1. Introduction
The quantity of land covered by various crops in a specific time span, referred to as a cropping pattern [
1], dictates the level of global agricultural production, which in turn influences the agricultural economy [
2]. Thus, small-scale farmers who employ various cropping patterns can play a critical role in the global food supply [
3]. However, cropping patterns and food security, particularly in Africa, are highly influenced by myriad factors such as climate change and variableness, inadequate agricultural inputs, insect pests and diseases, and other abiotic and biotic factors [
4].
The types of cropping patterns can include monocropping, crop rotation, and intercropping [
5], which are practiced for various reasons such as environmental conditions, profitability, adaptability to changing conditions, tolerance and resistance to insect pests and diseases, the requirement for specific technologies during growing or harvesting, and other elements in the production system [
6]. These cropping patterns possess several benefits and drawbacks. For instance, monocropping patterns ensure specialized crop production and expected higher earnings due to mass crop production. However, monocropping practice is characterized by, for example, a high risk of pests and low soil microbe diversity [
7]. By comparison, crop rotation minimizes the eroding of soil and improves soil fertility, which results in improved crop yields [
8]. Nonetheless, the necessary and regular crop diversification required in crop rotation may be a strain for the farmers if there are no readily available resources for its implementation. Intercropping also promotes soil fertility (reducing risk to climate stress), reduces pest risks, and maximizes land profit. Nevertheless, the different crops in an intercropping practice may require different uptakes of resources such as water and fertilizer, which may not be adequately utilized by the different crops [
5]. Intercropping comprises variants such as rows, relay, and mixed cropping, but the present study refers to the different groups of intercropping patterns as mixed cropping in general. Thus, the present study focused on two types of cropping patterns, i.e., monocropping patterns and mixed cropping patterns, that are commonly practiced in the study area.
The choice of crops for planting by farmers varies spatially and among farmers across the different agro-ecological systems [
9]. Hence, their accurate characterization is of paramount importance for policy making and implementation strategies necessary for addressing food and nutrition insecurity through precision agriculture. In addition, adequate characterization will improve understanding of the sustainability of food systems and how they are affected by climate. Their characterization is equally important for modeling and managing the abundance and spread of crop insect pests, diseases, and pollinators [
10,
11]. Nonetheless, small-scale farms (≤1.25 ha) and fragmentation of these cropping patterns in Africa, triggered by their high intra- and inter-seasonal variability, prohibit their accurate detection and characterization [
10,
12].
Currently, information on the spatial spread of these various cropping patterns is scarce, especially over larger regions. This makes reporting and decision making regarding pest and climate resilience of certain cropping patterns challenging. Moreover, traditional terrestrial surveys and assessment methods used to determine the commonly grown crops in an area are often inadequate, expensive, time consuming, and strenuous, and provide insufficient information necessary for precision agriculture, efficient utilization of resources, and effective pest management [
10,
12]. In contrast, the recent advancement in remote sensing technology provides coherent, timely, concise, and affordable data that can effectively capture the cropping variability at different spatiotemporal scales [
13]. In this regard, the usage of remotely sensed data for mapping and modeling cropping patterns and other agronomic practices is well documented in the literature. This has been evidenced by studies carried out on the mapping of various croplands [
14,
15,
16], crop types [
17,
18,
19], and cropping patterns [
20,
21,
22,
23], with the aim of investigating the relevance of different remote sensing systems and image classification methods for improving classification accuracy, reliability, and reproducibility of the results. Diverse imaging systems, analytical techniques, and spectral variables have also been explored for cropping pattern classification. These systems range from optical multispectral [
10,
24,
25] and hyperspectral imaging [
26,
27] to radar sensors [
28,
29] with high, medium, and coarse spatial resolutions. In addition, several studies have used varying remotely sensed variables, such as single date [
30] and multi-date (time-series) vegetation indices [
21] and phenometrics [
31,
32], to map cropping patterns. In these studies, various image classifications methods have also been applied, which range from parametric classifiers such as
K-means clustering [
19] and maximum likelihood [
22] to non-parametric or machine learning algorithms such as k-nearest neighbor (kNN) [
16], decision trees (DTs) [
21,
23], support vector machine (SVM) [
22], random forest (RF) [
15,
17], and fuzzy c-means clustering [
18]. The selection of these methods is mainly informed by the availability of training data, sample size, computation time, presence of high data dimensionality, and multicollinearity [
33]. One of the efficient methods for reducing data dimensionality and simultaneously handling multicollinearity is the guided regularized random forest (GRRF), [
34,
35]. Compared to the RF classifier [
36], GRRF is superior in selecting optimum uncorrelated variables [
37], whereas RF is a robust classifier because it adequately handles diverse scales, interactions, and nonlinearities, among other numerical and categorical variables [
36]. In other words, the GRRF is an efficient algorithm for selecting the most relevant predictors because its trees are grown in a sequential manner oriented towards the identification of the most important variables during the training step in a classification experiment [
38]. This type of training approach can lead to a high variance of predictions, as suggested by Deng and Runger [
37]. Hence, the GRRF should not be used as a classification method, but variables selected by the algorithm should be used as inputs in an efficient classification algorithm (e.g., RF) as predictors to evaluate their values in discriminating among features of interest, such as cropping patterns [
39].
