Evaluation of a Novel Molecular Marker Associated with the Tan Spot Disease Response in Wheat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Marker Selection
2.3. DNA Extraction and Genotypic Analysis
2.4. Sequence Analysis
2.5. Phenotypic and Statistical Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oliver, R.; Tan, K.-C.; Moffat, C. Necrotrophic pathogens of wheat. In Encyclopedia of Food Grains, 2nd ed.; Colin, W., Harold, C., Koushik, S., Jon, F., Eds.; Academic Press: Amsterdam, The Netherlands, 2016; pp. 273–278. ISBN 9780123947864. [Google Scholar] [CrossRef]
- Tan, K.-C.; Oliver, R.P.; Solomon, P.S.; Moffat, C.S. Proteinaceous necrotrophic effectors in fungal virulence. Funct. Plant. Biol. 2010, 37, 907–912. [Google Scholar] [CrossRef] [Green Version]
- See, P.T.; Marathamuthu, K.A.; Iagallo, E.M.; Oliver, R.P.; Moffat, C.S. Evaluating the importance of the tan spot ToxA–Tsn1 interaction in Australian wheat varieties. Plant. Pathol. 2018, 67, 1066–1075. [Google Scholar] [CrossRef] [Green Version]
- Manning, V.A.; Ciuffetti, L.M. Necrotrophic Effector Epistasis in the Pyrenophora tritici-repentis-Wheat Interaction. PLoS ONE 2015, 10, e0123548. [Google Scholar] [CrossRef] [Green Version]
- See, P.T.; Chen, K.; Marathamuthu, K.A.; Wood, B.; Schultz, N.; Shankar, M.; Moffat, C.S. Virulence assessment of an isolate collection of the tan spot pathogen Pyrenophora tritici-repentis in Australian wheat cultivars. Plant. Pathol. under review.
- Moffat, C.S.; Santana, F.M. Diseases Affecting Wheat: Tan Spot. In Integrated Disease Management of Wheat and Barley; Oliver, R., Ed.; Burleigh Dodds Science Publishing: Cambridge, UK, 2018. [Google Scholar]
- See, P.T.; Iagllo, E.M.; Oliver, R.P.; Moffat, C.S. Heterologous expression of the Pyrenophora tritici-repentis effector prpteins ToxA and ToxB, and the prevalence of effector sensitivity in Australian cereal crops. Front. Microbiol. 2019, 10, 182. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant. Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz, V.M.V.; Kilian, A.; Dierig, D.A. Development of DArT Marker Platforms and Genetic Diversity Assessment of the U.S. Collection of the New Oilseed Crop Lesquerella and Related Species. PLoS ONE 2013, 8, e64062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [Green Version]
