Genetic Variability, Correlation among Agronomic Traits, and Genetic Progress in a Sugarcane Diversity Panel
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Experimental Design and Phenotypic Data
2.3. Statistical Analysis of Phenotypic Data
3. Results
3.1. VCOV Models, Variance Components, and Heritabilities
3.2. Genotypic Correlations
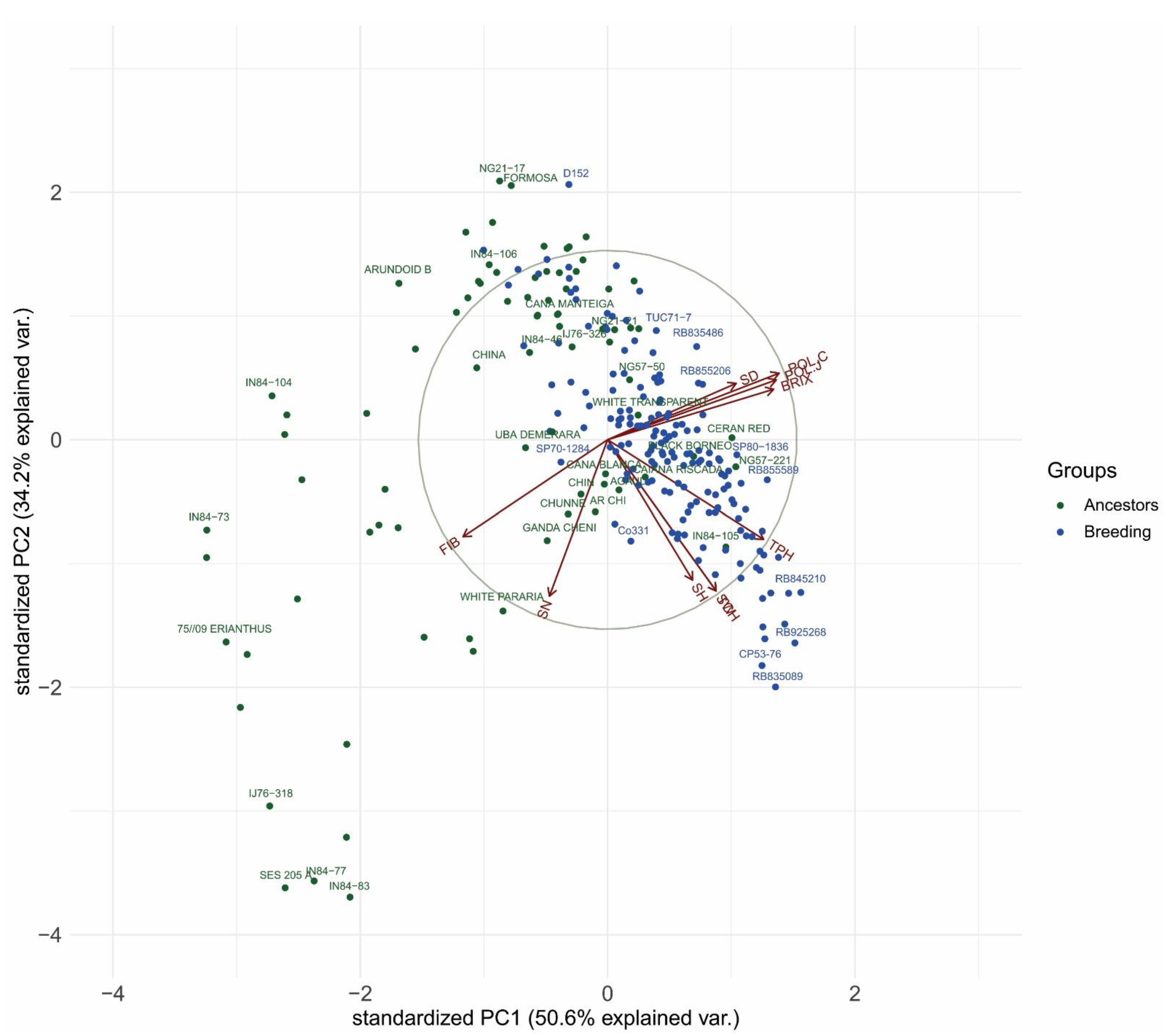
3.3. Biplot Analysis
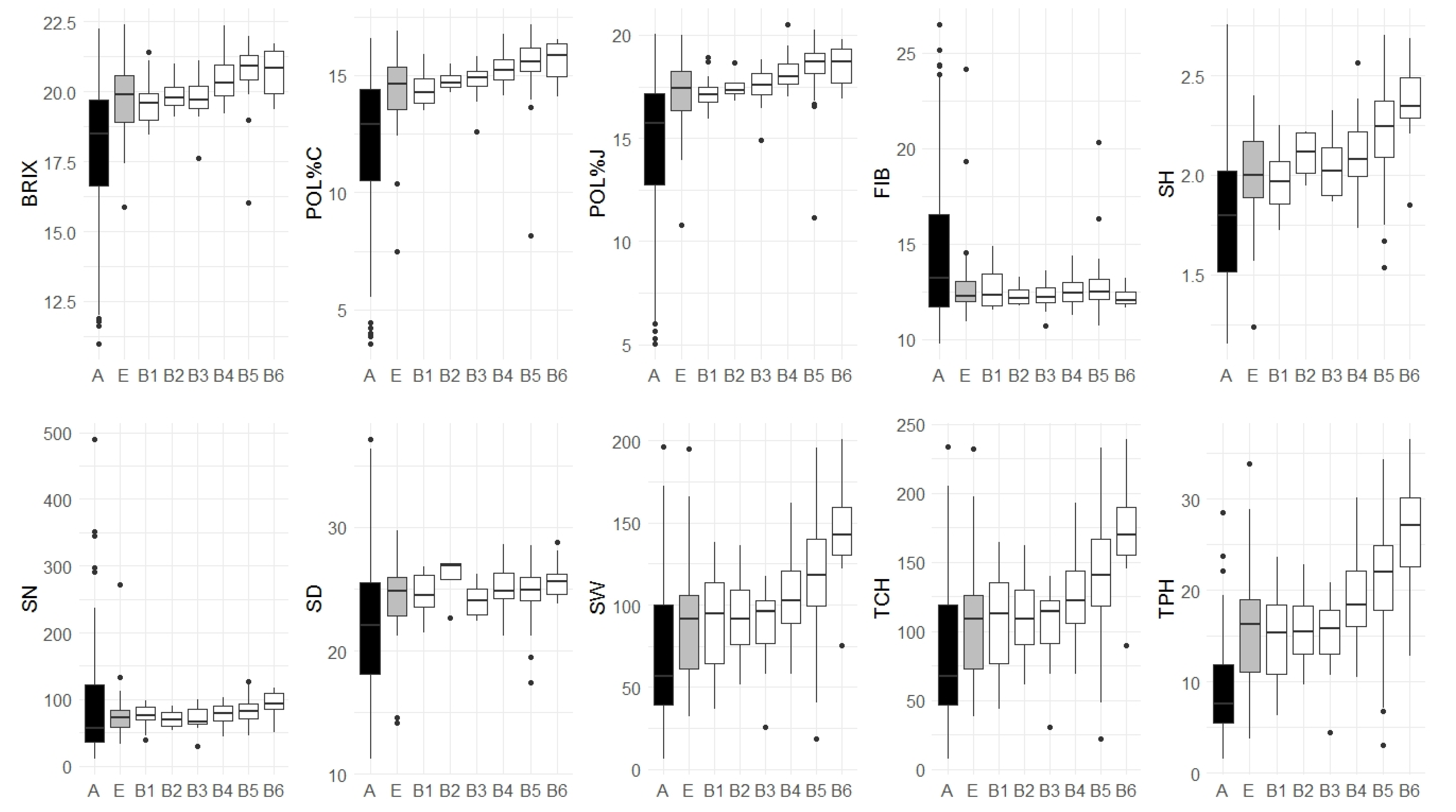
3.4. Association Panel Subsets
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Souza, A.P.; Grandis, A.; Leite, D.C.C.; Buckeridge, M.S. Sugarcane as a bioenergy source: History, performance, and perspectives for second-generation bioethanol. Bioenergy Res. 2014, 7, 24–35. [Google Scholar] [CrossRef]
- Sampaio, I.L.M.; Cardoso, T.F.; Souza, N.R.D.; Watanabe, M.D.B.; Carvalho, D.J.; Bonomi, A.; Junqueira, T.L. Electricity production from sugarcane straw recovered through bale system: Assessment of retrofit projects. Bioenergy Res. 2019. [Google Scholar] [CrossRef]
- de Oliveira Bordonal, R.; Carvalho, J.L.N.; Lal, R.; de Figueiredo, E.B.; de Oliveira, B.G.; La Scala, N. Sustainability of sugarcane production in brazil. A review. Agron. Sustain. Dev. 2018, 38. [Google Scholar] [CrossRef]
- Cursi, D.E.; Hoffmann, H.P.; Barbosa, G.V.S.; Bressiani, J.A.; Gazaffi, R.; Chapola, R.G.; Fernandes Junior, A.R.; Balsalobre, T.W.A.; Diniz, C.A.; Santos, J.M.; et al. History and current status of sugarcane breeding, germplasm development and molecular genetics in brazil. Sugar Tech 2021. [Google Scholar] [CrossRef]
- MME: Ministry of Mines and Energy. Resenha Energética Brasileira. Available online: http://antigo.mme.gov.br/documents/36208/948169/Resenha+Energ%C3%A9tica+Brasileira+-+edi%C3%A7%C3%A3o+2020/ab9143cc-b702-3700-d83a-65e76dc87a9e (accessed on 27 April 2021).