Despite the efforts in selecting optimum datasets and classification algorithms, challenges in mapping cropping patterns remain evident in Africa [
40]. These challenges include the inter- and intra-season changes in crop phenological cycles due to variability in farming practices or weather conditions, and highly fragmented landscapes on which the crops are grown [
18,
21,
41]. Moreover, accurate characterization of cropping patterns in heterogeneous landscapes often requires high (≤5 m) spatial resolution imagery [
42], whose accessibility can be constrained due to their cost implications. Previous studies have proposed the use of optical and radar datasets of medium spatial resolutions [
12], the use of crop phenological variables extracted from the relatively new satellite sensors such as Sentinel-2 (S2) [
43], the use of multisource remotely sensed and ancillary data to improve the quality and timeliness of in-season cropping patterns mapping [
44], together with the use of freely available multi-temporal remote sensing data [
10].
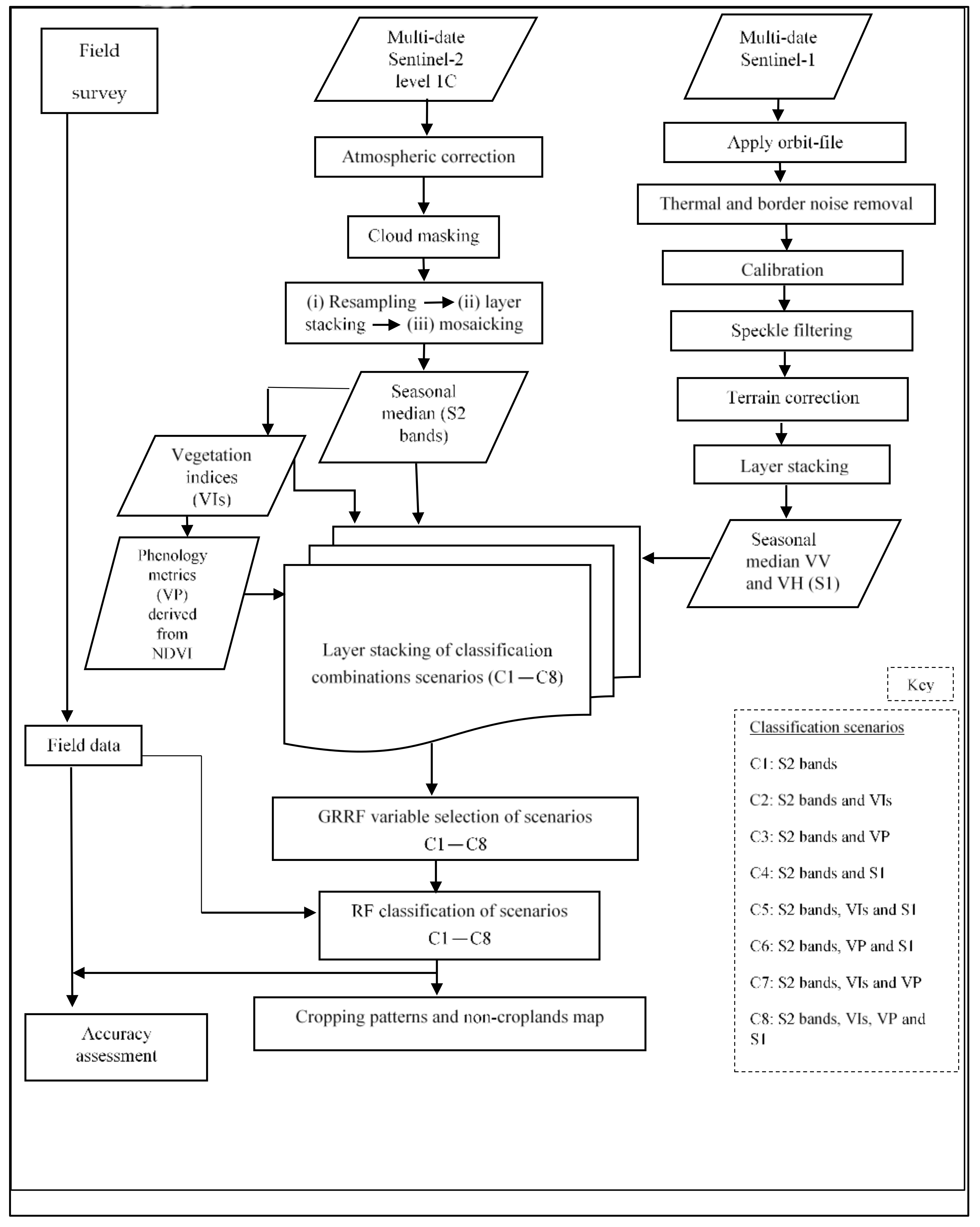
This study hypothesized that utilizing the unique advantages of multi-date and medium spatial resolution freely available S2 reflectance bands (S2 bands), their vegetation indices (VIs) and vegetation phenology (VP) derivatives, and Sentinel-1 (S1) backscatter data would improve cropping pattern mapping in heterogeneous landscapes using robust machine learning variable selection algorithms such as GRRF [
37] and classifiers such as RF [
36]. The tested null hypothesis was that the performance of mapping cropping patterns using eight different scenarios was not significantly different (
