Identification of Grain Size-Related QTLs in Korean japonica Rice Using Genome Resequencing and High-Throughput Image Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Experiments
2.2. Phenotypic Evaluation of Grain Size-Related Traits
2.3. Genetic Map Construction and QTL Mapping
2.4. Analysis of Whole-Genome Resequencing Data of Odae and Joun
2.5. Long-Read Sequencing of Odae and Joun
2.6. Selection of Candidate Genes Underlying Major QTLs
3. Results
3.1. Phenotypic Variation and Correlation Analysis
3.2. Genetic Map Construction
3.3. Identification of Grain Size-Related QTLs
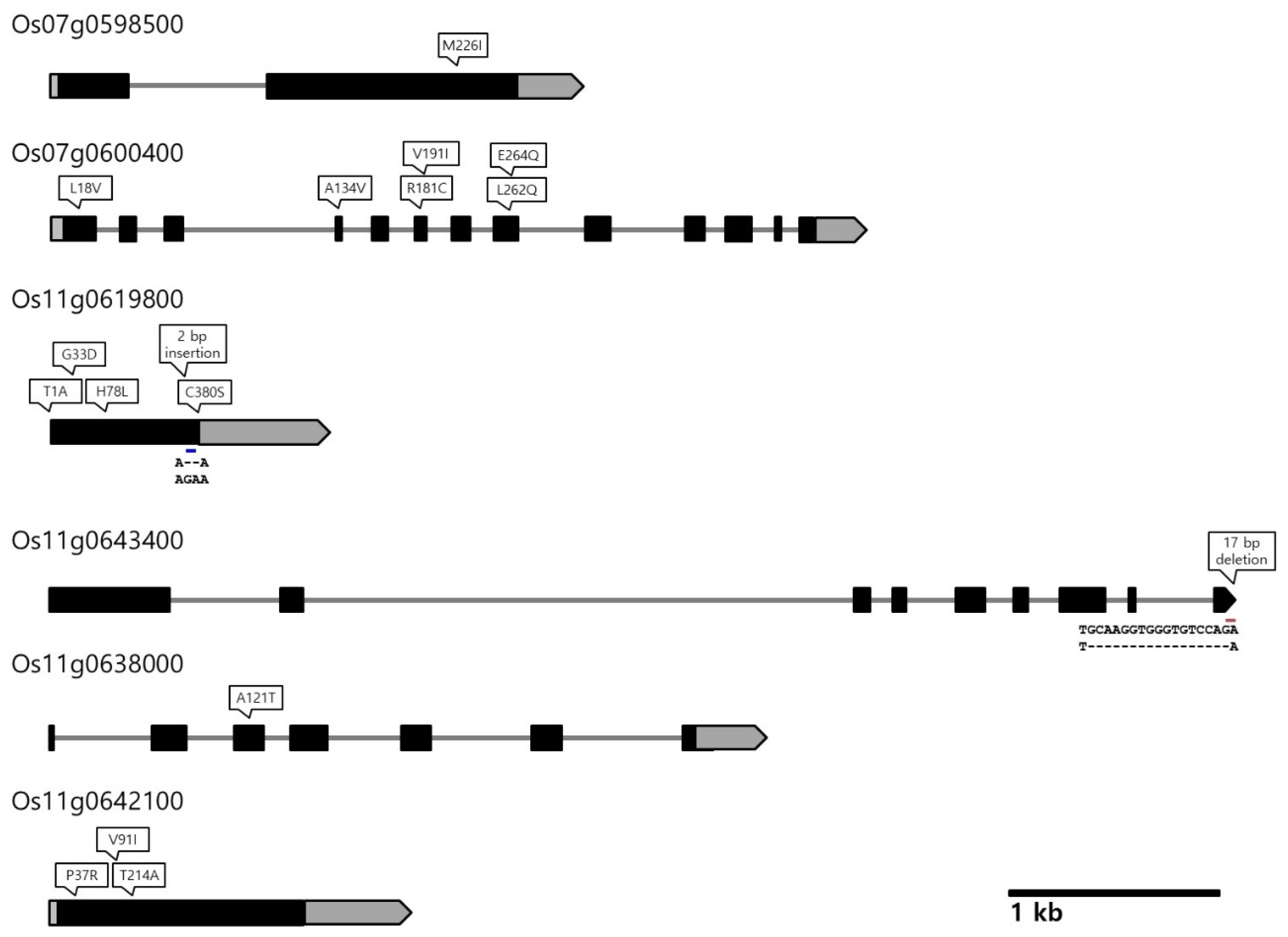
3.4. Analysis of Genome Sequencing Data of Odae and Joun and Selection of the Putative Candidate Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K. Impact of climate change on agriculture production and its sustainable solutions. Environ. Sustain. 2019, 2, 95–96. [Google Scholar] [CrossRef]
- Sakamoto, T.; Matsuoka, M. Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 2008, 11, 209–214. [Google Scholar] [CrossRef]
- Lee, C.M.; Park, J.; Kim, B.; Seo, J.; Lee, G.; Jang, S.; Koh, H.J. Influence of Multi-Gene Allele Combinations on Grain Size of Rice and Development of a Regression Equation Model to Predict Grain Parameters. Rice 2015, 8, 33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, R.; Jiang, L.; Zheng, J.; Wang, T.; Wang, H.; Huang, Y.; Hong, Z. Genetic bases of rice grain shape: So many genes, so little known. Trends Plant Sci. 2013, 18, 218–226. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Duan, P.; Li, Y. Control of grain size in rice. Plant Reprod. 2018, 31, 237–251. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Huang, K.; Wang, D.; Duan, P.; Zhang, B.; Xu, R.; Li, N.; Li, Y. WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 2017, 91, 849–860. [Google Scholar] [CrossRef]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2015, 2, 15203. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Lin, Y.S.; Song, X.J.; Shen, J.B.; Huang, W.; Shan, J.X.; Zhu, M.Z.; Jiang, L.; Gao, J.P.; Lin, H.X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012, 22, 1666–1680. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef]
