Bacillus-Based Probiotic Treatment Modified Bacteriobiome Diversity in Duck Feces
Abstract
1. Introduction
2. Materials and Methods
2.1. Duck Breed and Experimental Design
2.2. Sample Collection
2.3. Extraction of Total Nucleic Acid from Feces
2.4. 16S rRNA Gene Amplification and Sequencing
2.5. Bioinformatic and Statistical Analyses
3. Results
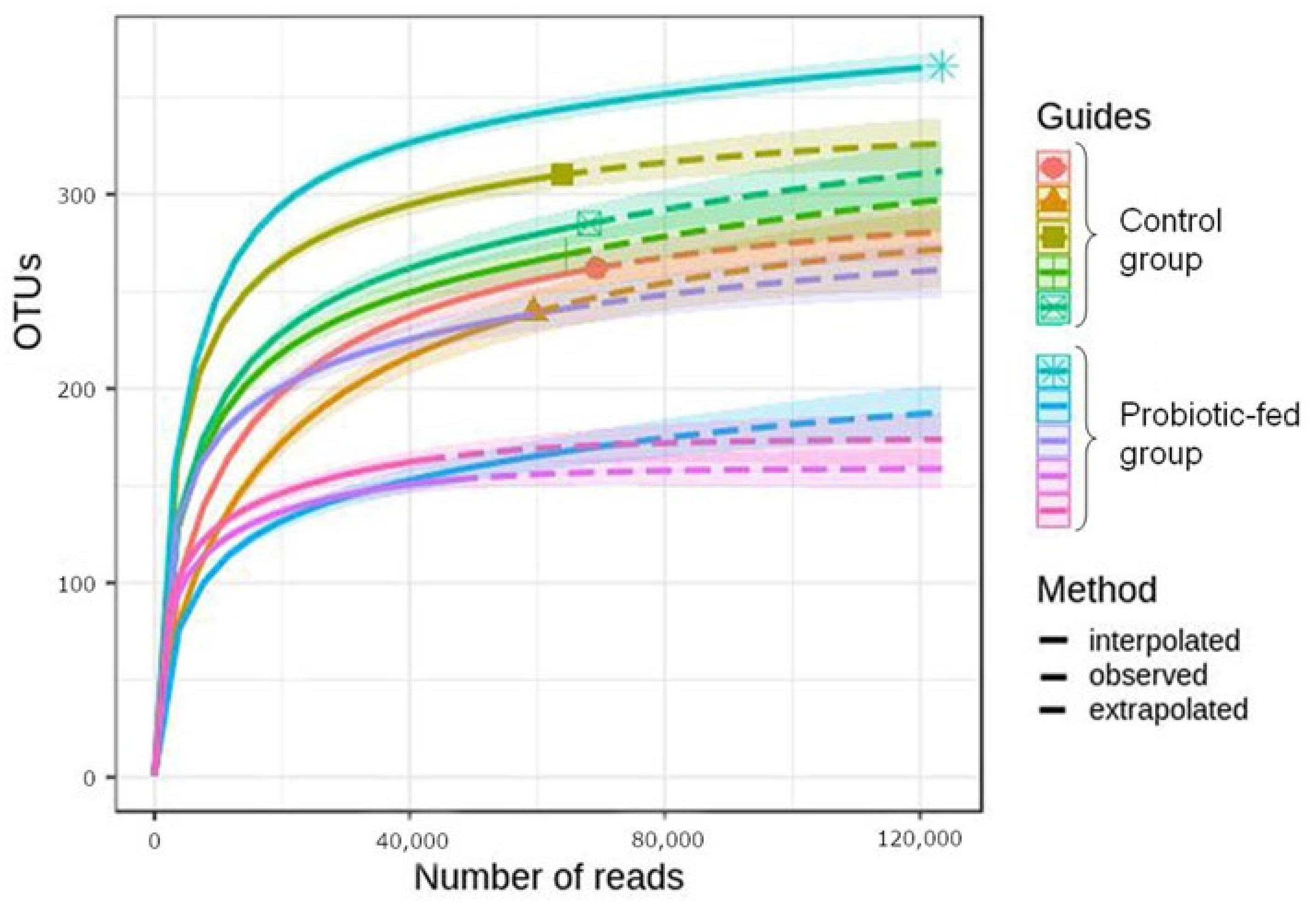