p ≤ 0.05) using McNemar’s test [
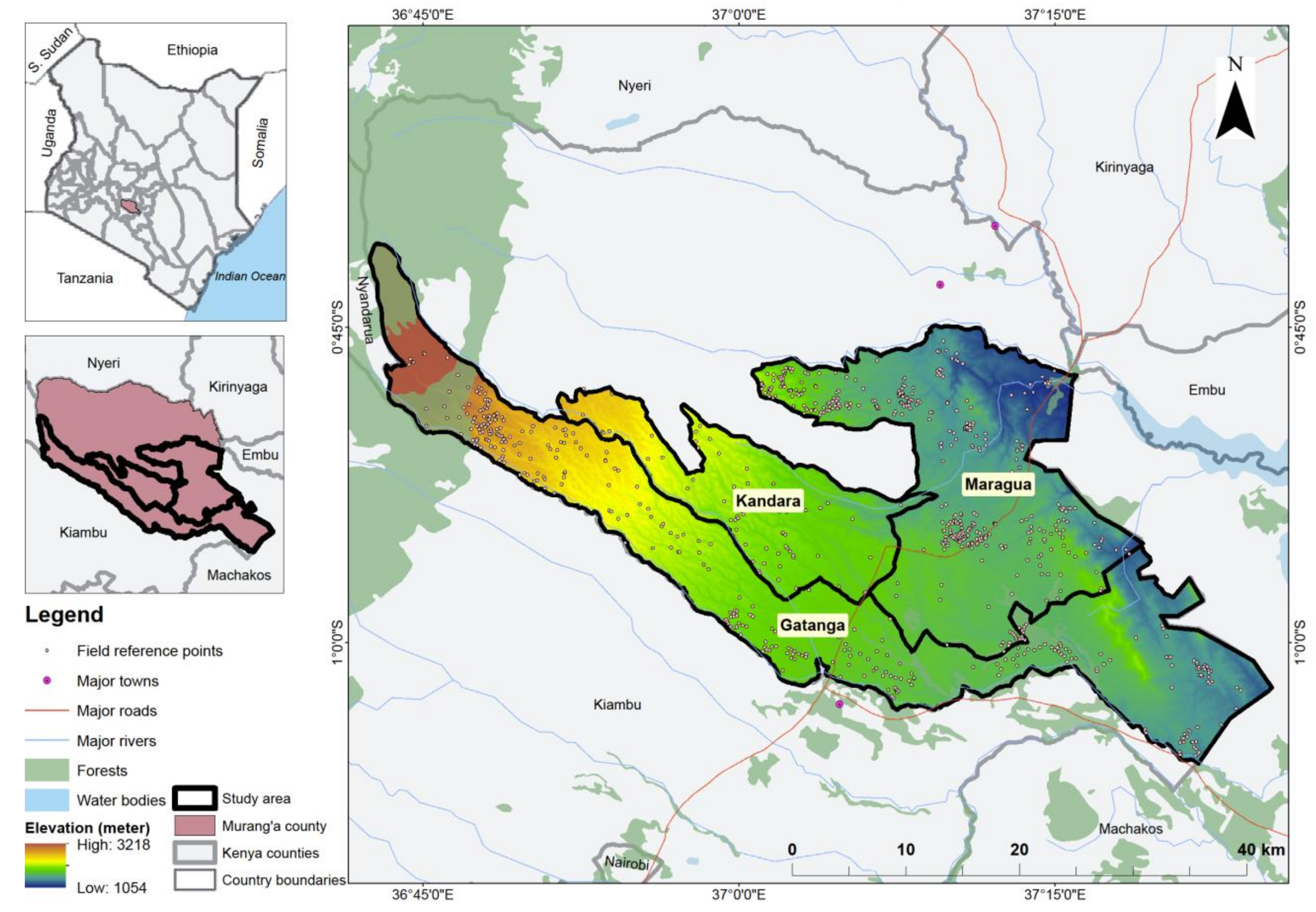
45]; these scenarios were: (i) only S2 bands; (ii) S2 bands and VIs; (iii) S2 bands and VP; (iv) S2 bands and S1; (v) S2 bands, VIs, and S1; (vi) S2 bands, VP, and S1; (vii) S2 bands, VIs, and VP; and (viii) S2 bands, VIs, VP, and S1. Therefore, this study’s objective was to evaluate the strengths of the freely available multi-date medium resolution S2 bands, their VIs and VP variables, and S1 backscatter data for mapping cropping patterns in a heterogeneous agro-natural production system in Murang’a County, Kenya. Specifically, the performance of the eight multi-sensor classification scenarios for delineating cropping patterns utilizing a robust feature selection and classification algorithm (i.e., GRRF and RF, respectively) was assessed and compared.
5. Discussion
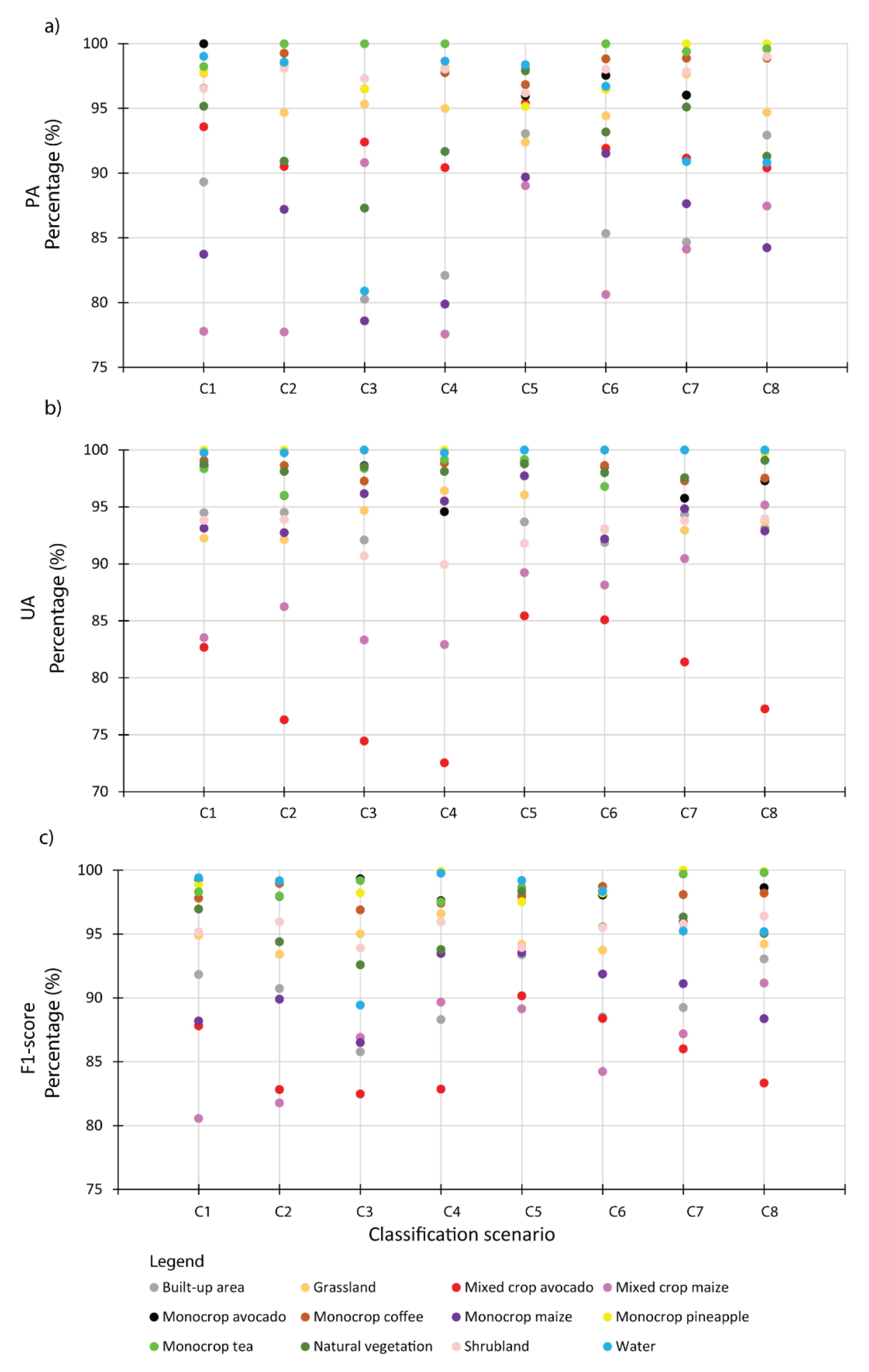
This study leveraged the synergetic advantage of integrating the freely available medium resolution multi-date and multi-sensor variables for classifying cropping patterns in a heterogeneous agro-natural landscape in Kenya when high-resolution imagery is not available. The performance of eight classification scenarios that resulted from combining S2 bands, their VIs and VP variables, and S1 SAR backscatter imagery was tested. Seven cropping patterns were classified. The study demonstrated that cropping patterns in a heterogeneous landscape in Murang’a, Kenya, can be accurately (OA > 90% and Kappa > 0.88) mapped using the freely available multi-date medium resolution S2 bands, their VIs and VP, and S1 backscatter data by employing a robust GRRF feature selection algorithm and RF classifier. In summary, the objective and the tested hypothesis were successfully achieved.