- Bernardo, A.N.; Bradbury, P.J.; Ma, H.; Hu, S.; Bowden, R.L.; Buckler, E.S.; Bai, G. Discovery and mapping of single feature polymorphisms in wheat using Affymetrix arrays. BMC Genom. 2009, 10, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheben, A.; Verpaalen, B.; Lawley, C.T.; Chan, C.-K.K.; Bayer, P.E.; Batley, J.; Edwards, D. CropSNPdb: A database of SNP array data for Brassica crops and hexaploid bread wheat. Plant. J. 2019, 98, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Andorf, C.M.; Cannon, E.K.; Portwood, J.L., 2nd; Gardiner, J.M.; Harper, L.C.; Schaeffer, M.L.; Braun, B.L.; Campbell, D.A.; Vinnakota, A.G.; Sribalusu, V.V.; et al. MaizeGDB update: New tools, data and interface for the maize model organism database. Nucleic Acids Res. 2016, 44, D1195–D1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganal, M.W.; Altmann, T.; Röder, M.S. SNP identification in crop plants. Curr. Opin. Plant. Biol. 2009, 12, 211–217. [Google Scholar] [CrossRef]
- Grewal, S.; Hubbart-Edwards, S.; Yang, C.; Devi, U.; Baker, L.; Heath, J.; Ashling, S.; Scholefield, D.; Howells, C.; Yarde, J.; et al. Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific KASP genotyping assays. Plant. Biotechnol. J. 2020, 18, 743–755. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Yu, J.; Liao, B.; Cisse, N.; Zhang, X. Development of Highly Informative Genome-Wide Single Sequence Repeat Markers for Breeding Applications in Sesame and Construction of a Web Resource: SisatBase. Front. Plant. Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Deng, P.; Wang, M.; Feng, K.; Cui, L.; Tong, W.; Song, W.; Nie, X. Genome-wide characterization of microsatellites in Triticeae species: Abundance, distribution and evolution. Sci. Rep. 2016, 6, 32224. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, L.; Xu, Y.; Chen, C.; Rong, T.; Ali, F.; Zhou, S.; Wu, F.; Liu, Y.; Wang, J.; et al. Development and characterization of simple sequence repeat markers providing genome-wide coverage and high resolution in maize. DNA Res. 2013, 20, 497–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blake, V.C.; Woodhouse, M.R.; Lazo, G.R.; Odell, S.G.; Wight, C.P.; Tinker, N.A.; Wang, Y.; Gu, Y.Q.; Birkett, C.L.; Jannink, J.-L.; et al. GrainGenes: Centralized small grain resources and digital platform for geneticists and breeders. Database 2019, 2019. [Google Scholar] [CrossRef]
