Abstract
Rubber (Hevea brasiliensis, (Willd. Ex Adr. de Juss) Muell. Arg, Euphorbiaceae) is an important commercial latex-producing plant. Commercially, rubber is reproduced from a limited number of grifting genotypes. New promising genotypes have been selected to replace traditional genotypes. In addition, rubber has been promoted to recuperate Amazon soils degraded by extensive cattle ranching. Arbuscular mycorrhizal (AM) symbiosis is an important alternative for improving plant nutrition in rubber trees and recuperating degraded soils, but AM fungal communities on different plantations and in rubber genotypes are unknown. Spore abundance, root colonization and AM fungal community composition were evaluated in rubber roots of Colombian and introduced genotypes cultivated in degraded soils with different plantation types. Traditional (spore isolation and description; clearing and staining roots) and molecular techniques (Illumina sequencing) were used to assess AM fungi. Rubber roots hosted a diverse AM fungal community of 135 virtual taxa (VT) in 13 genera. The genus Glomus represented 66% of the total AM fungal community. Rubber genotype did not affect the arbuscular mycorrhization, hosting similar AM fungal communities. The composition of the AM fungal community on old and young rubber plantations was different. Diversity in AM fungi in rubber roots is an important characteristic for restoring degraded soils.
1. Introduction
Rubber (Hevea brasiliensis, (Willd. Ex Adr. de Juss) Muell. Arg, Euphorbiaceae) is a native species from the Amazon region and an important commercial plant species. It is cultivated in more than 40 countries for latex production [1]. Global consumption of natural rubber is increasing. Between 2000 and 2018, rubber consumption increased 96.23% [2].
Global rubber production is based on the cultivation of a few rubber clones that are reproduced from grifting stumps. Although it secures homogeneity in latex production, the lack of genetic diversity increases susceptibility to phytosanitary problems and reduces rubber latex yielding. Producers are creating new plantations in areas that were covered by natural tropical forests [3], avoiding common diseases such as the South American leaf blight (SALB), caused by Pseudocercospora ulei (Henn.), with negative consequences for natural ecosystems. Others are developing new rubber genotypes with more resistance to diseases and good latex production [4] for cultivation in traditional areas. The genetic improvement of commercial rubber is not enough to guarantee good latex production because cultivated plants require good nutrition. Commercial plantations require regular fertilization during the 25-year production cycle, which increases latex production cost.
Although Asiatic countries are the main latex rubber producers, rubber is still an important crop in Amazonian countries. In the Amazon region of Colombia, 4534 Ha are cultivated with rubber, producing 183 tons of latex per year [5]. In Colombia, rubber production is based mainly on three introduced genotypes—IAN 873, IAN 710, and FX 3864 [6]. These rubber genotypes have been planted in areas with a high incidence of SALB, resulting in a high presence of the disease and low local latex production [4].
In the Colombian Amazon, rubber has been promoted as a key crop recuperating degraded areas after years of extensive cattle ranching. Therefore, around 70 Ha of rubber trees were planted on degraded pastures with the purpose of transforming these unproductive areas into sustainable productive ones. Naturally, most of the Amazon basin has low-fertility soils [7], in which phosphorus (P) is usually the limiting soil nutrient. Degraded Amazon soils have very low P levels that are sometimes undetectable by traditional P analyses (e.g., less than 0.09 mg/Kg soil by Bray II). Although rubber is a native species from the Amazon region, it can experience P deficiencies, which manifest as lower leaf production in young rubber trees, brown color on leaf edges, less growth, and lower latex production in mature rubber trees [8]. Low-cost alternatives that improve soil conditions for rubber plantations are becoming an important issue for producers.
Arbuscular mycorrhization seems to be a feasible low-cost alternative that improves latex production locally because it occurs naturally in rubber [9,10]. Arbuscular mycorrhiza is a symbiosis between plant roots and Glomeromycota obligate endosymbiont fungi [11], occurring in about 80% of all plants. Arbuscular mycorrhiza is identified as the main plant–root association in crop species that mobilizes nutrients to plants, especially P, improving nutrition even in low fertile soils [12]. This association also reduces stress in host plants produced by water or nutrient limitations or the presence of contaminants or pathogens [13]. Although it seems to be the best alternative for a more sustainable and chipper rubber tree agriculture, there are still some unsolved issues that limit our knowledge on how grifting plants react to arbuscular mycorrhizal associations.