- D’Hont, A.; Glaszmann, J.C. Sugarcane genome analysis with molecular markers: A first decade research. Proc. Int. Soc. Sugar-Cane Technol. 2001, 24, 556–559. [Google Scholar]
- Vieira, M.L.C.; Almeida, C.B.; Oliveira, C.A.; Tacuatiá, L.O.; Munhoz, C.F.; Cauz-Santos, L.A.; Pinto, L.R.; Monteiro-Vitorello, C.B.; Xavier, M.A.; Forni-Martins, E.R. Revisiting meiosis in sugarcane: Chromosomal irregularities and the prevalence of bivalent configurations. Front. Genet. 2018, 9, 213. [Google Scholar] [CrossRef]
- Sforça, D.A.; Vautrin, S.; Cardoso-Silva, C.B.; Mancini, M.C.; Romero-da Cruz, M.V.; da Silva Pereira, G.; Conte, M.; Bellec, A.; Dahmer, N.; Fourment, J.; et al. Gene duplication in the sugarcane genome: A case study of allele interactions and evolutionary patterns in two genic regions. Front. Plant Sci. 2019, 10, 553. [Google Scholar] [CrossRef] [PubMed]
- Souza, G.M.; Van Sluys, M.-A.; Lembke, C.G.; Lee, H.; Margarido, G.R.A.; Hotta, C.T.; Gaiarsa, J.W.; Diniz, A.L.; de Medeiros Oliveira, M.; de Siqueira Ferreira, S.; et al. Assembly of the 373k Gene space of the polyploid sugarcane genome reveals reservoirs of functional diversity in the world’s leading biomass crop. Gigascience 2019, 8. [Google Scholar] [CrossRef]
- Daniels, J.; Roach, B.T. Taxonomy and evolution. In Sugarcane Improvement through Breeding; Heinz, D.J., Ed.; Elsevier: Amsterdan, The Netherlands, 1987; pp. 7–84. [Google Scholar]
- Cheavegatti-Gianotto, A.; de Abreu, H.M.C.; Arruda, P.; Bespalhok Filho, J.C.; Burnquist, W.L.; Creste, S.; di Ciero, L.; Ferro, J.A.; de Oliveira Figueira, A.V.; de Sousa Filgueiras, T.; et al. Sugarcane (Saccharum X officinarum): A reference study for the regulation of genetically modified cultivars in brazil. Trop. Plant Biol. 2011, 4, 62–89. [Google Scholar] [CrossRef]
- Morais, L.K.; Aguiar, M.S.; Silva, P.A.; Câmara, T.M.M.; Cursi, D.E.; Júnior, A.R.F.; Chapola, R.G.; Carneiro, M.S.; Filho, J.C.B. Breeding of sugarcane. In Industrial Crops: Breeding for Bioenergy and Bioproducts; Cruz, V.M.V., Dierig, D.A., Eds.; Springer: New York, NY, USA, 2015. [Google Scholar]
- Pompidor, N.; Charron, C.; Hervouet, C.; Bocs, S.; Droc, G.; Rivallan, R.; Manez, A.; Mitros, T.; Swaminathan, K.; Glaszmann, J.-C.; et al. Three founding ancestral genomes involved in the origin of sugarcane. Ann. Bot. 2021. [Google Scholar] [CrossRef]
- Gouy, M.; Rousselle, Y.; Thong Chane, A.; Anglade, A.; Royaert, S.; Nibouche, S.; Costet, L. Genome wide association mapping of agro-morphological and disease resistance traits in sugarcane. Euphytica 2015, 202, 269–284. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Zou, C.; Lu, Y.; Xie, C.; Zhang, X.; Prasanna, B.M.; Olsen, M.S. Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 2017, 68, 2641–2666. [Google Scholar] [CrossRef]
- Costa Coutinho, H.; Dias Turetta, A.; Guimarães Monteiro, J.; Simões de Castro, S.; Pietrafesa, J. Participatory sustainability assessment for sugarcane expansion in Goiás, Brazil. Sustainability 2017, 9, 1573. [Google Scholar] [CrossRef]
- Dal-Bianco, M.; Carneiro, M.S.; Hotta, C.T.; Chapola, R.G.; Hoffmann, H.P.; Garcia, A.A.F.; Souza, G.M. Sugarcane improvement: How far can we go? Curr. Opin. Biotechnol. 2012, 23, 265–270. [Google Scholar] [CrossRef]
- Bechara, R.; Gomez, A.; Saint-Antonin, V.; Schweitzer, J.-M.; Maréchal, F.; Ensinas, A. Review of design works for the conversion of sugarcane to first and second-generation ethanol and electricity. Renew. Sustain. Energy Rev. 2018, 91, 152–164. [Google Scholar] [CrossRef]