- Hu, Z.; Lu, S.J.; Wang, M.J.; He, H.; Sun, L.; Wang, H.; Liu, X.H.; Jiang, L.; Sun, J.L.; Xin, X.; et al. A Novel QTL qTGW3 Encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that Interacts with OsARF4 to Negatively Regulate Grain Size and Weight in Rice. Mol. Plant 2018, 11, 736–749. [Google Scholar] [CrossRef]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef]
- Shi, C.L.; Dong, N.Q.; Guo, T.; Ye, W.W.; Shan, J.X.; Lin, H.X. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 2020, 103, 1174–1188. [Google Scholar] [CrossRef]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Qian, Q. Gene Network of Grain Size and Number in Rice. In Rice Genomics, Genetics and Breeding; Springer: Berlin/Heidelberg, Germany, 2018; pp. 191–206. [Google Scholar]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J.; et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, R.; Li, Y. Molecular Networks of Seed Size Control in Plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Li, J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 2014, 48, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Garris, A.J.; Tai, T.H.; Coburn, J.; Kresovich, S.; McCouch, S. Genetic structure and diversity in Oryza sativa L. Genetics 2005, 169, 1631–1638. [Google Scholar] [CrossRef]
- Hori, K.; Yamamoto, T.; Yano, M. Genetic dissection of agronomically important traits in closely related temperate japonica rice cultivars. Breed. Sci. 2017, 67, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, D.; Bohra, A.; Thudi, M.; Varshney, R.K. Fine mapping and gene cloning in the post-NGS era: Advances and prospects. Theor. Appl. Genet. 2020, 133, 1791–1810. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.L.; Grondin, A.; Courtois, B.; Gantet, P. Next-Generation Sequencing Accelerates Crop Gene Discovery. Trends Plant Sci. 2019, 24, 263–274. [Google Scholar] [CrossRef]
- Cheon, K.-S.; Baek, J.; Cho, Y.-I.; Jeong, Y.-M.; Lee, Y.-Y.; Oh, J.; Won, Y.J.; Kang, D.-Y.; Oh, H.; Kim, S.L.; et al. Single Nucleotide Polymorphism (SNP) Discovery and Kompetitive Allele-Specific PCR (KASP) Marker Development with Korean japonica Rice Varieties. Plant Breed. Biotechnol. 2018, 6, 391–403. [Google Scholar] [CrossRef]
- Cheon, K.S.; Jeong, Y.M.; Oh, H.; Oh, J.; Kang, D.Y.; Kim, N.; Lee, E.; Baek, J.; Kim, S.L.; Choi, I.; et al. Development of 454 New Kompetitive Allele-Specific PCR (KASP) Markers for Temperate japonica Rice Varieties. Plants 2020, 9, 1531. [Google Scholar] [CrossRef]