Satellite-based data acquired over four seasons (i.e., hot and cool dry, in addition to short and long rainy season) that were assumed to have captured all the changes in inter- and intra-vegetation dynamics were utilized. The use of optical satellite time-series data increased the chances of acquiring cloud-free imagery and hence resulted in higher chances of improved classification results, corroborating results of other previous studies [
82,
83]. Persistent cloud coverage can considerably affect the quality of optical S2 imagery [
84]. S2 and S1 have varied revisit periods of 5 and 12 days, respectively, and these differences were accounted for using seasonal median composites. Furthermore, the performance of the classification experiments using an error matrix that takes into account the estimated area proportion of each class [
80] was assessed. This is a robust and more reliable classification accuracy assessment method compared to the traditional classification confusion matrix that considers the number of mapped instances of each class.
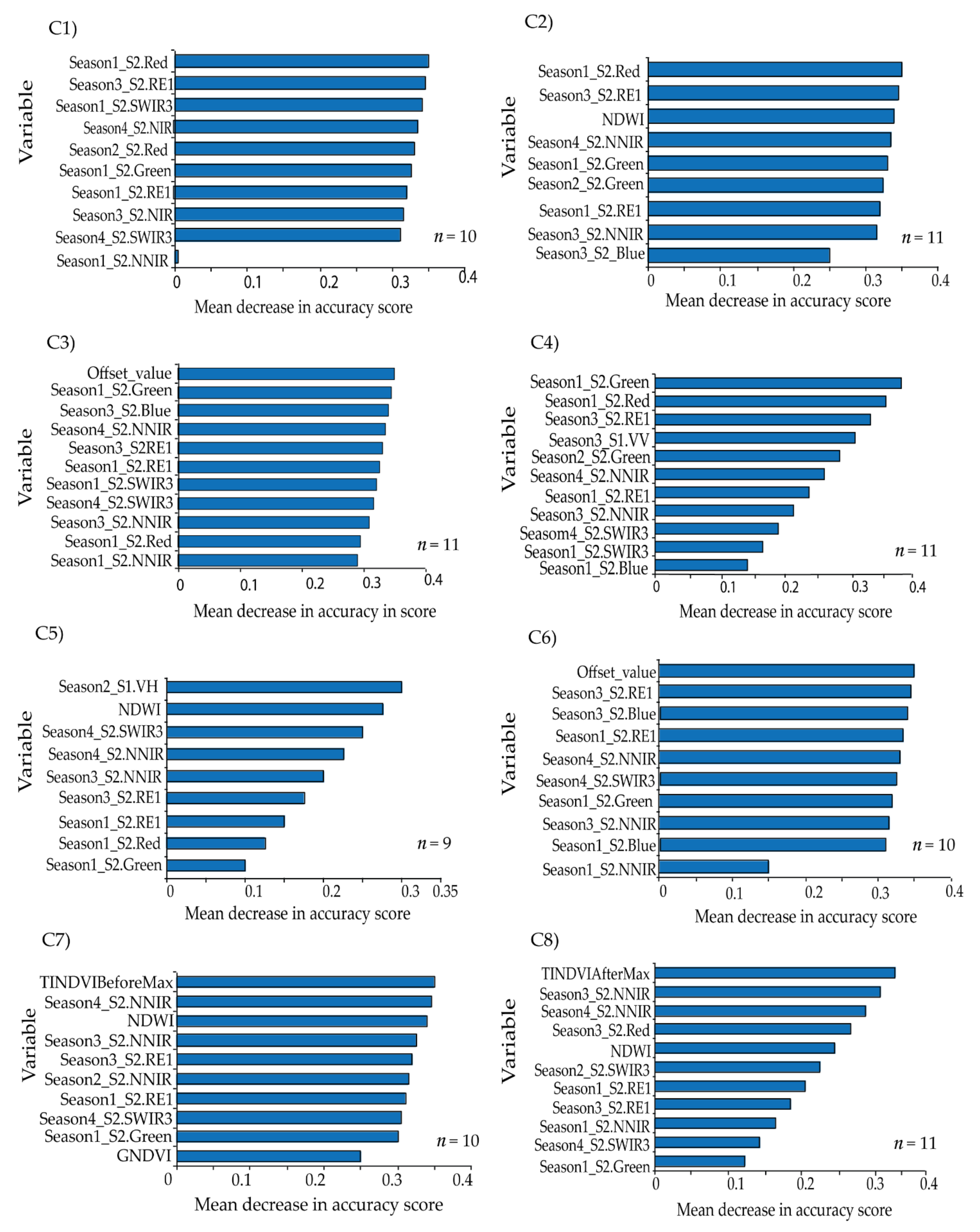
The overall classification accuracies achieved were above 90% for all the tested scenarios, whereas Kappa ranged from 0.89 to 0.93 across the tested scenarios. This could be attributed to the use of the multi-date imageries and the selection of the most relevant variables using GRRF. GRRF has the capability of reducing multidimensionality and the expected multicollinearity by maintaining the most relevant variables for the analysis. Therefore, the most important variables selected across the eight scenarios were S2 bands of