3.1. Taxonomic Richness and Structure of Duck Fecal Bacterial Assemblages
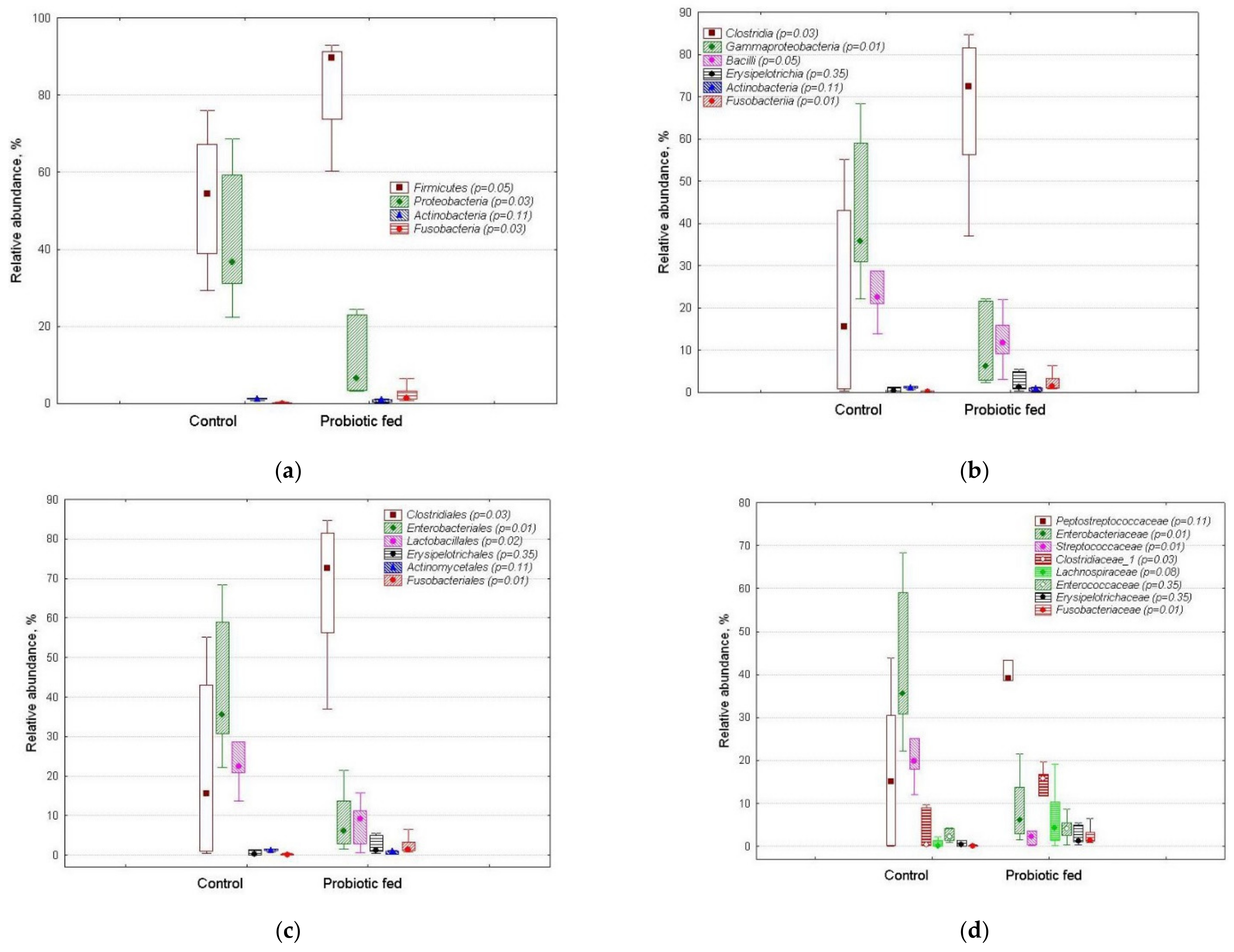
3.2. OTUs’ Relative Abundance in Duck Fecal Bacterial Assemblages
3.3. Biodiversity Indices of the Duck Fecal Bacterial Assemblages
3.4. Production Performance of Ducks
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. 2021. Available online: http://www.fao.org/faostat/en/#data/QA (accessed on 20 April 2021).