- Diniz, A.L.; Ferreira, S.S.; Ten-Caten, F.; Margarido, G.R.; Dos Santos, J.M.; Barbosa, G.V.D.S.; Carneiro, M.S.; Souza, G.M. Genomic resources for energy cane breeding in the post genomics era. Comput. Struct. Biotechnol. J. 2019, 17, 1404–1414. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.D.H.; Gonçalves, T.A.; Uchima, C.A.; dos Reis, L.; Fontana, R.C.; Squina, F.M.; Dillon, A.J.P.; Camassola, M. Comparison of the production of enzymes to cell wall hydrolysis using different carbon sources by Penicillium echinulatum strains and its hydrolysis potential for lignocelullosic biomass. Process Biochem. 2018, 66, 162–170. [Google Scholar] [CrossRef]
- da Silva, D.S.; Dantzger, M.; Assis, M.A.; Gallardo, J.C.M.; Teixeira, G.S.; Missawa, S.K.; Domingues, R.R.; Carazzolle, M.F.; Lunardi, I.; Leme, A.F.P.; et al. Lignocellulolytic Characterization and Comparative Secretome Analysis of a Trichoderma erinaceum strain isolated from decaying sugarcane straw. Fungal Biol. 2019, 123, 330–340. [Google Scholar] [CrossRef] [PubMed]
- de Faria, L.C.; Melo, P.G.S.; Pereira, H.S.; Peloso, M.J.D.; Brás, A.J.B.P.; Moreira, J.A.A.; de Carvalho, H.W.L.; Melo, L.C. Genetic progress during 22 years of improvement of carioca-type common bean in brazil. Field Crops Res. 2013, 142, 68–74. [Google Scholar] [CrossRef]
- Dumont, T.; Thong-Chane, A.; Barau, L.; Siegmund, B.; Hoarau, J.-Y. Genetic variabilities and genetic gains for yield components in regional sugarcane breeding programmes on Réunion Island. Sugar Tech 2019, 21, 868–878. [Google Scholar] [CrossRef]
- Oury, F.-X.; Godin, C.; Mailliard, A.; Chassin, A.; Gardet, O.; Giraud, A.; Heumez, E.; Morlais, J.-Y.; Rolland, B.; Rousset, M.; et al. A Study of genetic progress due to selection reveals a negative effect of climate change on bread wheat yield in france. Eur. J. Agron. 2012, 40, 28–38. [Google Scholar] [CrossRef]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the rate of genetic gain in public-sector plant breeding programs: Lessons from the breeder’s equation. Züchter Genet. Breed. Res. 2019, 132, 627–645. [Google Scholar] [CrossRef]
- Santana, P.N.D.; Reis, A.J.D.S.; Chaves, L.J. Combining ability of sugarcane genotypes based on the selection rates of single cross families. Crop Breed. Appl. Biotechnol. 2017, 17, 47–53. [Google Scholar] [CrossRef]
- Todd, J.R.; Sandhu, H.; Binder, J.; Arundale, R.; Gordon, V.; Song, J.; Glaz, B.; Wang, J. Fiber composition of a diversity panel of the world collection of sugarcane (Saccharum spp.) and related grasses. Bragantia 2017, 77, 48–61. [Google Scholar] [CrossRef]
- Barreto, F.Z.; Rosa, J.R.B.F.; Balsalobre, T.W.A.; Pastina, M.M.; Silva, R.R.; Hoffmann, H.P.; de Souza, A.P.; Garcia, A.A.F.; Carneiro, M.S. A Genome-wide association study identified loci for yield component traits in sugarcane (Saccharum spp.). PLoS ONE 2019, 14, e0219843. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, C.; Balsalobre, T.W.A.; Carneiro, M.S. Molecular diversity and genetic structure of Saccharum complex accessions. PLoS ONE 2020, 15, e0233211. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Song, J.; Todd, J.; Peng, Z.; Paudel, D.; Luo, Z.; Ma, X.; You, Q.; Hanson, E.; Zhao, Z.; et al. Target Enrichment sequencing of 307 germplasm accessions identified ancestry of ancient and modern hybrids and signatures of adaptation and selection in sugarcane (Saccharum spp.), a “sweet” crop with “bitter” genomes. Plant Biotechnol. J. 2019, 17, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, T.W.; Mancini, M.C.; Pereira, G.D.S.; Anoni, C.O.; Barreto, F.Z.; Hoffmann, H.P.; de Souza, A.P.; Garcia, A.A.; Carneiro, M.S. Mixed modeling of yield components and brown rust resistance in sugarcane families. Agron. J. 2016, 108, 1824–1837. [Google Scholar] [CrossRef]