NNIR, SWIR3, RE1, Red, Green, and Blue; VI bands of NDWI and GNDVI, VP bands of Offset_value, TINDVIBeforeMax and TINDVIAfterMax; and S1 bands of VV and VH. The selection of NNIR can be explained by the narrowness of the NIR waveband region at 865 nm, which is known to be less contaminated by water vapor and represents the NIR plateau for vegetation while also being sensitive to some soil chemical properties [
51]. Moreover, the reflectance in the SWIR3 band is sensitive to the internal leaf structure [
85], whereas the red edge band (RE1), in addition to visible bands of red, green, and blue, are also sensitive to vegetation chlorophyll and other biochemical contents [
86,
87,
88]. The dominance of S2 bands in the GRRF-selected variables in all the tested scenarios confirmed the strength and importance of the raw S2 bands in discriminating vegetation land use/cover features [
89].
Furthermore, the VIs were also useful in specific crop type identification [
90] by measuring the photosynthetic size of specific plant canopies, which could improve the individual cropping patterns classification [
91]. The selection of the VIs, i.e., NDWI and GNDVI, can be attributed to the sensitivity of the NIR band in the NDWI to leaf internal structure and leaf dry matter content, whereas the SWIR3 band in the index is sensitive to the vegetation water content and the spongy mesophyll structure in vegetation canopies, reflecting biochemical metrics of vegetation [
58]. On the other hand, the GNDVI is more sensitive to the chlorophyll content of the plant because it constitutes the green channel in lieu of the red band [
63]. Challenges in mapping cropping patterns using VIs could arise due to multicollinearity in the VIs [