- Jaiswal, S.; Sheoran, S.; Arora, V.; Angadi, U.B.; Iquebal, M.A.; Raghav, N.; Aneja, B.; Kumar, D.; Singh, R.; Sharma, P.; et al. Putative Microsatellite DNA Marker-Based Wheat Genomic Resource for Varietal Improvement and Management. Front. Plant. Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.J.; Shi, J.R.; Singh, S.; Fickus, E.W.; Costa, J.M.; Lewis, J.; Gill, B.S.; Ward, R.; Cregan, P.B. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 2005, 110, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Choudhury, D.R.; Singh, A.K.; Kumar, S.; Srinivasan, K.; Tyagi, R.K.; Singh, N.K.; Singh, R. Comparison of SSR and SNP markers in estimation of genetic diversity and population structure of Indian rice varieties. PLoS ONE 2013, 8, e84136. [Google Scholar] [CrossRef] [Green Version]
- Chao, S.; Zhang, W.; Akhunov, E.; Sherman, J.; Ma, Y.; Luo, M.-C.; Dubcovsky, J. Analysis of gene-derived SNP marker polymorphism in US wheat (Triticum aestivum L.) cultivars. Mol. Breed. 2009, 23, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Russell, J.; Booth, A.; Fuller, J.; Harrower, B.; Hedley, P.; Machray, G.; Powell, W. A comparison of sequence-based polymorphism and haplotype content in transcribed and anonymous regions of the barley genome. Genome 2004, 47, 389–398. [Google Scholar] [CrossRef]
- Peleg, Z.; Fahima, T.; Krugman, T.; Abbo, S.; Yakir, D.; Korol, A.B.; Saranga, Y. Genomic dissection of drought resistance in durum wheat x wild emmer wheat recombinant inbreed line population. Plant. Cell Environ. 2009, 32, 758–779. [Google Scholar] [CrossRef]
- Pinto, R.S.; Reynolds, M.P.; Mathews, K.L.; McIntyre, C.L.; Olivares-Villegas, J.J.; Chapman, S.C. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Appl. Genet. 2010, 121, 1001–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondal, S.; Mason, R.E.; Huggins, T.; Hays, D.B. QTL on wheat (Triticum aestivum L.) chromosomes 1B, 3D and 5A are associated with constitutive production of leaf cuticular wax and may contribute to lower leaf temperatures under heat stress. Euphytica 2015, 201, 123–130. [Google Scholar] [CrossRef]