- Oviedo-Rondón, E.O. Holistic view of intestinal health in poultry. Anim. Feed Sci. Technol. 2019, 250, 1–8. [Google Scholar] [CrossRef]
- Patterson, J.A.; Burkholder, K.M. Application of prebiotics and probiotics in poultry production. Poult. Sci. 2003, 82, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Sorokulova, I.B. A comparative study of the biological properties of Biosporin and other commercial Bacillus-based preparations. Mikrobiol. Zhournal 1997, 59, 43–49. (In Russian) [Google Scholar]
- Mingmongkolchai, S.; Panbangred, W. Bacillus probiotics: An alternative to antibiotics for livestock production. J. Appl. Microbiol. 2018, 124, 1334–1346. [Google Scholar] [CrossRef]
- Kogut, M.H. The effect of microbiome modulation on the intestinal health of poultry. Anim. Feed Sci. Technol. 2019, 250, 32–40. [Google Scholar] [CrossRef]
- Franciosini, M.P.; Costarelli, S.; Cobellis, G.; Trabalza-Marinucci, M. Effects of dietary Lactobacillus acidophilus and Bacillus subtilis on laying performance, egg quality, blood biochemistry and immune response of organic laying hens. J. Anim. Physiol. Anim. Nutr. 2016, 100, 977–987. [Google Scholar] [CrossRef]
- Zamanizadeh, A.; Mirakzehi, M.T.; Agah, M.J.; Saleh, H.; Baranzehi, T. A comparison of two probiotics Aspergillus oryzae and Saccharomyces cerevisiae on productive performance, egg quality, small intestinal morphology, and gene expression in laying Japanese quail. Ital. J. Anim. Sci. 2021, 20, 232–242. [Google Scholar] [CrossRef]
- Wang, H.; Ha, B.D.; Kim, I.H. Effects of probiotics complex supplementation in low nutrient density diet on growth performance, nutrient digestibility, faecal microbial, and faecal noxious gas emission in growing pigs. Ital. J. Anim. Sci. 2020, 20, 163–170. [Google Scholar] [CrossRef]
- Liu, X.; Liu, W.; Deng, Y.; He, C.; Xiao, B.; Guo, S.; Zhou, X.; Tang, S.; Qu, X. Use of encapsulated Bacillus subtilis and essential oils to improve antioxidant and immune status of blood and production and hatching performance of laying hens. Ital. J. Anim. Sci. 2020, 19, 1583–1591. [Google Scholar] [CrossRef]
- Park, I.; Lee, Y.; Goo, D.; Zimmerman, N.P.; Smith, A.H.; Rehberger, T.; Lillehoj, H.S. The effects of dietary Bacillus subtilis supplementation, as an alternative to antibiotics, on growth performance, intestinal immunity, and epithelial barrier integrity in broiler chickens infected with Eimeria maxima. Poult. Sci. 2020, 99, 725–733. [Google Scholar] [CrossRef]
- Liu, X.; Peng, C.; Qu, X.; Guo, S.; Chen, J.F.; He, C.; Zhou, X.; Zhu, S. Effects of Bacillus subtilis C-3102 on production, hatching performance, egg quality, serum antioxidant capacity and immune response of laying breeders. J. Anim. Physiol. Anim. Nutr. 2019, 103, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Hao, G.; Wang, B.; Li, N.; Li, R.; Wei, L.; Chai, T. Dietary Administration of Bacillus subtilis Enhances Growth Performance, Immune Response and Disease Resistance in Cherry Valley Ducks. Front. Microbiol. 2016, 7, 1975. [Google Scholar] [CrossRef] [PubMed]
- Khattab, A.; El Basuini, M.; El-Ratel, I.T.; Fouda, S.F. Dietary probiotics as a strategy for improving growth performance, intestinal efficacy, immunity, and antioxidant capacity of white Pekin ducks fed with different levels of CP. Poult. Sci. 2021, 100, 100898. [Google Scholar] [CrossRef] [PubMed]
- Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the Protection of Animals Used for Scientific Purposes. Available online: https://data.europa.eu/eli/dir/2010/63/oj (accessed on 27 April 2021).