- Edwards, D.; Batley, J.; Snowdon, R.J. Accessing complex crop genomes with next-generation sequencing. Züchter Genet. Breed. Res. 2013, 126, 1–11. [Google Scholar] [CrossRef]
- Sadras, V.O.; Rebetzke, G.J.; Edmeades, G.O. The phenotype and the components of phenotypic variance of crop traits. Field Crops Res. 2013, 154, 255–259. [Google Scholar] [CrossRef]
- Malosetti, M.; Ribaut, J.-M.; van Eeuwijk, F.A. The statistical analysis of multi-environment data: Modeling genotype-by-environment interaction and its genetic basis. Front. Physiol. 2013, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Beyene, Y.; Kassa, S.; Pérez, P.; Hickey, J.M.; Chen, C.; de los Campos, G.; Burgueño, J.; Windhausen, V.S.; Buckler, E.; et al. Genomic prediction in maize breeding populations with genotyping-by-sequencing. G3 (Bethesda) 2013, 3, 1903–1926. [Google Scholar] [CrossRef]
- Beaulieu, J.; Doerksen, T.; Clément, S.; MacKay, J.; Bousquet, J. Accuracy of genomic selection models in a large population of open-pollinated families in white spruce. Heredity (Edinburgh) 2014, 113, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Fickett, N.; Gutierrez, A.; Verma, M.; Pontif, M.; Hale, A.; Kimbeng, C.; Baisakh, N. Genome-wide association mapping identifies markers associated with cane yield components and sucrose traits in the Louisiana sugarcane core collection. Genomics 2019, 111, 1794–1801. [Google Scholar] [CrossRef]
- Cardoso-Silva, C.B.; Costa, E.A.; Mancini, M.C.; Balsalobre, T.W.A.; Canesin, L.E.C.; Pinto, L.R.; Carneiro, M.S.; Garcia, A.A.F.; de Souza, A.P.; Vicentini, R. De novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS ONE 2014, 9, e88462. [Google Scholar] [CrossRef]
- Balsalobre, T.W.A.; da Silva Pereira, G.; Margarido, G.R.A.; Gazaffi, R.; Barreto, F.Z.; Anoni, C.O.; Cardoso-Silva, C.B.; Costa, E.A.; Mancini, M.C.; Hoffmann, H.P.; et al. GBS-based single dosage markers for linkage and qtl mapping allow gene mining for yield-related traits in sugarcane. BMC Genom. 2017, 18, 72. [Google Scholar] [CrossRef]
- Manual de instruções CONSECANA. State of Sao Paulo Sugarcane, Sugar and Alcohol Growers Council. 2006. Available online: http://www.oricana.com.br/novosite/manual_consecana.pdf (accessed on 27 April 2021).
- Piepho, H.P.; Büchse, A.; Richter, C. A Mixed Modelling Approach for Randomized Experiments with Repeated Measures. J Agron. Crop Sci. 2004, 190, 230–247. [Google Scholar] [CrossRef]
- VSN International. Genstat for Windows 21st Edition. VSN International: Hemel Hempstead, UK, 2020. Available online: https://www.vsni.co.uk/software/genstat (accessed on 27 April 2021).
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Pastina, M.M.; Malosetti, M.; Gazaffi, R.; Mollinari, M.; Margarido, G.R.A.; Oliveira, K.M.; Pinto, L.R.; Souza, A.P.; van Eeuwijk, F.A.; Garcia, A.A.F. A mixed model QTL analysis for sugarcane multiple-harvest-location trial data. Züchter Genet. Breed. Res. 2012, 124, 835–849. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martínez, C.T. Estimating and interpreting heritability for plant breeding: An update. In Plant Breeding Reviews; John Wiley & Sons, Inc.: Oxford, UK, 2010; pp. 9–112. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 27 April 2021).