- Cheon, K.-S.; Jeong, Y.-M.; Lee, Y.-Y.; Oh, J.; Kang, D.-Y.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; Baek, J.; et al. Kompetitive Allele-Specific PCR Marker Development and Quantitative Trait Locus Mapping for Bakanae Disease Resistance in Korean japonica Rice Varieties. Plant Breed. Biotechnol. 2019, 7, 208–219. [Google Scholar] [CrossRef]
- Kang, D.Y.; Cheon, K.S.; Oh, J.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; Choi, I.; Baek, J.; Kim, K.H.; et al. Rice Genome Resequencing Reveals a Major Quantitative Trait Locus for Resistance to Bakanae Disease Caused by Fusarium fujikuroi. Int. J. Mol. Sci. 2019, 20, 2598. [Google Scholar] [CrossRef]
- Cheon, K.S.; Won, Y.J.; Jeong, Y.M.; Lee, Y.Y.; Kang, D.Y.; Oh, J.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; et al. QTL mapping for pre-harvest sprouting resistance in japonica rice varieties utilizing genome re-sequencing. Mol. Genet. Genom. 2020, 295, 1129–1140. [Google Scholar] [CrossRef]
- Baek, J.; Lee, E.; Kim, N.; Kim, S.L.; Choi, I.; Ji, H.; Chung, Y.S.; Choi, M.S.; Moon, J.K.; Kim, K.H. High Throughput Phenotyping for Various Traits on Soybean Seeds Using Image Analysis. Sensors 2020, 20, 248. [Google Scholar] [CrossRef] [PubMed]
- Lorieux, M. MapDisto: Fast and efficient computation of genetic linkage maps. Mol. Breed. 2012, 30, 1231–1235. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Basten, C.J.; Weir, B.S.; Zeng, Z.-B. QTL Cartographer: A Reference Manual and Tutorial for QTL Mapping; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 1996. [Google Scholar]
- Kumagai, M.; Nishikawa, D.; Kawahara, Y.; Wakimoto, H.; Itoh, R.; Tabei, N.; Tanaka, T.; Itoh, T. TASUKE+: A web-based platform for exploring GWAS results and large-scale resequencing data. DNA Res. 2020, 27, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.C.; Iwamoto, M.; Abe, T.; et al. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Cingolani, P.; Patel, V.M.; Coon, M.; Nguyen, T.; Land, S.J.; Ruden, D.M.; Lu, X. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front. Genet. 2012, 3, 35. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Marcos, J.F.; Dal Pra, M.; Giulini, A.; Costa, L.M.; Gavazzi, G.; Cordelier, S.; Sellam, O.; Tatout, C.; Paul, W.; Perez, P.; et al. empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 2007, 19, 196–210. [Google Scholar] [CrossRef]