- Probiotic Plus LLC. Available online: https://www.probiotic-plus.ru (accessed on 27 April 2021). (in Russian).
- Federal Service for Proprietary Rights. Patent RU 2,437,563C1, 27 December 2011.
- QIAGEN. Quick-Start Protocol. DNeasy PowerSoil Kit. 2016. Available online: https://www.qiagen.com/au/resources/resourcedetail?id=91cf8513-a8ec-4f45-921e-8938c3a5490c&lang=en (accessed on 27 April 2021).
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Igolkina, A.A.; Grekhov, G.A.; Pershina, E.V.; Samosorova, G.G.; Leunova, V.M.; Semenov, A.N.; Baturina, O.A.; Kabilov, M.R.; Andronov, E.E. Identifying components of mixed and contaminated soil samples by detecting specific signatures of control 16S rRNA libraries. Ecol. Ind. 2018, 94, 446–453. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon reads. bioRxiv 2016. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Fauth, E.; Bernardo, J.; Camara, M.; Resetarits, W.J.; Van Buskirk, J.A.; McCollum, S. Simplifying the Jargon of Community Ecology: A Conceptual Approach. Am. Nat. 1996, 147, 282–286. [Google Scholar] [CrossRef]
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Hughes, J.B.; Hellmann, J.J. The Application of Rarefaction Techniques to Molecular Inventories of Microbial Diversity. Methods Enzymol. 2005, 397, 292–308. [Google Scholar] [CrossRef]
- Vasaï, F.; Ricaud, K.B.; Cauquil, L.; Daniel, P.; Peillod, C.; Gontier, K.; Tizaoui, A.; Bouchez, O.; Combes, S.; Davail, S. Lactobacillus sakei modulates mule duck microbiota in ileum and ceca during overfeeding. Poult. Sci. 2014, 93, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Best, A.A.; Porter, A.L.; Fraley, S.M.; Fraley, G.S. Characterization of Gut Microbiome Dynamics in Developing Pekin Ducks and Impact of Management System. Front. Microbiol. 2017, 4, 2125. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, L.; He, M.; Shen, J.; Li, G.; Tao, Z.; Wu, R.; Lu, L. Different rearing conditions alter gut microbiota composition and host physiology in Shaoxing ducks. Sci. Rep. 2018, 8, 7387. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zheng, M.; Lin, F.; Cheng, X.; Xiao, S.; Chen, S.; Chen, S. Impacts of novel duck reovirus infection on the composition of intestinal microbiota of Muscovy ducklings. Microb. Pathog. 2019, 137, 103764. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lyu, W.; Lu, L.; Shi, X.; Li, N.; Wang, W.; Xiao, Y. Biogeography of microbiome and short-chain fatty acids in the gastrointestinal tract of duck. Poult. Sci. 2020, 99, 4016–4027. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.A.; Berryman, D.E.; Murphy, E.R.; Carroll, R.K.; Busken, J.; List, E.O.; Broach, W.H. Heterogeneity spacers in 16S rDNA primers improve analysis of mouse gut microbiomes via greater nucleotide diversity. Biotechniques 2019, 67, 55–62. [Google Scholar] [CrossRef]
- Pandit, R.J.; Hinsu, A.T.; Patel, N.V.; Koringa, P.G.; Jakhesara, S.J.; Thakkar, J.R.; Shah, T.M.; Limon, G.; Psifidi, A.; Guitian, J.; et al. Microbial diversity and community composition of caecal microbiota in commercial and indigenous Indian chickens determined using 16s rDNA amplicon sequencing. Microbiome 2018, 6, 115. [Google Scholar] [CrossRef]
- Gaastra, W.; Kusters, J.G.; van Duijkeren, E.; Lipman, L.J.A. Escherichia fergusonii. Vet. Microbiol. 2014, 172, 7–12. [Google Scholar] [CrossRef]
- Li, M.; Gu, C.; Zhang, W.; Li, S.; Liu, J.; Qin, C.; Su, J.; Cheng, G.; Hu, X. Isolation and characterization of Streptococcus gallolyticus subsp. pasteurianus causing meningitis in ducks. Vet. Microbiol. 2013, 162, 93036. [Google Scholar] [CrossRef]
- Cheng, M.P.; Domingo, M.C.; Lévesque, S.; Yansouni, C.P. A case report of a deep surgical site infection with Terrisporobacter glycolicus/T. Mayombei and review of the literature. BMC Infect. Dis. 2016, 29, 529. [Google Scholar] [CrossRef]
- Müller, V.; Frerichs, J. Acetogenic Bacteria; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Groher, A.; Weuster-Botz, D. General medium for the autotrophic cultivation of acetogens. Bioprocess Biosyst. Eng. 2016, 39, 1645. [Google Scholar] [CrossRef] [PubMed]
- Kollarcikova, M.; Kubasova, T.; Karasova, D.; Crhanova, M.; Cejkova, D.; Sisak, F.; Rychlik, I. Use of 16S rRNA gene sequencing for prediction of new opportunistic pathogens in chicken ileal and cecal microbiota. Poult. Sci. 2019, 98, 2347–2353. [Google Scholar] [CrossRef]
- Wang, W.; Zheng, S.; Li, L.; Yang, Y.; Liu, Y.; Wang, A.; Sharshov, K.; Li, Y. Comparative metagenomics of the gut microbiota in wild greylag geese (Anser anser) and ruddy shelducks (Tadorna ferruginea). MicrobiologyOpen 2019, 8, e00725. [Google Scholar] [CrossRef]
- Miller, D.A.; Suen, G.; Bruce, D.; Copeland, A.; Cheng, J.F.; Detter, C.; Goodwin, L.A.; Han, C.S.; Hauser, L.J.; Land, M.L.; et al. Complete genome sequence of the cellulose-degrading bacterium Cellulosilyticum lentocellum. J. Bacteriol. 2011, 193, 2357–2358. [Google Scholar] [CrossRef]