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of genetic diversity assessment in crop plants and its recent advances: An overview of its analytical perspectives. Genet. Res. Int. 2015, 2015, 431487. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ferro, J.; Arruda, P. The Brazilian Experience of Sugarcane Ethanol Industry. Vitro Cell. Dev. Biol. Plant 2009, 45, 372–381. [Google Scholar] [CrossRef]
- Margarido, G.R.A.; Pastina, M.M.; Souza, A.P.; Garcia, A.A.F. Multi-trait multi-environment quantitative trait loci mapping for a sugarcane commercial cross provides insights on the inheritance of important traits. Mol. Breed. 2015, 35, 175. [Google Scholar] [CrossRef]
- Breseghello, F.; Coelho, A.S.G. Traditional and Modern Plant Breeding Methods with Examples in Rice (Oryza Sativa L.). J. Agric. Food Chem. 2013, 61, 8277–8286. [Google Scholar] [CrossRef]
- Welham, S.J.; Gogel, B.J.; Smith, A.B.; Thompson, R.; Cullis, B.R. A comparison of analysis methods for late-stage variety evaluation trials: Comparison of methods for variety trials. Aust. N. Z. J. Stat. 2010, 52, 125–149. [Google Scholar] [CrossRef]
- Nayak, S.N.; Song, J.; Villa, A.; Pathak, B.; Ayala-Silva, T.; Yang, X.; Todd, J.; Glynn, N.C.; Kuhn, D.N.; Glaz, B.; et al. Promoting utilization of Saccharum spp. Genetic resources through genetic diversity analysis and core collection construction. PLoS ONE 2014, 9, e110856. [Google Scholar] [CrossRef]
- Byrne, P.F.; Volk, G.M.; Gardner, C.; Gore, M.A.; Simon, P.W.; Smith, S. Sustaining the future of plant breeding: The critical role of the USDA-ARS National Plant Germplasm System. Crop Sci. 2018, 58, 451–468. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Longman: Harlow, UK, 1996. [Google Scholar]
- Piepho, H.-P.; Möhring, J. Computing heritability and selection response from unbalanced plant breeding trials. Genetics 2007, 177, 1881–1888. [Google Scholar] [CrossRef]
- Chaudhary, R.R. Genetic variability and heritability in sugarcane. Nepal Agric. Res. J. 2001, 4, 56–59. [Google Scholar] [CrossRef]
- Kimbeng, C.A.; Zhou, M.M.; da Silva, J.A. Genotype × environment interactions and resource allocation in sugarcane yield trials in the Rio Grande valley region of Texas. J. Am. Soc. Sugar Cane Technol. 2009, 29, 11–24. [Google Scholar]
- Gouy, M.; Rousselle, Y.; Bastianelli, D.; Lecomte, P.; Bonnal, L.; Roques, D.; Efile, J.-C.; Rocher, S.; Daugrois, J.; Toubi, L.; et al. Experimental assessment of the accuracy of genomic selection in sugarcane. Züchter Genet. Breed. Res. 2013, 126, 2575–2586. [Google Scholar] [CrossRef]
- Bourke, P.M.; Voorrips, R.E.; Visser, R.G.F.; Maliepaard, C. Tools for genetic studies in experimental populations of polyploids. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Ram, B.; Chaudhary, B.S.; Yadav, D.K. General and specific selection indices for single stool stages of selection in sugarcane. Euphytica 1997, 95, 39–44. [Google Scholar] [CrossRef]
- Cellon, C.; Amadeu, R.R.; Olmstead, J.W.; Mattia, M.R.; Ferrao, L.F.V.; Munoz, P.R. Estimation of genetic parameters and prediction of breeding values in an autotetraploid blueberry breeding population with extensive pedigree data. Euphytica 2018, 214. [Google Scholar] [CrossRef]
- Ming, R.; Liu, S.C.; Moore, P.H.; Irvine, J.E.; Paterson, A.H. QTL analysis in a complex autopolyploid: Genetic control of sugar content in sugarcane. Genome Res. 2001, 11, 2075–2084. [Google Scholar] [CrossRef]