- Awlachew, Z.T.; Singh, R.; Kaur, S.; Bains, N.S.; Chhuneja, P. Transfer and mapping of the heat tolerance component traits of Aegilops speltoides in tetraploid wheat Triticum durum. Mol. Breed. 2016, 36, 78. [Google Scholar] [CrossRef]
- Wiśniewska, H.; Surma, M.; Krystkowiak, K.; Adamski, T.; Kuczyńska, A.; Ogrodowicz, P.; Mikołajczak, K.; Belter, J.; Majka, M.; Kaczmarek, Z.; et al. Simultaneous selection for yield-related traits and susceptibility to Fusarium head blight in spring wheat RIL population. Breed. Sci 2016, 66, 281–292. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Bonnett, D.; Ellis, M.; He, X.; Heslot, N.; Dreisigacker, S.; Gao, C.; Singh, P. Characterization of Fusarium head blight resistance in a CIMMYT synthetic-derived bread wheat line. Euphytica 2016, 208, 367–375. [Google Scholar] [CrossRef]
- Miranda, L.M.; Murphy, J.P.; Marshall, D.; Leath, S. Pm34: A new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2006, 113, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Friesen, T.L.; Simons, K.J.; Xu, S.S.; Faris, J.D. Development, identification, and validation of markers for marker-assisted selection against the Stagonospora nodorum toxin sensitivity genes Tsn1 and Snn2 in wheat. Mol. Breed. 2009, 23, 35–49. [Google Scholar] [CrossRef]
- Kumar, S.; Röder, M.S.; Tripathi, S.B.; Kumar, S.; Chand, R.; Joshi, A.K.; Kumar, U. Mendelization and fine mapping of a bread wheat spot blotch disease resistance QTL. Mol. Breed. 2015, 35, 218. [Google Scholar] [CrossRef]
- Singh, V.; Singh, G.; Chaudhury, A.; Ojha, A.; Tyagi, B.S.; Chowdhary, A.K.; Sheoran, S. Phenotyping at hot spots and tagging of QTLs conferring spot blotch resistance in bread wheat. Mol. Biol Rep. 2016, 43, 1293–1303. [Google Scholar] [CrossRef]
- Han, B.; Wang, C.; Tang, Z.; Ren, Y.; Li, Y.; Zhang, D.; Dong, Y.; Zhao, X. Genome-Wide Analysis of Microsatellite Markers Based on Sequenced Database in Chinese Spring Wheat (Triticum aestivum L.). PLoS ONE 2015, 10, e0141540. [Google Scholar] [CrossRef] [Green Version]