- Wachemo, A.C.; Tong, H.Y.; Hairong, Z.X.; Korai, R.M.; Li, X. Continuous dynamics in anaerobic reactor during bioconversion of rice straw: Rate of substance utilization biomethane production and changes in microbial community structure. Sci. Total Environ. 2019, 687, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jia, Y.; Liu, C.; Ding, L.; Xia, Z. RNA-Seq transcriptome analysis of breast muscle in Pekin ducks supplemented with the dietary probiotic Clostridium butyricum. BMC Genom. 2018, 28, 844. [Google Scholar] [CrossRef] [PubMed]
- Even, M.; Davail, S.; Rey, M.; Tavernier, A.; Houssier, M.; Bernadet, M.D.; Gontier, K.; Pascal, G.; Ricaud, K. Probiotics Strains Modulate Gut Microbiota and Lipid Metabolism in Mule Ducks. Open Microbiol. J. 2018, 12, 71–93. [Google Scholar] [CrossRef] [PubMed]
- Wizna, Z.; Abbas, M.H.; Mahata, M.E.; Fauzano, R. Effect of Bacillus amyloliquefaciens as a Probiotic on Growth Performance Parameters of Pitalah Ducks. Int. J. Poult. Sci. 2017, 16, 147–153. [Google Scholar] [CrossRef]
- Li, W.F.; Rajput, I.R.; Xu, X.; Li, Y.L.; Lei, J.; Huang, Q.; Wang, M.Q. Effects of Probiotic (Bacillus subtilis) on Laying Performance, Blood Biochemical Properties and Intestinal Microflora of Shaoxing Ducks. Int. J. Poult. Sci. 2011, 10, 583–589. [Google Scholar] [CrossRef]
- Xing, Y.; Wang, S.; Fan, J.; Oso, A.O.; Kim, S.W.; Xiao, D.; Yang, T.; Liu, G.; Jiang, G.; Li, Z.; et al. Effects of dietary supplementation with lysine-yielding Bacillus subtilis on gut morphology, cecal microflora, and intestinal immune response of Linwu ducks. J. Anim. Sci. 2015, 93, 3449–3457. [Google Scholar] [CrossRef] [PubMed]
| Taxon Level | Taxonomic Attribution | |||
|---|---|---|---|---|
| All OTUs | Dominant a OTUs | |||
| Both Groups | Control Group | Probiotic-Fed Group | ||
| Phylum | 15 | 4 | 3 | 3 |
| Class | 36 | 6 | 4 | 6 |
| Order | 63 | 6 | 4 | 6 |
| Family | 137 | 9 | 4 | 9 |
| Genus | 251 | 12 | 5 | 12 |
| OTU | 567 | 13 | 5 | 13 |
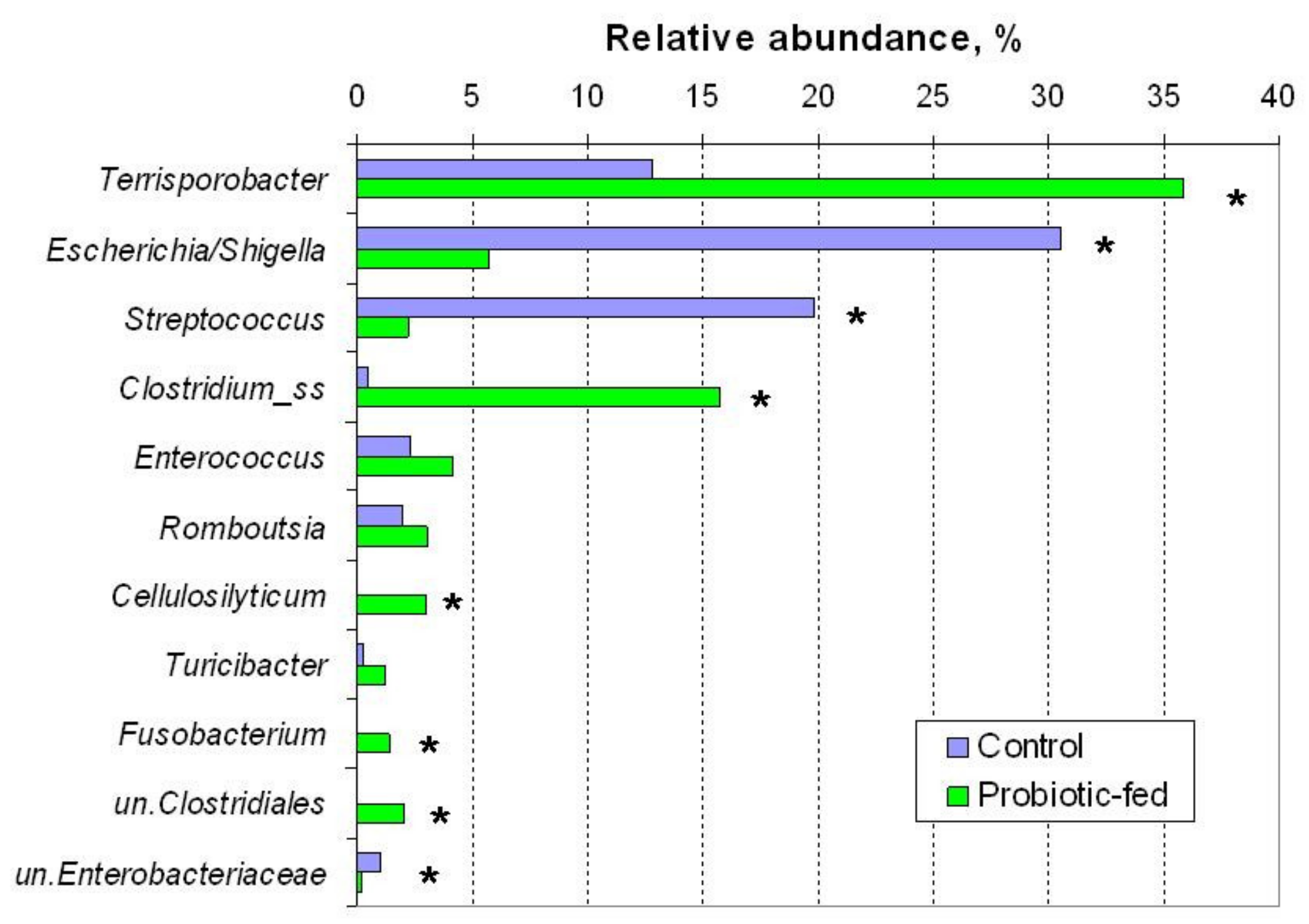
| OTU | Control Group | Probiotic-Fed Group | p-Value | |
|---|---|---|---|---|
| 1 | Escherichia/ Shigella sp. 1 | 30.6 | 5.7 | 0.012 |
| 2 | Terrisporobacter sp. | 12.9 | 36.4 | 0.037 |
| 3 | Streptococcus sp. | 20.0 | 2.2 | 0.012 |
| 6 | Enterococcus cecorum | 1.2 | 3.8 | 0.210 |
| 7 | unc. Clostridiaceae_1 2 | 0.2 | 3.7 | 0.012 |
| 8 | Clostridium_ss3sp. | 0.3 | 1.5 | 0.295 |
