 17–31. [Google Scholar] [CrossRef]
- Pang, Y.; Chen, K.; Wang, X.; Wang, W.; Xu, J.; Ali, J.; Li, Z. Simultaneous improvement and genetic dissection of salt tolerance of rice (Oryza sativa L.) by designed QTL pyramiding. Front. Plant Sci. 2017, 8, 1275. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, J.; Zhao, H.; Liu, H.; Sun, J.; Guo, L.; Zou, D. Genetic structure, linkage disequilibrium and association mapping of salt tolerance in japonica rice germplasm at the seedling stage. Mol. Breed. 2015, 35, 152. [Google Scholar] [CrossRef]
- Yu, J.; Zao, W.; He, Q.; Kim, T.S.; Park, Y.J. Genome-wide association study and gene set analysis for understanding candidate genes involved in salt tolerance at the rice seedling stage. Mol. Genet. Genom. 2017, 292, 1391–1403. [Google Scholar] [CrossRef]
- Puram, V.R.R.; Ontoy, J.; Linscombe, S.; Subudhi, P.K. Genetic dissection of seedling stage salinity tolerance in rice using introgression lines of a salt tolerant landrace nona bokra. J. Hered. 2017, 108, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.X.; Zhu, M.Z.; Yano, M.; Gao, J.P.; Liang, Z.W.; Su, W.A.; Hu, X.H.; Ren, Z.H.; Chao, D.Y. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 2004, 108, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef]
- Huang, X.Y.; Chao, D.Y.; Gao, J.P.; Zhu, M.Z.; Shi, M.; Lin, H.X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes. Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, Y.; Wu, W.; Zhan, X.; Anis, G.B.; Rahman, M.H.; Hong, Y.; Riaz, A.; Zhu, A.; Cao, Y.; et al. qSE7 is a major quantitative trait locus (QTL) influencing stigma exsertion rate in rice (Oryza sativa L.). Sci. Rep. 2018, 8, 14523. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, J.; Bao, Y.; Wu, Y.; Zhang, H. Quantitative trait loci controlling rice seed germination under salt stress. Euphytica 2011, 178, 297–307. [Google Scholar] [CrossRef]
- Hoang, T.M.L.; Tran, T.N.; Nguyen, T.K.T.; Williams, B.; Wurm, P.; Bellairs, S.; Mundree, S. Improvement of salinity stress tolerance in rice: Challenges and opportunities. Agronomy 2016, 6, 54. [Google Scholar] [CrossRef]
- Wang, Z.-F.; Wang, J.-F.; Bao, Y.-M.; Wu, Y.-Y.; Su, X.; Zhang, H.-S. Inheritance of rice seed germination ability under salt stress. Rice Sci. 2010, 17, 105–110. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, W.; Tong, W.; He, Q.; Yoon, M.Y.; Li, F.P.; Choi, B.; Heo, E.B.; Kim, K.W.; Park, Y.J. A genome-wide association study reveals candidate genes related to salt tolerance in rice (Oryza sativa) at the germination stage. Int. J. Mol. Sci. 2018, 19, 3145. [Google Scholar] [CrossRef]
- Atwell, S.; Huang, Y.S.; Vilhjálmsson, B.J.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.M.; Hu, T.T.; et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef]
- Sauvage, C.; Segura, V.; Bauchet, G.; Stevens, R.; Do, P.T.; Nikoloski, Z.; Fernie, A.R.; Causse, M. Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol. 2014, 165, 1120–1132. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Breseghello, F.; Sorrells, M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Liu, K.; Wang, B.; Tian, Y.; Ge, Y.; Zhang, Y.; Tang, W.; Chen, G.; Yu, J.; Wu, W.; et al. Genome-wide association study identifies QTLs conferring salt tolerance in rice. Plant Breed. 2020, 139, 73–82. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, X.; Zhao, Y.; Khan, N.U.; Zhao, Z.; Zhang, Y.; Wen, X.; Tang, F.; Wang, F.; Li, Z. Genetic basis and identification of candidate genes for salt tolerance in rice by GWAS. Sci. Rep. 2020, 10, 9958. [Google Scholar] [CrossRef]
- Rohila, J.S.; Edwards, J.D.; Tran, G.D.; Jackson, A.K.; McClung, A.M. Identification of Superior Alleles for Seedling Stage Salt Tolerance in the USDA Rice Mini-Core Collection. Plants 2019, 8, 472. [Google Scholar] [CrossRef]
- Peng, L.; Sun, S.; Yang, B.; Zhao, J.; Li, W.; Huang, Z.; Li, Z.; He, Y.; Wang, Z. Genome-wide association study reveals that the cupin domain protein OsCDP3.10 regulates seed vigour in rice. Plant Biotechnol. J. 2022, 20, 485–498. [Google Scholar] [CrossRef]
- Ji, S.L.; Jiang, L.; Wang, Y.H.; Zhang, W.W.; Liu, X.; Liu, S.J.; Chen, L.M.; Zhai, H.Q.; Wan, J.M. Quantitative trait loci mapping and stability for low temperature germination ability of rice. Plant Breed. 2009, 128, 387–392. [Google Scholar] [CrossRef]