- Nazareno, A.G.; dos Reis, M.S. The same but different: Monomorphic microsatellite markers as a new tool for genetic analysis. Am. J. Bot 2011, 98, e265-267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manifesto, M.; Schlatter, R.A.; Hopp, H.; Suárez, Y.E.; Dubcovsky, J. Quantitative Evaluation of Genetic Diversity in Wheat Germplasm Using Molecular Markers. Crop Sci. 2001, 41, 682–690. [Google Scholar] [CrossRef] [Green Version]
- Lamari, L.; Bernier, C.C. Evaluation of wheat lines and varieties to tan spot Pyrenophora triciti-repentis based on lesion type. Can. J. Plant. Pathol. 1989, 32, 4–10. [Google Scholar] [CrossRef]
- Moffat, C.S.; See, P.T.; Oliver, R.P. Generation of a ToxA knockout strain of the wheat tan spot pathogen Pyrenophora tritici-repentis. Mol. Plant. Pathol. 2014, 15, 918–926. [Google Scholar] [CrossRef]
- Simons, K.; Abate, Z.; Chao, S.; Zhang, W.; Rouse, M.; Jin, Y.; Elias, E.; Dubcovsky, J. Genetic mapping of stem rust resistance gene Sr13 in tetraploid wheat (Triticum turgidum ssp. durum L.). Theor. Appl. Genet. 2011, 122, 649–658. [Google Scholar] [CrossRef] [Green Version]
- Periyannan, S.K.; Qamar, Z.U.; Bansal, U.K.; Bariana, H.S. Development and validation of molecular markers linked with stem rust resistance gene Sr13 in durum wheat. Crop. Pasture Sci. 2014, 65, 74–79, 76. [Google Scholar] [CrossRef]
- A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [CrossRef]
- Friesen, T.L.; Ali, S.; Kianian, S.; Francl, L.J.; Rasmussen, J.B. Role of host sensitivity to ptr ToxA in development of tan spot of wheat. Phytopathology 2003, 93, 397–401. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.G.; Xu, S.S.; Faris, J.D.; Nevo, E.; Friesen, T.L. Seedling Resistance to Tan Spot and Stagonospora nodorum Leaf Blotch in Wild Emmer Wheat (Triticum dicoccoides). Plant. Dis. 2008, 92, 1229–1236. [Google Scholar] [CrossRef] [Green Version]
- Faris, J.D.; Abeysekara, N.S.; McClean, P.E.; Xu, S.S.; Friesen, T.L. Tan spot susceptibility governed by the Tsn1 locus and race-nonspecific resistance quantitative trait loci in a population derived from the wheat lines Salamouni and Katepwa. Mol. Breed. 2012, 30, 1669–1678. [Google Scholar] [CrossRef]