Grifting plants as commercial rubber are still physiologically poorly understood. Some authors consider them as individual plants that interact, while others consider them as hybrid plants with unique and individual autonomy [14]. The latter is based on the observed root-top relationship of grafting plants, in which upward water and mineral nutrient supply, downward flow of photosynthates, and root-top interchange of hormonal signals change [15]. Symbiotic associations such as arbuscular mycorrhiza might offer insights into how gifting plants behave. If grifting plants act as different individuals downward and upward, arbuscular mycorrhizal (AM) associations will not be affected by upward genotypic variations. However, if grifting plants act as hybrids, upward genotypic variations will affect root affinity by AM fungi and therefore AM associations among different clones.
Ecologically, plants with a high affinity for arbuscular mycorrhizal (AM) fungi benefit themselves and the plant community around them [16]. Therefore, AM symbiosis might play an important role in recovering degraded soils [17]. Different results support or deny the importance of rubber as an AM host plant in the restoration of environments. Based on soil spore-borne AM fungi, Feldmann et al. [18] found higher AM fungal diversity in natural rubber stands than in commercial monoclonal plantations. These differences were attributed to crop management practices on rubber plantations. However, Herrmann et al. [9] found highly diverse communities of AM fungi on commercial plantations of rubber cultivated in Thailand despite the high levels of applied fertilizers.
Since arbuscular mycorrhiza occurs at the root level, it is not well understood whether genomic variation or environmental and abiotic factors affect more AM symbiosis in rubber. Rubber production in the Amazon region offers a good environment for addressing these issues. Since 2009, the Sinchi Institute has selected promising introduced and Colombian rubber genotypes that are less susceptible to common pathogens and have good potential for latex production [19]. Sterling and Rodriguez [19] compared plant growth, resistance to phytopathogens, and potential latex production between promising rubber genotypes and the introduced genotype IAN 873 (control) indicating that new genotypes performed better than IAN 873. For a better understanding of arbuscular mycorrhizal association in rubber, this study (i) established differences in the AM fungal community associated with rubber roots cultivated on different plantation types, (ii) determined the influence of soil variables on the AM fungal community associated with these rubber plantations, and (iii) evaluated the root colonization and AM fungal community composition of Colombian and introduced rubber genotypes cultivated in degraded soils of the Colombian Amazon region.
2. Materials and Methods
2.1. Sites and Samples
Samples were collected in five municipalities of the Caquetá state, Colombian Amazon region: Albania, Belen de los Andaquíes, Florencia, El Paujil, and San Vicente del Caguán. The study area was located at 1°17′15″ N to 02°02′41″ N, and 74°54′39″ W to 75°54′2″ W (Figure 1). The elevation in this area ranges between 245 and 559 m above sea level. The average annual rainfall is around 3000 mm, with a dryer period between December and February. The average annual temperature is up to 24 °C [20]. The area corresponded to the upper Amazon basin and is influenced by the Andes mountain chain. Soils in the study area are classified as Inceptisols and Oxisols (USDA soil classification). The area is covered by introduced pastures for extensive cattle ranching with different levels of degradation, commercial rubber plantations, and some relicts of natural secondary forests.
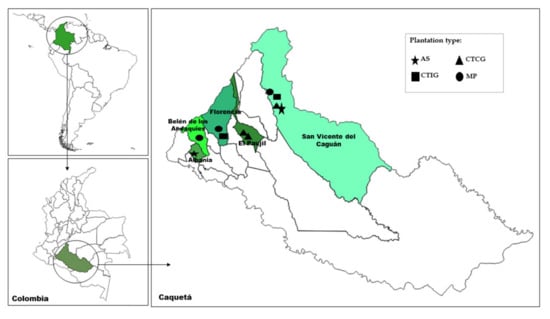
Figure 1.
Map with the location of the study area. Scale: 1:2,000,000.
Four types of rubber plantations were visited to collect soil and root samples—three monoclonal plantations (MPs) between 30 and 50 years old with introduced rubber genotypes; two six-year-old agroforestry systems (ASs) in which introduced genotypes of rubber were intercropped with copoazu fruit trees (Theobroma grandiflomum (Willd. Ex Spreng.) K.Schum.); three nine-year-old clonal trials with introduced genotypes (CTIGs); and three one-year-old clonal trials with Colombian rubber genotypes (CTCGs), plus the introduced genotype IAN 873 (traditional cultivar) as a control for comparisons (Table S1, Supplementary Materials).
Rubber plantations differed in their agronomic management. MPs were cleaned periodically of weeds and fertilized once per year with an inorganic NPK fertilizer. ASs received weed control every three months and fertilization twice per year with organic fertilizers. Additionally, two commercial insecticides were applied every 15 days to copoazu trees when insects were causing phytosanitary problems [21]. CTCGs and CTIGs received weed control every three months and fertilization with a mix of organic and inorganic fertilizers every six months. Phytosanitary controls were not performed on the latter two plantation types [22].