- Huang, J.; Lu, G.; Liu, L.; Raihan, M.S.; Xu, J.; Jian, L.; Zhao, L.; Tran, T.M.; Zhang, Q.; Liu, J.; et al. The Kernel Size-Related Quantitative Trait Locus qKW9 Encodes a Pentatricopeptide Repeat Protein That Aaffects Photosynthesis and Grain Filling. Plant Physiol. 2020, 183, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- He, S.-L. Functional Characterization of a WD-Repeat Protein Gene (OsWD1) in Rice. Am. J. Agric. For. 2018, 6, 18–27. [Google Scholar] [CrossRef]
- Yang, L.; Liu, H.; Zhao, J.; Pan, Y.; Cheng, S.; Lietzow, C.D.; Wen, C.; Zhang, X.; Weng, Y. LITTLELEAF (LL) encodes a WD40 repeat domain-containing protein associated with organ size variation in cucumber. Plant J. 2018, 95, 834–847. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Li, J.; Walker, J.C. Overexpression of a serine carboxypeptidase increases carpel number and seed production in Arabidopsis thaliana. Food Energy Secur. 2012, 1, 61–69. [Google Scholar] [CrossRef]
- Li, S.; Liu, W.; Zhang, X.; Liu, Y.; Li, N.; Li, Y. Roles of the Arabidopsis G protein gamma subunit AGG3 and its rice homologs GS3 and DEP1 in seed and organ size control. Plant Signal. Behav. 2012, 7, 1357–1359. [Google Scholar] [CrossRef] [PubMed][Green Version]
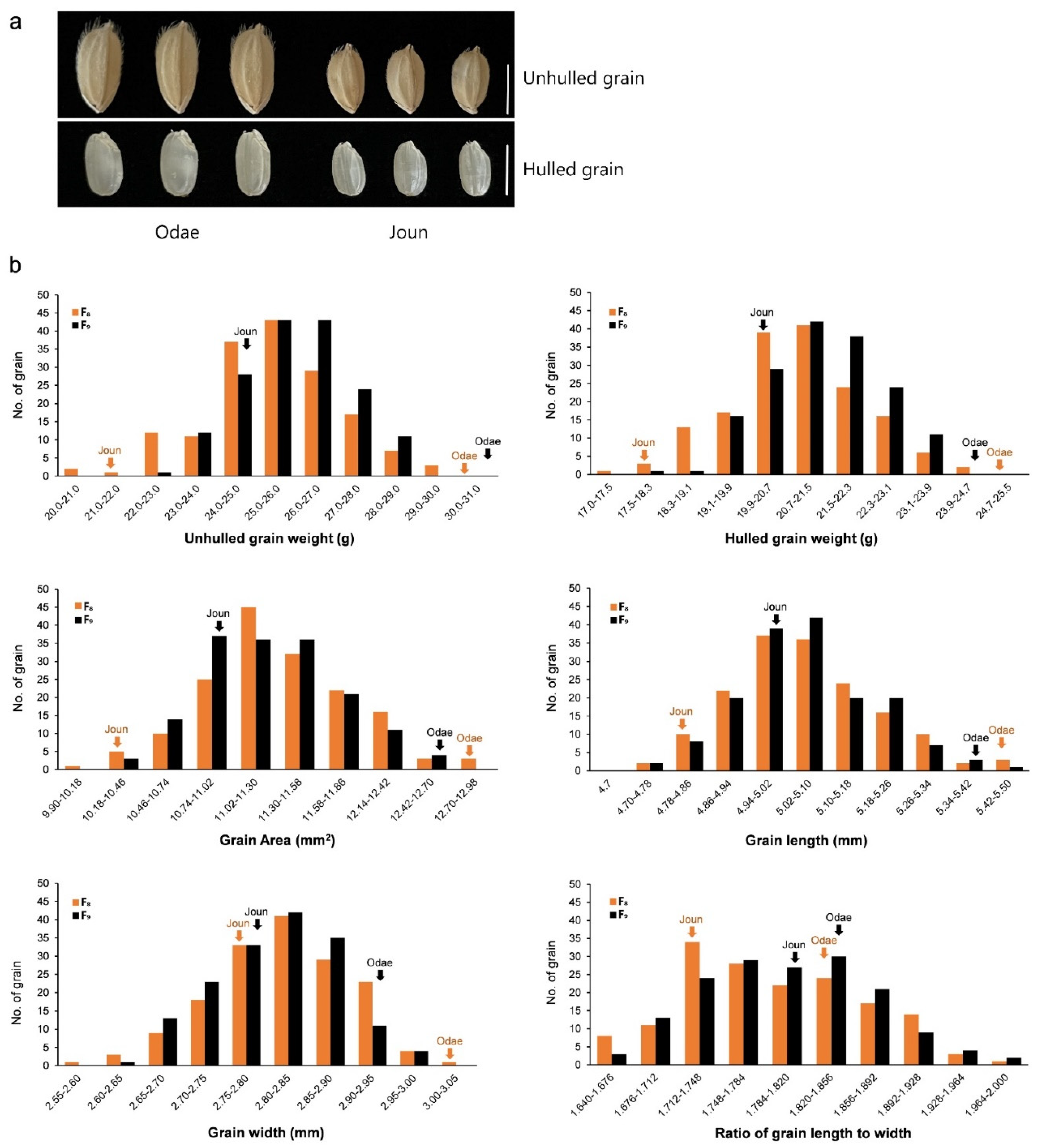

| UGW F8 | HGW F8 | GA F8 | GL F8 | GW F8 | RLW F8 | UGW F9 | HGW F9 | GA F9 | GL F9 | GW F9 | RLW F9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UGW F8 | ||||||||||||
| HGW F8 | 0.968 *** | |||||||||||
| GA F8 | 0.853 *** | 0.875 *** | ||||||||||
| GL F8 | 0.437 *** | 0.454 *** | 0.696 *** | |||||||||
| GW F8 | 0.755 *** | 0.767 *** | 0.697 *** | −0.030 | ||||||||
| RLW F8 | −0.223 ** | −0.220 ** | −0.006 | 0.714 *** | −0.721 *** | |||||||
| UGW F9 | 0.705 *** | 0.697 *** | 0.666 *** | 0.356 *** | 0.573 *** | −0.151 | ||||||
| HGW F9 | 0.699 *** | 0.703 *** | 0.652 *** | 0.341 *** | 0.569 *** | −0.159 * | 0.988 *** | |||||
| GA F9 | 0.658 *** | 0.676 *** | 0.756 *** | 0.524 *** | 0.526 *** | −0.007 | 0.834 *** | 0.806 *** | ||||
| GL F9 | 0.328 *** | 0.340 *** | 0.553 *** | 0.895 *** | −0.125 | 0.707 *** | 0.441 *** | 0.421 *** | 0.654 *** | |||
| GW F9 | 0.548 *** | 0.560 *** | 0.458 *** | −0.179 * | 0.812 *** | −0.693 *** | 0.673 *** | 0.655 *** | 0.681 *** | −0.108 | ||
| RLW F9 | −0.158 * | −0.158 * | 0.049 | 0.711 *** | −0.640 *** | 0.941 *** | −0.168 * | −0.169 * | −0.037 | 0.732 *** | −0.756 *** |