30], but this was catered for by using the GRRF algorithm to select the most important VIs in the respective scenarios.
Time-series phenology variables have been found to be the best at differentiating temporal and spectral variability of crop growth [
92]. The selection of the VP variables of Offset_value, TINDVIBeforeMax, and TINDVIAfterMax can be explained by the sensitivity of Offset_value to land use and land cover differences, whereas TINDVIBeforeMax and TINDVIAfterMax describe the pre- and post-anthesis stages, respectively, which can differ among different plants [
66]. The specific phenological states of the different crops and vegetation were not physically observed in the present because the aim was to utilize the uniqueness of the phenological profiles in a broader sense, as a function of cropping patterns to discriminate among them. Earlier studies, such as that of Makori et al. [
93], have utilized VP variables in a landscape of mixed vegetation species that included perennial and annual plants to model some features of interest. It is speculated that a phenological profile of a mixed cropping pattern (e.g., avocado and maize) would considerably differ from that of a monocropping pattern (e.g., avocado). However, the mixed cropping pattern classes were mapped with the least individual (F1-score) accuracy, which could be due to the confounding VP variables of, for instance, a mixture of perennial (avocado) and annual (maize) cropping pattern classes. This is one of the limitations of the present study; therefore, future studies could develop advanced methods for differentiating among the VP variables across mixed vegetation classes.