On each plantation, two types of samples were collected—soil samples and rubber tree root samples. A composed topsoil sample of about 500 g from 5 sub-samples of 100 g was collected to evaluate (i) the physicochemical composition of the soils where rubber tree was growing, (ii) the relative abundance of soil-borne AM fungal spores, and (iii) the AM fungal community composition at the family level. Soil samples were dried at room temperature to reduce the percentage of humidity and transported to the laboratory in plastic bags.
Root samples were used to estimate (i) the percentage of AM root colonization and (ii) the AM fungal community composition of rubber roots. Between 5 to 15 fine root samples of individual rubber trees were collected in each site from the bulky roots present in the first 20 cm of soil depth. The length of fine roots collected from each plant ranged between 3 and 10 cm approximately. A total of 255 root samples were collected. Root samples were transported in paper bags to the laboratory where root samples were divided in two: one-half of each sample was fixed in a Formalin-alcohol solution (Formaldehyde-ethanol-water 1:1:2) to estimate the percentage of AM root colonization. From the second half, a subset of 160 samples was stored at −70 °C and used to estimate the AM fungal community composition with molecular approaches. The subset included root samples of at least one replicate of each plantation type, with root samples of all rubber tree genotypes.
The number of soil and root samples that were collected and analyzed is summarized in Table S1, Supplementary Materials.
2.2. Soil Physicochemical Analysis
Soil samples were processed to estimate the following soil physicochemical properties: soil texture (Bouyoucos); pH (1:1 in water); percentage of organic carbon (with potassium dichromate solution); cation exchange capacity (CEC), Ca, Mg, K, and Na expressed in mg/kg of dry soil (with 1N ammonium acetate at pH = 7); and available phosphorus (P) expressed as mg/kg of dry soil (Bray II).
2.3. Extraction of AM Spores from Field Soils
About 25 g of each soil sample was used to extract AM fungal spores present in each soil. Five replicates of each soil sample were processed. Spores were obtained by wet sieving, followed by centrifugation in a saturated sucrose solution [23]. Isolated soil-borne AM fungal spores were placed in Petri dishes and counted directly with a stereoscope (SZ40 Olympus, Allentown, PA, USA). Distinctive AM fungal spore morphotypes were mounted on slides with lactoglycerol (LG) (Lactic acid: Glycerol: water 1:2:1) and LG + Melzer’s reagent (Micro-science, London, UK) (1:1 v/v) and observed under a compound microscope (BX53 Olympus, Allentown, PA, USA) in order to classify the spores into particular AM fungal families.
2.4. AM Fungal Colonization of Rubber Roots
Root samples were processed by clearing and staining roots with a solution of 0.05% trypan blue in lactoglycerol [24]. The percentage of AM root colonization was estimated by the gridline intersect method [25], obtaining the total percentage of AM root colonization in 100 root intersections. The procedure was repeated thrice per sample.
2.5. Molecular Analysis of Rubber Roots
DNA was obtained from 0.2 g of dried roots using the PowerMax® Soil DNA Isolation Kit (Qiagen, Germantown, MD, USA). This DNA was quantified with Nanodrop® (Thermo Fisher Scientific, Madison, WI, USA) before sequencing. The AM fungal DNA was amplified from samples using AM fungal specific primers for the small-subunit (SSU) ribosomal RNA gene—WANDA [26] and AML2 [27]. The PCR mixture contained 5 µL of 5XHOT FirePol Blend Master Mix (Solis Biodyne, Tartu, Estonia); 0.5 µL of each 20 µM primer; 1 µL of template DNA; and nuclease-free water to reach a total reaction volume of 25 µL. The PCR was performed following the thermocycling conditions reported by Davison et al. [28]: 95 °C for 15 min, 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 5 min. PCR products and the amplification success were checked on 1% agarose gel. Both negative (distilled water) and positive (synthetic double-stranded DNA with relevant priming sites) controls were included in the PCR to secure the quality of the PCR products. A DNA concentration of 5 ng/μL was used for the library preparation. A library was prepared for each sample. The library preparation was carried outusing a modified Illumina 16S rRNA protocol, standardized by Asper Biogene. Each library was ligated with Illumina adaptors using the TruSeq DNA PCR-free library prep kit (Illumina Inc., San Diego, CA, USA). The second PCR used a reaction mix composed of 5 μL of PCR product of fist PCR, 5 μL of Nextera XT index 1 primer (N7xxx), 5 μL of Nextera XT index 2 primer (E5xxx), and 15 μL of 2 × KAPA HiFi Hotstart PCR mix (without KAPA primer). The PCR program was run as follows: 3 min at 95 °C, followed by 7 cycles of 30 s at 95 °C, 30 s at 55 °C, and 30 s at 72 °C, with a final extension of 5 min at 72 °C and held at 4 °C. Libraries were sequenced on the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) using a 2 × 300 bp paired-read sequencing approach at Asper Biogene laboratory (Tartu, Estonia). A total of 22,810,852 raw sequences and 17,632,788 clean sequences were obtained, which corresponded to 77.3% of the total sequences. The raw sequences are available under request.