- Hoarau, J.-Y.; Grivet, L.; Offmann, B.; Raboin, L.-M.; Diorflar, J.-P.; Payet, J.; Hellmann, M.; D’Hont, A.; Glaszmann, J.-C. Genetic dissection of a modern sugarcane cultivar (Saccharum spp.): II. Detection of QTLs for yield components. Züchter Genet. Breed. Res. 2002, 105, 1027–1037. [Google Scholar] [CrossRef]
- Aitken, K.S.; Hermann, S.; Karno, K.; Bonnett, G.D.; McIntyre, L.C.; Jackson, P.A. Genetic control of yield related stalk traits in sugarcane. Züchter Genet. Breed. Res. 2008, 117, 1191–1203. [Google Scholar] [CrossRef]
- Tew, T.L.; Cobill, R.M. Genetic improvement of sugarcane (Saccharum spp.) as an energy crop. In Genetic Improvement of Bioenergy Crops; Springer: New York, NY, USA, 2008; pp. 273–294. [Google Scholar]
- Matsuoka, S.; Kennedy, A.J.; Santos, E.G.D.D.; Tomazela, A.L.; Rubio, L.C.S. Energy Cane: Its concept, development, characteristics, and prospects. Adv. Bot. 2014, 2014, 1–13. [Google Scholar] [CrossRef]
- Aitken, K.; Li, J.; Piperidis, G.; Qing, C.; Yuanhong, F.; Jackson, P. Worldwide genetic diversity of the wild species Saccharum spontaneum and level of diversity captured within sugarcane breeding programs. Crop. Sci. 2018, 58, 218–229. [Google Scholar] [CrossRef]
- Silva, J.A.G.D.; Costa, P.M.D.A.; Marconi, T.G.; Barreto, E.J.D.S.; Solís-Gracia, N.; Park, J.W.; Glynn, N.C. Agronomic and molecular characterization of wild germplasm Saccharum Spontaneum for sugarcane and energycane breeding purposes. Sci. Agric. 2018, 75, 329–338. [Google Scholar] [CrossRef]
- Aitken, K.S.; Jackson, P.A.; McIntyre, C.L. Quantitative Trait Loci identified for sugar related traits in a sugarcane (Saccharum spp.) cultivar x Saccharum officinarum population. Züchter Genet. Breed. Res. 2006, 112, 1306–1317. [Google Scholar] [CrossRef]
- Todd, J.; Wang, J.; Glaz, B.; Sood, S.; Ayala-Silva, T.; Nayak, S.N.; Glynn, N.C.; Gutierrez, O.A.; Kuhn, D.N.; Tahir, M.; et al. Phenotypic Characterization of the Miami World Collection of Sugarcane (Saccharum spp.) and Related Grasses for selecting a representative core. Genet. Resour. Crop. Evol. 2014, 61, 1581–1596. [Google Scholar] [CrossRef]
- Pardey, P.G.; Beddow, J.M.; Hurley, T.M.; Beatty, T.K.M.; Eidman, V.R. A Bounds analysis of world food futures: Global agriculture through to 2050. Aust. J. Agric. Resour. Econ. 2014, 58, 571–589. [Google Scholar] [CrossRef]
- de Souza Barbosa, G.V.; dos Santos, J.M.; Diniz, C.A.; Cursi, D.E.; Hoffmann, H.P. Energy Cane Breeding. In Sugarcane Biorefinery, Technology and Perspectives; Elsevier: Amsterdam, The Netherlands, 2020; pp. 103–116. [Google Scholar]
- Cowling, W.A.; Li, L.; Siddique, K.H.M.; Henryon, M.; Berg, P.; Banks, R.G.; Kinghorn, B.P. Evolving Gene Banks: Improving diverse populations of crop and exotic germplasm with optimal contribution selection. J. Exp. Bot. 2016, 68, 1927–1939. [Google Scholar] [CrossRef] [PubMed]
- Raboin, L.-M.; Pauquet, J.; Butterfield, M.; D’Hont, A.; Glaszmann, J.-C. Analysis of genome-wide linkage disequilibrium in the highly polyploid sugarcane. Züchter Genet. Breed. Res. 2008, 116, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Jackson, P.A.; Hermann, S.; Kilian, A.; Heller-Uszynska, K.; Deomano, E. Simultaneously accounting for population structure, genotype by environment interaction, and spatial variation in marker–trait associations in sugarcane. Genome 2010, 53, 973–981. [Google Scholar] [CrossRef] [PubMed]

| BRIX (°Brix) | POL%C | POL%J | FIB (%) | SH (m) | SN | SD (mm) | SW (Kg) | TCH (t/ha) | TPH (t/ha) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Ranges (min/max) | 10.98/22.41 | 3.54/17.18 | 5.04/20.52 | 9.76/26.50 | 1.15/2.75 | 10.83/490.01 | 11.22/37.16 | 6.36/201.02 | 7.60/239.30 | 1.56/36.55 |