Regarding the contribution of S1 backscatter data, the sensitivity of VH polarization to vegetation could have influenced the performance of the most accurate classification scenario (C5), i.e., GRFF-selected variables in S2 bands, VIs, and S1, with an overall accuracy of 94.33% and Kappa coefficient of 0.93. Interestingly, the sensitivity of VV polarization to soil moisture [
94] could have rendered the underperformance of scenario C4 i.e., GRFF-selected variables in S2 bands and S1 compared to all other scenarios. This is contrary to other previous studies, such as that of Tricht et al. [
95], which reported an improvement in classification accuracy when combining S2 and S1 datasets.
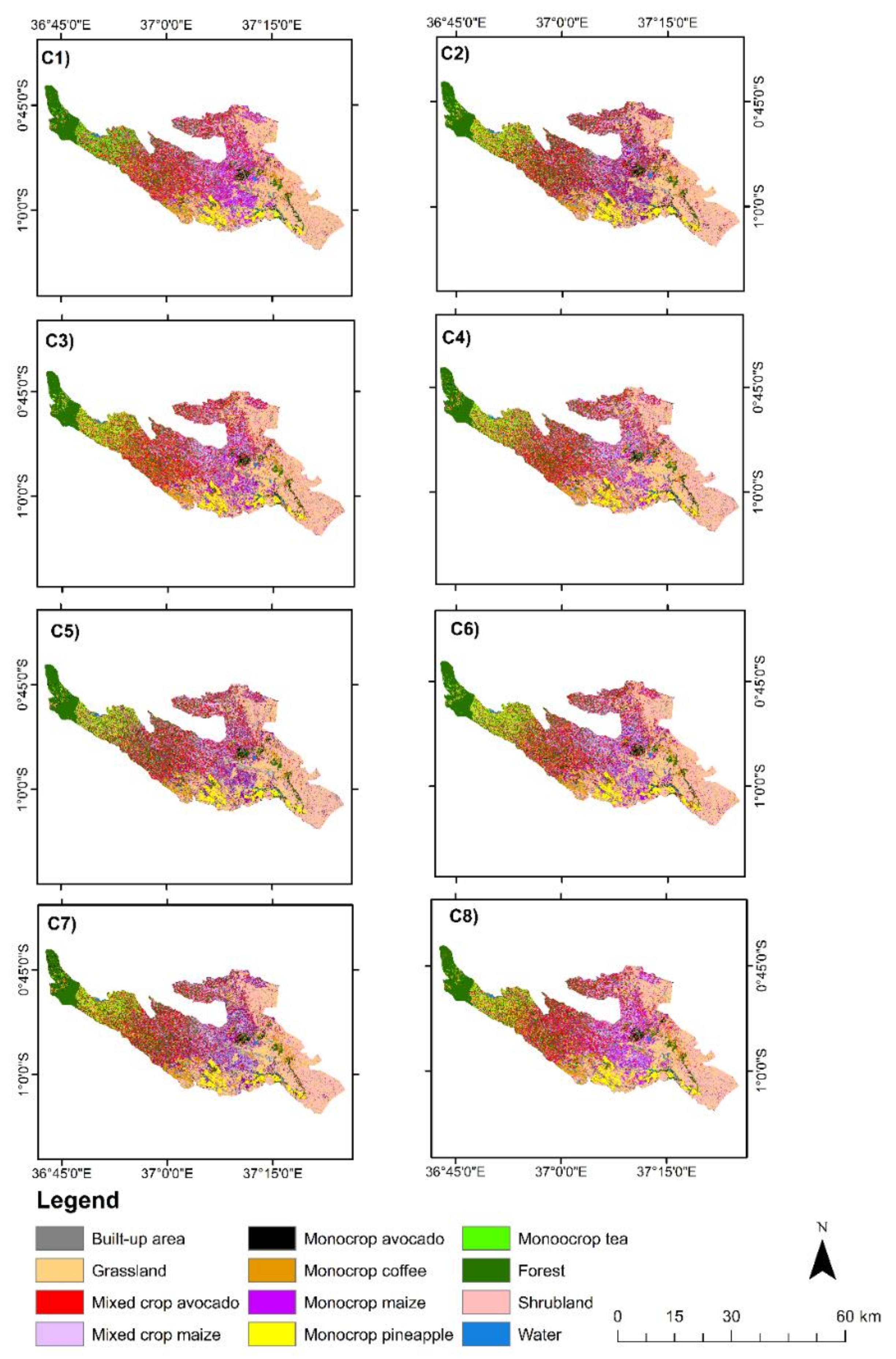
Cropping patterns including monocrop avocado, mixed crop avocado, monocrop maize, mixed crop maize, monocrop tea, monocrop pineapple, and monocrop coffee, grown in a highly complex, heterogeneous, and dynamic agro-natural setup in Murang’a County, Kenya, were accurately mapped. In the field, it was observed that most of the mixed crop fields were small-scale avocado and small-scale maize fields that were mixed with other crops such as common bean, banana, and macadamia. It is speculated that the farmers intend to maximize the profit of their land by mixing the crops in the same piece of land, particularly due to the uncertainty in rainfall trend and invasion of insect pests, such as the fall armyworm, which could damage their maize crop [
96]. High class-wise UA, PA, and F1-scores were especially observed in the mapping of monocropping patterns of avocado, coffee, tea, and pineapple. Presumably, this could be due to the high spectral uniformity of the monocropping patterns, which also resulted in less intra-class variability. However, mixed crop avocado and mixed crop maize cropping pattern classes had lower F1-scores ranging from 80% to 91% compared to other cropping pattern classes in the present study. This can be explained by high inter-class variability associated with the different vegetation compositions, including non-croplands such as forest, grasslands, and shrublands; hence, a high rate of misclassification was found within the farms [
27]. In terms of acreage estimation from the most accurate scenario (C5), i.e., GRRF-selected variables in S2 bands, VIs, and S1, monocrop maize had the greatest acreage (15,052.61 ha) followed by mixed crop avocado (14,404.16 ha). This could be explained by the fact that maize is a staple crop in Kenya, and avocado farming is gaining popularity among small-scale farmers in Murang’a County due to its increasing export value in Kenya [
49,
97]. Overall, all the classes in the tested scenarios provided good agreement when compared with high spatial resolution Google Earth imagery. The insignificant statistical differences from McNemar’s test between the tested classification scenarios could be explained by the use of related samples (i.e., the same training data used in the classification of all the scenarios) [