- Li, N.; Zheng, H.; Cui, J.; Wang, J.; Liu, H.; Sun, J.; Liu, T.; Zhao, H.; Lai, Y.; Zou, D. Genome-wide association study and candidate gene analysis of alkalinity tolerance in japonica rice germplasm at the seedling stage. Rice 2019, 12, 24. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Wang, Q.; Xie, W.; Xing, H.; Yan, J.; Meng, X.; Li, X.; Fu, X.; Xu, J.; Lian, X.; Yu, S.; et al. Genetic architecture of natural variation in rice Chlorophyll content revealed by a genome-wide association study. Mol. Plant 2015, 8, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.S.; He, W.M.; Ji, J.J.; Zhang, C.; Guo, Y.; Yang, T.L. LDBlockShow: A fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 2021, 22, bbaa227. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Cai, Z.; Xia, K.; Wang, Y.; Duan, J.; Zhang, M. Identification and analysis of eight peptide transporter homologs in rice. Plant Sci. 2010, 179, 374–382. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Subudhi, P.K. Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice 2016, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. Cell Mol. Biol. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Yu, R.M.; Wong, M.M.; Jack, R.W.; Kong, R.Y. Structure, evolution and expression of a second subfamily of protein phosphatase 2A catalytic subunit genes in the rice plant (Oryza sativa L.). Planta 2005, 222, 757–768. [Google Scholar] [CrossRef]
- Bañuelos, M.A.; Garciadeblas, B.; Cubero, B.; Rodríguez-Navarro, A. Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 2002, 130, 784–795. [Google Scholar] [CrossRef]
- Huang, J.; Wang, M.M.; Bao, Y.M.; Sun, S.J.; Pan, L.J.; Zhang, H.S. SRWD: A novel WD40 protein subfamily regulated by salt stress in rice (Oryza sativa L.). Gene 2008, 424, 71–79. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Zhan, X.; Shen, Q.; Wang, X.; Hong, Y. The sulfoquinovosyltransferase-like enzyme SQD2.2 is Involved in flavonoid glycosylation, regulating sugar metabolism and seed setting in rice. Sci. Rep. 2017, 7, 4685. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.Q.; Sun, Y.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C.; et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.C.; Zhang, S.M.; Wang, L.P.; Wang, M.D.; Zhang, H. Overexpression of GST gene accelerates the growth of transgenic Arabidopsis under salt stress. J. Plant Physiol. Mol. Biol. 2004, 30, 517–522. [Google Scholar] [CrossRef]
- Lima-Melo, Y.; Carvalho, F.E.; Martins, M.O.; Passaia, G.; Sousa, R.H.; Neto, M.C.; Margis-Pinheiro, M.; Silveira, J.A. Mitochondrial GPX1 silencing triggers differential photosynthesis impairment in response to salinity in rice plants. J. Integr. Plant Biol. 2016, 58, 737–748. [Google Scholar] [CrossRef]
- Zhang, R.; Hussain, S.; Wang, Y.; Liu, Y.; Li, Q.; Chen, Y.; Wei, H.; Gao, P.; Dai, Q. Comprehensive Evaluation of Salt Tolerance in Rice (Oryza sativa L.) Germplasm at the Germination Stage. Agronomy 2021, 11, 1569. [Google Scholar] [CrossRef]
- Shi, Y.; Gao, L.; Wu, Z.; Zhang, X.; Wang, M.; Zhang, C.; Zhang, F.; Zhou, Y.; Li, Z. Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol. 2017, 17, 92. [Google Scholar] [CrossRef]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Zhai, L.; Zheng, T.; Wang, X.; Wang, Y.; Chen, K.; Wang, S.; Wang, Y.; Xu, J.; Li, Z. QTL mapping and candidate gene analysis of peduncle vascular bundle related traits in rice by genome-wide association study. Rice 2018, 11, 13. [Google Scholar] [CrossRef]
- Zhan, X.; Shen, Q.; Chen, J.; Yang, P.; Wang, X.; Hong, Y. Rice sulfoquinovosyltransferase SQD2.1 mediates flavonoid glycosylation and enhances tolerance to osmotic stress. Plant Cell Environ. 2019, 42, 2215–2230. [Google Scholar] [CrossRef]
- Jain, M.; Ghanashyam, C.; Bhattacharjee, A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genom. 2010, 11, 73. [Google Scholar] [CrossRef]
- Moons, A. Osgstu3 and osgtu4, encoding tau class glutathione S-transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots. FEBS Lett. 2003, 553, 427–432. [Google Scholar] [CrossRef]
- Roxas, V.P.; Smith, R.K., Jr.; Allen, E.R.; Allen, R.D. Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 1997, 15, 988–991. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).