- Shankar, M.; Mather, D.; Jorgensen, D.; Golzar, H.; Chalmers, K.; Hollaway, G.; McLean, M.; Neate, S.; Loughman, R. Germplasm Enhancement for Resistance to Pyrenophora Tritici-Repentis in Wheat. In Advances in Wheat Genetics: From Genome to Field, Proceedings of the 12th International Wheat Genetics Symposium; Ogihara, Y., Takumi, S., Handa, H., Eds.; Springer: Tokyo, Japan, 2015; pp. 193–199. [Google Scholar]
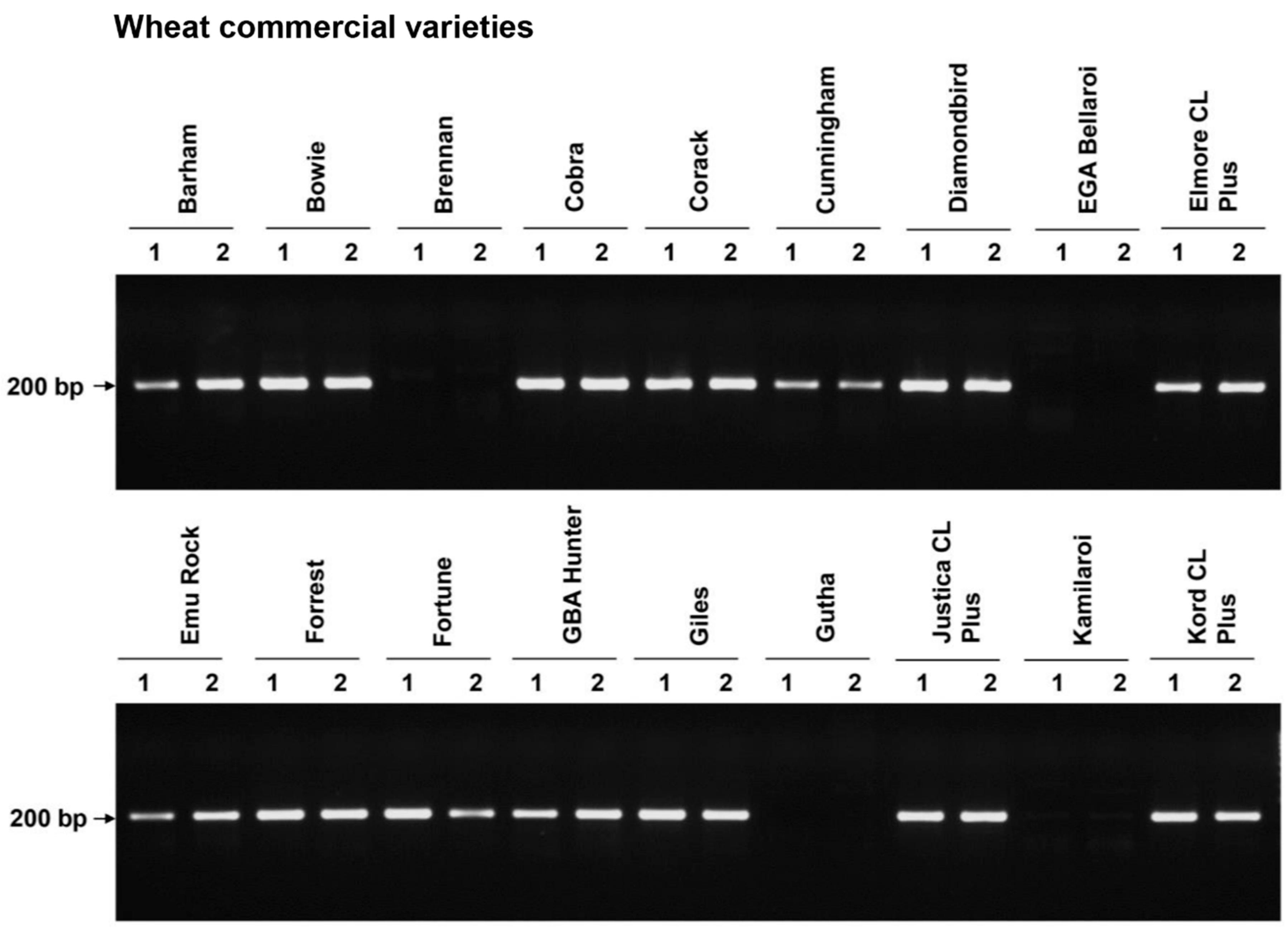
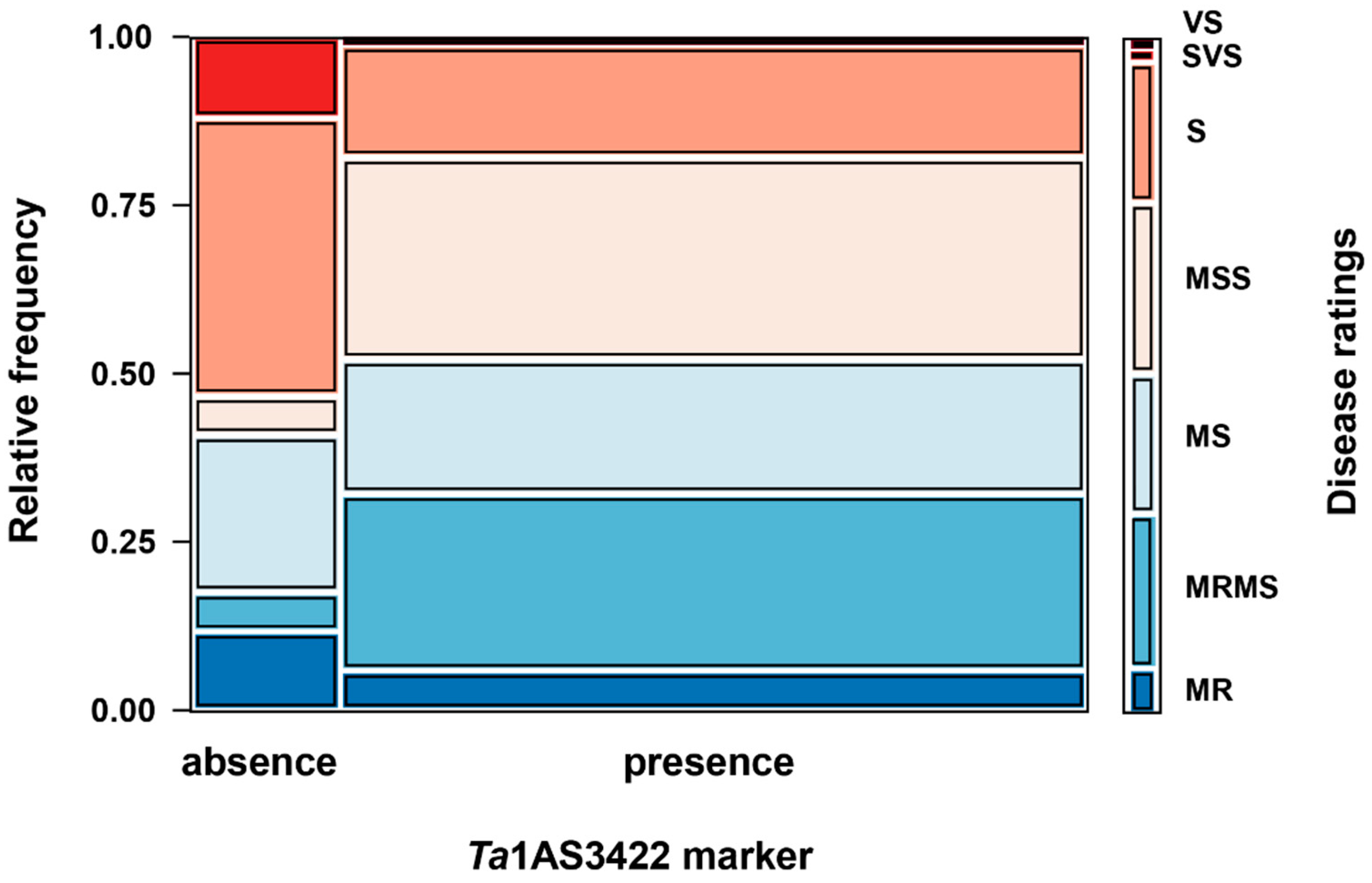
| Genotype | Number of Varieties | Disease Scores (Mean ± SD) |
|---|---|---|
| Seedling growth stage | ||
| Presence of Ta1AS3422 marker | 30 | 2.8 ± 0.7 * |
| Absence of Ta1AS3422 marker | 10 | 3.6 ± 0.8 |
| Tillering growth stage | ||
| Presence of Ta1AS3422 marker | 9 | 2.4 ± 0.4 * |
| Absence of Ta1AS3422 marker | 6 | 3.6 ± 0.8 |
| Variety | Disease Rating | Presence/Absence of Ta1AS3422 Marker |
|---|---|---|
| Hexaploid wheat varieties | ||
| Corack | MR | + |
| Einstein | MR | + |
| Hydra * | MR | + |
| Whistler | MR | + |
| Wyalkatchem *,+ | MR | + |
| Adagio SF | MRMS | + |
| Arrow LRPB | MRMS | + |
| Beaufort LRPB | MRMS | + |
| Buchanan | MRMS | + |
| Cobra LRPB * | MRMS | + |
| Cosmick * | MRMS | + |
| DS Pascal | MRMS | + |
| EGA Bonnie Rock *,+ | MRMS | + |
| Emu Rock | MRMS | + |
| Forrest | MRMS | + |
| GBA Hunter | MRMS | + |
| Impress CL Plus * | MRMS | + |
| King Rock * | MRMS | + |
| Kittyhawk LRPB | MRMS | + |
| Mace *,+ | MRMS | + |
| Marombi | MRMS | + |
| Ninja | MRMS | + |
| Scepter | MRMS | + |
| Sunlamb | MRMS | + |
| Tenfour | MRMS | + |
| Yenda | MRMS | + |
| Zen *,+ | MRMS | + |
| Carnamah * | MS | + |
| Cunningham | MS | + |
| Dart * | MS | + |
| Fortune | MS | + |
| Kennedy * | MS | + |
| Lancer *,+ | MS | + |
| Mansfield | MS | + |
| Mitch | MS | + |
| Naparoo | MS | + |
| Scenario SF | MS | + |
| SQP Revenue | MS | + |
| Strzelecki | MS | + |
| Sunbri | MS | + |
| Sunsoft 98 | MS | + |
| Westonia * | MS | + |
| Wylah | MS | + |
| Annuello * | MSS | + |
| Barham | MSS | + |
| Calingiri * | MSS | + |
| Catalina LRPB | MSS | + |
| Cobalt | MSS | + |
| Diamondbird | MSS | + |
| Estoc * | MSS | + |
| Gauntlet LRPB * | MSS | + |
| Gazelle LRPB *,+ | MSS | + |
| GBA Sapphire | MSS | + |
| Giles | MSS | + |
| Impala LRPB * | MSS | + |