45]. The use of the same training data in all the classification scenarios was necessary for this study to enable the unbiased comparison of the performance of the classification scenarios. Another reason for the insignificant differences among the eight classification scenarios could also be due to the fact that all the scenarios included S2 bands.
In summary, this study’s approach for mapping cropping patterns performed significantly better than most pre-existing approaches for classifying land use/land cover and cropping systems in agro-ecological landscapes. For instance, Kyalo et al. [
10], who mapped maize-based cropping systems in a study area in Kenya using bio-temporal RapidEye bands and VIs, achieved about 85% mapping accuracy. Ochungo et al. [
89], who fused single-date S1 and S2 datasets to map different land use/land cover features in Kenya, obtained an OA of 86%. Because the readily available S2 and S1 datasets, and a semi-automated protocol to map smallholder farmer cropping patterns were utilized, it is expected that these results are repeatable, and could be used to promptly provide feedback to different stakeholders, including farmers themselves, when high spatial resolution imagery is not readily available due to cost implications.
6. Conclusions
This study investigated the synergetic advantage of integrating multi-date freely available medium spatial resolution S2 bands, their VIs and VP derivatives, and S1 backscatter data for mapping cropping patterns using GRRF and RF machine learning algorithms for relevant variable selection and cropping pattern classification, respectively, in an agro-natural heterogeneous landscape in Kenya. The study also used the area under class method for assessing classification accuracy, which provided an opportunity to obtain an insight into the acreage of the various cropping patterns. The best performing classification scenario was GRRF-selected variables of S2, VIs, and S1 combination with OA = 94.33% and Kappa = 0.93. The selected variables in this scenario were VH, NDWI, SWIR, NNIR, RE1, Red, and Green bands. In general, the mixed cropping patterns of avocado and maize had the lowest accuracies compared to the monocropping patterns of tea, pineapple, maize, avocado, and coffee. Future studies could examine the use of more advanced algorithms such as artificial intelligence to improve the mapping accuracy of mixed cropping pattern classes.
Overall, this study’s approach could be extended to other locations of similar agro-ecological conditions. Moreover, the study’s output could also be used as input parameters for the prediction of the abundance and spread of crop insect pests, diseases, and pollinators.