| Trait 1 | QTL | Chr. 2 | QTL Interval (cM) 3 | Interval-Flanking Markers | F8 (2019) 4 | F9 (2020) 4 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Left | Right | LOD | Add. | PVE 5 | LOD | Add. | PVE 5 | ||||
| UGW | qUGW2.1 | 2 | 80.1–92.3 | KJ02_036 | KJ02_042 | 6.5 | −0.61 | 14 | 3.6 | −0.33 | 6 |
| qUGW2.2 | 2 | 98.6–104.8 | KJ02_050 | KJ02_052 | 7.6 | −0.63 | 14 | ||||
| qUGW3 | 3 | 0–2.4 | KJ03_015 | KJ03_016 | 6.5 | 0.55 | 11 | ||||
| qUGW7 | 7 | 124.3–130.7 | KJ07_071 | KJ07_074 | 9.0 | 0.65 | 16 | 14.4 | 0.65 | 24 | |
| qUGW11 | 11 | 84.7–91.9 | KJ11_092 | KJ11_095 | 3.3 | 0.42 | 6 | 9.6 | 0.52 | 15 | |
| HGW | qHGW2.1 | 2 | 81.2–93.3 | KJ02_036 | KJ02_042 | 7.3 | −0.52 | 15 | 3.8 | −0.29 | 6 |
| qHGW2.2 | 2 | 95.7–98.6 | KJ02_045 | KJ02_050 | 7.8 | −0.50 | 13 | ||||
| qHGW3 | 3 | 0–2.1 | KJ03_015 | KJ03_016 | 7.9 | 0.50 | 13 | ||||
| qHGW7.1 | 7 | 62.2–72.6 | KJ07_026 | KJ07_029 | 3.1 | 0.30 | 5 | ||||
| qHGW7.2 | 7 | 123.1–131.4 | KJ07_071 | KJ07_074 | 6.7 | 0.45 | 11 | 12.3 | 0.52 | 21 | |
| qHGW11 | 11 | 84.6–92.3 | KJ11_092 | KJ11_095 | 3.3 | 0.33 | 6 | 8.3 | 0.42 | 13 | |
| GA | qGA3 | 3 | 0–4.6 | KJ03_015 | KJ03_016 | 3.1 | 0.09 | 5 | |||
| qGA7 | 7 | 123.5–135.2 | KJ07_071 | KJ07_074 | 6.3 | 0.18 | 16 | 8.7 | 0.17 | 18 | |
| qGA11 | 11 | 86.6–94.4 | KJ11_092 | KJ11_095 | 3.7 | 0.12 | 7 | 8.7 | 0.16 | 16 | |
| GL | qGL1.1 | 1 | 5.5–13.3 | KJ01_007 | KJ01_009 | 4.0 | 0.04 | 7 | |||
| qGL1.2 | 1 | 190.9–197.6 | KJ01_112 | KJ01_116 | 2.8 | −0.03 | 4 | ||||
| qGL6.1 | 6 | 44.3–58.4 | KJ06_034 | KJ06_068 | 4.3 | 0.05 | 10 | 10.0 | 0.07 | 23 | |
| qGL6.2 | 6 | 87–110.3 | KJ06_078 | KJ06_084 | 5.5 | 0.07 | 22 | ||||
| qGL7 | 7 | 123–135.5 | KJ07_071 | KJ07_074 | 3.4 | 0.03 | 6 | ||||
| qGL10.1 | 10 | 53.3–62.9 | KJ10_030 | KJ10_039 | 3.59 | 0.04 | 7 | ||||
| qGL10.2 | 10 | 63.9–71.9 | KJ10_039 | KJ10_047 | 7.5 | 0.05 | 15 | ||||
| GW | qGW6 | 6 | 44.6–60.2 | KJ06_034 | KJ06_068 | 3.6 | −0.03 | 9 | 4.9 | 0.03 | 12 |
| qGW8 | 8 | 58.8–68.3 | KJ08_060 | KJ08_064 | 3.4 | −0.02 | 7 | 5.4 | −0.02 | 10 | |
| qGW9 | 9 | 4.7–12.7 | KJ09_002 | KJ09_024 | 4.3 | −0.03 | 10 | ||||
| qGW11 | 11 | 80.6–88.8 | KJ11_092 | KJ11_095 | 4.6 | 0.02 | 8 | ||||
| RLW | qRLW1 | 1 | 0.4–9.7 | KJ01_001 | KJ01_007 | 4.6 | 0.02 | 8 | 4.6 | 0.02 | 7 |