| 10 | Cellulosilyticum sp. | 0.1 | 1.3 | 0.094 |
| 11 | Turicibacter sanguinis | 0.3 | 1.2 | 0.403 |
| 12 | unc. Clostridiales | 0.01 | 1.9 | 0.094 |
| 13 | Clostridium_ss3sp. | 0.04 | 1.04 | 0.210 |
| 14 | Fusobacterium sp. | 0.11 | 1.41 | 0.012 |
| 15 | Cellulosilyticum lentocellum | 0.04 | 1.48 | 0.037 |
| 19 | Romboutsia sedimentorum | 2.0 | 3.2 | 0.403 |
| Index | Control Group | Probiotic-Fed Group | p-Value |
|---|---|---|---|
| Total number of identified OTUs | 208 | 114 | 0.095 |
| Dominance (D) | 0.31 | 0.17 | 0.222 |
| Simpson (1-D) | 0.69 | 0.83 | 0.222 |
| Shannon | 1.92 | 2.49 | 0.151 |
| Evenness 1 | 0.02 | 0.06 | 0.032 |
| Brillouin | 1.91 | 2.49 | 0.151 |
| Menhinick 1 | 0.85 | 0.51 | 0.032 |
| Margalef | 19 | 9 | 0.095 |
| Equitability | 0.33 | 0.49 | 0.095 |
| Fisher-alpha | 27 | 13 | 0.095 |
| Berger-Parker | 0.46 | 0.36 | 0.841 |
| Chao-1 | 246 | 127 | 0.095 |
| Characteristic | Control Group (n = 16) 2 | Probiotic-Fed Group (n = 16) | p-Value 1 |
|---|---|---|---|
| Living mass of a 1-day-old duck, g/bird | 57.8 ± 5.8 2 | 57.9 ± 6.3 | 0.931 |
| Living mass of a 60-day-old duck, g/bird | 2772 ± 222 | 3007 ± 141 | 0.001 |
| Average daily gain, g/bird per day | 45.2 ±3.7 | 49.2 ± 2.3 | 0.001 |
| Feed intake, kg/kg bird mass | 3.35 | 2.85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naumova, N.B.; Alikina, T.Y.; Zolotova, N.S.; Konev, A.V.; Pleshakova, V.I.; Lescheva, N.A.; Kabilov, M.R. Bacillus-Based Probiotic Treatment Modified Bacteriobiome Diversity in Duck Feces. Agriculture 2021, 11, 406. https://doi.org/10.3390/agriculture11050406
Naumova NB, Alikina TY, Zolotova NS, Konev AV, Pleshakova VI, Lescheva NA, Kabilov MR. Bacillus-Based Probiotic Treatment Modified Bacteriobiome Diversity in Duck Feces. Agriculture. 2021; 11(5):406. https://doi.org/10.3390/agriculture11050406
Chicago/Turabian StyleNaumova, Natalia B., Tatiana Y. Alikina, Natalia S. Zolotova, Alexey V. Konev, Valentina I. Pleshakova, Nadezhda A. Lescheva, and Marsel R. Kabilov. 2021. "Bacillus-Based Probiotic Treatment Modified Bacteriobiome Diversity in Duck Feces" Agriculture 11, no. 5: 406. https://doi.org/10.3390/agriculture11050406
APA StyleNaumova, N. B., Alikina, T. Y., Zolotova, N. S., Konev, A. V., Pleshakova, V. I., Lescheva, N. A., & Kabilov, M. R. (2021). Bacillus-Based Probiotic Treatment Modified Bacteriobiome Diversity in Duck Feces. Agriculture, 11(5), 406. https://doi.org/10.3390/agriculture11050406