| Kord CL Plus | MSS | + |
| Livingston | MSS | + |
| Merinda | MSS | + |
| QALBIS | MSS | + |
| Shield * | MSS | + |
| Spitfire *,+ | MSS | + |
| Sunguard *,+ | MSS | + |
| Sunstate | MSS | + |
| Suntop * | MSS | + |
| Trojan LRPB | MSS | + |
| Viking LRPB | MSS | + |
| Waagan | MSS | + |
| Wallup * | MSS | + |
| Bowie | S | + |
| DS Darwin | S | + |
| EGA Gregory * | S | + |
| EGA Wylie * | S | + |
| Elmore CL Plus | S | + |
| Grenade CL plus * | S | + |
| Jade | S | + |
| Justica CL Plus | S | + |
| Kellalac | S | + |
| Lorikeet | S | + |
| Merlin *,+ | S | + |
| Peake | S | + |
| Rosella | S | + |
| Sunbrook | S | + |
| Axe * | VS | + |
| Magenta *,+ | MR | - |
| Tennant | MR | - |
| Sunvex | MRMS | - |
| Brennan | MS | - |
| Perenjori | MS | - |
| Petrel | MS | - |
| Supreme *,+ | MS | - |
| Arrino | MSS | - |
| EGA Eagle Rock *,+ | S | - |
| Frame * | S | - |
| Gutha | S | - |
| Harper * | S | - |
| Machete *,+ | S | - |
| Scout * | S | - |
| Stiletto * | S | - |
| Phantom LRPB *,+ | SVS | - |
| Yitpi *,+ | SVS | - |
| Tetraploid wheat varieties | ||
| DBA Aurora | MR | - |
| DBA Lillaroi | MRMS | - |
| Dural | N/A | - |
| Durati | N/A | - |
| EGA Bellaroi | MR | - |
| Jandaroi | MRMS | - |
| Kamilaroi | MRMS | - |
| Tamaroi | MRMS | - |
| WID802 | N/A | - |
| Wollaroi | MRMS | - |
| Yallaroi | MRMS | - |
| Yawa | MR | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
See, P.T.; Moffat, C.S. Evaluation of a Novel Molecular Marker Associated with the Tan Spot Disease Response in Wheat. Agriculture 2021, 11, 513. https://doi.org/10.3390/agriculture11060513
See PT, Moffat CS. Evaluation of a Novel Molecular Marker Associated with the Tan Spot Disease Response in Wheat. Agriculture. 2021; 11(6):513. https://doi.org/10.3390/agriculture11060513
Chicago/Turabian StyleSee, Pao Theen, and Caroline S. Moffat. 2021. "Evaluation of a Novel Molecular Marker Associated with the Tan Spot Disease Response in Wheat" Agriculture 11, no. 6: 513. https://doi.org/10.3390/agriculture11060513
APA StyleSee, P. T., & Moffat, C. S. (2021). Evaluation of a Novel Molecular Marker Associated with the Tan Spot Disease Response in Wheat. Agriculture, 11(6), 513. https://doi.org/10.3390/agriculture11060513