| qRLW6 | 6 | 47.6–57.6 | KJ06_034 | KJ06_068 | 9.8 | 0.03 | 20 | 14.1 | 0.04 | 26 | |
| qRLW10.1 | 10 | 53.8–57 | KJ10_030 | KJ10_034 | 7.3 | 0.03 | 13 | 9.0 | 0.03 | 15 | |
| qRLW10.2 | 10 | 58.3–66.7 | KJ10_034 | KJ10_039 | 7.6 | 0.03 | 13 | 11.8 | 0.03 | 19 | |
| Cluster | Chr. 1 | QTLs | Associated Traits | Physical Position (Mbp) | Gene ID | Putative Candidate Genes with Functional Annotations from RAP-DB 2 | Ref 3 |
|---|---|---|---|---|---|---|---|
| 2 | 7 | qUGW7, qHGW7.2, qGA7, qGL7 | UGW, HGW, GA, GL | 24.1–24.8 | Os07g0598500 Os07g0600400 | Pentatricopeptide repeat (PPR) domain- containing protein WD40/YVTN repeat-like domain-containing protein | [51,52] [53,54] |
| 3 | 11 | qUGW11, qHGW11, qGA11, qGW11 | UGW, HGW, GA, GW | 23.7–25.5 | Os11g0619800 Os11g0643400 Os11g0638000 Os11g0642100 | Kelch-related domain-containing protein Serine carboxypeptidase (SCP) family protein GTP-binding protein engA (G protein) Cyclin-like F-box domain-containing protein | [14,15] [12,55] [56] [10] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, Y.; Won, Y.J.; Lee, C.; Cheon, K.-S.; Oh, H.; Lee, G.-S.; Baek, J.; Yoon, I.S.; Kim, S.L.; Cha, Y.-S.; et al. Identification of Grain Size-Related QTLs in Korean japonica Rice Using Genome Resequencing and High-Throughput Image Analysis. Agriculture 2022, 12, 51. https://doi.org/10.3390/agriculture12010051
Shin Y, Won YJ, Lee C, Cheon K-S, Oh H, Lee G-S, Baek J, Yoon IS, Kim SL, Cha Y-S, et al. Identification of Grain Size-Related QTLs in Korean japonica Rice Using Genome Resequencing and High-Throughput Image Analysis. Agriculture. 2022; 12(1):51. https://doi.org/10.3390/agriculture12010051
Chicago/Turabian StyleShin, Yunji, Yong Jae Won, Chaewon Lee, Kyeong-Seong Cheon, Hyoja Oh, Gang-Seob Lee, Jeongho Baek, In Sun Yoon, Song Lim Kim, Young-Soon Cha, and et al. 2022. "Identification of Grain Size-Related QTLs in Korean japonica Rice Using Genome Resequencing and High-Throughput Image Analysis" Agriculture 12, no. 1: 51. https://doi.org/10.3390/agriculture12010051
APA StyleShin, Y., Won, Y. J., Lee, C., Cheon, K.-S., Oh, H., Lee, G.-S., Baek, J., Yoon, I. S., Kim, S. L., Cha, Y.-S., Kim, K.-H., & Ji, H. (2022). Identification of Grain Size-Related QTLs in Korean japonica Rice Using Genome Resequencing and High-Throughput Image Analysis. Agriculture, 12(1), 51. https://doi.org/10.3390/agriculture12010051