| Average | 19.48 | 13.98 | 16.79 | 13.45 | 2.00 | 82.67 | 23.79 | 131.30 | 156.20 | 19.83 |
| Phi_1 | 0.96 | 0.96 | 0.96 | 0.95 | 0.93 | 0.94 | 0.97 | 0.92 | 0.92 | 0.95 |
| 4.79 | 7.62 | 8.84 | 13.90 | 0.11 | 1987.00 | 19.98 | 3103.00 | 4397.00 | 109.40 | |
| 3.90 | 7.62 | 8.84 | 6.11 | 0.15 | 4060.00 | 16.15 | 2754.00 | 3902.00 | 108.90 | |
| 5.12 | 7.62 | 8.84 | 11.48 | 0.07 | 3570.00 | 13.55 | 557.00 | 789.20 | 15.08 | |
| 1.14 | 2.02 | 2.32 | 3.14 | 0.06 | 267.50 | 3.48 | 1180.00 | 1673.00 | 35.99 | |
| 0.85 | 1.16 | 1.57 | 1.29 | 0.05 | 707.60 | 4.71 | 1036.00 | 1468.00 | 41.01 | |
| 0.77 | 2.99 | 3.02 | 12.45 | 0.03 | 1943.00 | 2.85 | 591.30 | 837.80 | 19.64 | |
| CVG(1) | 0.11 | 0.19 | 0.17 | 0.27 | 0.16 | 0.53 | 0.18 | 0.42 | 0.42 | 0.52 |
| CVG(2) | 0.10 | 0.19 | 0.17 | 0.18 | 0.19 | 0.77 | 0.16 | 0.40 | 0.40 | 0.52 |
| CVG(3) | 0.11 | 0.19 | 0.17 | 0.25 | 0.13 | 0.72 | 0.15 | 0.18 | 0.18 | 0.19 |
| CVR(1) | 0.05 | 0.10 | 0.09 | 0.13 | 0.12 | 0.19 | 0.07 | 0.26 | 0.26 | 0.30 |
| CVR(2) | 0.04 | 0.07 | 0.07 | 0.08 | 0.11 | 0.32 | 0.09 | 0.24 | 0.24 | 0.32 |
| CVR(3) | 0.04 | 0.12 | 0.10 | 0.26 | 0.08 | 0.53 | 0.07 | 0.18 | 0.18 | 0.22 |
| 0.80 | 0.79 | 0.79 | 0.81 | 0.64 | 0.88 | 0.85 | 0.72 | 0.72 | 0.75 | |
| 0.82 | 0.86 | 0.85 | 0.82 | 0.75 | 0.85 | 0.77 | 0.72 | 0.72 | 0.72 | |
| 0.87 | 0.71 | 0.74 | 0.48 | 0.70 | 0.64 | 0.82 | 0.48 | 0.48 | 0.43 |
| Trait | Linear Regression | R² (%) | p-value | GP (%) |
|---|---|---|---|---|
| BRIX (°Brix) | y = 19.30 + 0.27x | 17.66 | 6.41 × 10−7 | 0.69 |
| POL%C | y = 14.13 + 0.26x | 13.44 | 1.79 × 10−5 | 0.92 |
| POL%J | y = 16.82 + 0.31x | 15.51 | 3.55 × 20−6 | 0.92 |
| FIB (%) | y = 12.50 + 0.01x | 0.03 | 0.84 ns | 0.04 |
| SH (m) | y = 1.87 + 0.06x | 19.69 | 1.24 × 10−7 | 1.60 |
| SN | y = 67.00 + 3.27x | 6.80 | 2.71 × 10−3 | 2.44 |
| SD (mm) | y = 24.39 + 0.11x | 0.94 | 0.27 ns | 0.23 |
| SW (Kg) | y = 72.06 + 9.42x | 15.96 | 2.48 × 10−6 | 6.53 |
| TCH (t/ha) | y = 85.77 + 11.22x | 15.97 | 2.48 × 10−6 | 6.54 |
| TPH (t/ha) | y = 11.40 + 2.08x | 21.15 | 3.70 × 10−8 | 9.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barreto, F.Z.; Balsalobre, T.W.A.; Chapola, R.G.; Garcia, A.A.F.; Souza, A.P.; Hoffmann, H.P.; Gazaffi, R.; Carneiro, M.S. Genetic Variability, Correlation among Agronomic Traits, and Genetic Progress in a Sugarcane Diversity Panel. Agriculture 2021, 11, 533. https://doi.org/10.3390/agriculture11060533
Barreto FZ, Balsalobre TWA, Chapola RG, Garcia AAF, Souza AP, Hoffmann HP, Gazaffi R, Carneiro MS. Genetic Variability, Correlation among Agronomic Traits, and Genetic Progress in a Sugarcane Diversity Panel. Agriculture. 2021; 11(6):533. https://doi.org/10.3390/agriculture11060533
Chicago/Turabian StyleBarreto, Fernanda Zatti, Thiago Willian Almeida Balsalobre, Roberto Giacomini Chapola, Antonio Augusto Franco Garcia, Anete Pereira Souza, Hermann Paulo Hoffmann, Rodrigo Gazaffi, and Monalisa Sampaio Carneiro. 2021. "Genetic Variability, Correlation among Agronomic Traits, and Genetic Progress in a Sugarcane Diversity Panel" Agriculture 11, no. 6: 533. https://doi.org/10.3390/agriculture11060533
APA StyleBarreto, F. Z., Balsalobre, T. W. A., Chapola, R. G., Garcia, A. A. F., Souza, A. P., Hoffmann, H. P., Gazaffi, R., & Carneiro, M. S. (2021). Genetic Variability, Correlation among Agronomic Traits, and Genetic Progress in a Sugarcane Diversity Panel. Agriculture, 11(6), 533. https://doi.org/10.3390/agriculture11060533