2.6. Bioinformatics
Demultiplexed paired-end reads were analyzed according to the bioinformatics steps described by Vasar et al. [29]. Primer sequences were matched allowing one mismatch in forward or reverse chains. Removal of barcode and primer sequences was conducted using the PEAR v. 0.9.8. program. Singletons were removed, and amplicons between 170 and 540 pb in length were included in the analysis. Amplicons longer than 540 bp were cut at that length and conserved for further analysis. Chimeric sequences were detected and removed using UCHIME v7.0.1090 [30] in the reference database mode using the default parameters and MaarjAM database (https://maarjam.botany.ut.ee, accessed on 8 January 2021, status June 2019, 384 virtual taxa (VT)). Sequence alignment was performed using the MAFFT v. 7 multiple sequence alignment web service in JALVIEW version 2.8 [31], subjected to a neighbor-joining phylogenetic analysis in TOPALi v2.5 [32] using the default parameters with the MaarjAM and the International Nucleotide Sequence Database Collaboration (INSDC). Retained reads were subjected to a BLAST+ search v 2.8.1 [33] using 97% identity and 95% alignment length thresholds. Representative sequences of Glomeromycota virtual taxa (VT) were deposited in the NCBI GenBank under the accession numbers (MW900266 to MW900422).
2.7. Statistical Analysis
Differences in the abundance of soil-borne spores and in the percentage of AM root colonization among the plantation types were assessed using linear mixed-effects models (LMEs) (plantation identity as a random effect) (function lme from the R package v. 3.5.1. nlme). When significant differences were obtained, a Tukey honest significant difference (HSD) test was performed with a p < 0.05 value to discriminate significant differences. Additionally, comparisons in the percentage of root colonization and richness between the Colombian and introduced rubber tree genotypes within the same plantation type were assessed using a Kruskal-Wallis test from the R program v. 3.5.1.
Sampling efficacy was assessed with species accumulation curves (method “exact”) using the functions accumcomp and ggplot from the R packages BiodiversityR and ggplot2, respectively [34,35]. The AM fungal richness between the plantation types and between the genotype types was visualized with Venn diagrams using the function ggvenn from R package ggven [36]. The AM fungal richness (S) and diversity indices (Shannon-Wiener, InvSimpson, and Pielou’s evenness) were estimated using the functions Specnumber and diversity from the R package vegan [37]. The Shannon index (H’) was exponentially transformed to obtain a variable (expH’) that satisfied the replication principle, following Herrmann et al. [9] recommendations. An LME (function lme from the R package nlme) [38] was adjusted to analyze the effect of the plantation type or genotype type on ecological indices. The effect of the plot (plantation identity or genotype identity) was included as a random effect in each model for the nested study design. The differences in mean variables among the plantation types or between the genotype types were analyzed with the Fisher’s least significant difference LSD post hoc test (α = 0.05).
A nested permitational multivariate analysis of variance PERMANOVA using the function nested.npmanova from the R package BiodiversityR [34] was performed to compare the AM fungal community composition among the plantations and between the genotypes. A non-metric multidimensional scaling ((NMDS), 50 iterations) was used (Bray-Curtis distance) and plotted with two axes and standard deviations ellipses (α = 0.05) to visualize changes in the AM fungal community composition among the plantation types. The functions metaMDS and ordiellipse (R package vegan), and ggplot and theme (R package ggplot2) were used [34,37]. The indicator species value analysis was used to study the strength of the relationship between the AM fungal VT and the rubber plantations or the AM fungal VT and the rubber genotypes (function indval from the R package labdsv [39]). An indicator value species of at least 0.25 was considered as a good indicator taxon [9].
Soil physicochemical variables were analyzed with LME and the means pairs were compared with Fisher’s LSD post hoc test (α = 0.05). Spearman’s correlation was used to relate the AM fungal diversity with the soil variables. The AM fungal community composition data were Hellinger transformed and included with all soil variables in a redundancy analysis (RDA) in order to assess the effect of soil variables on the AM fungal community composition (function rda from the R package vegan) [37]. Sequential ANOVA tests (1000 iterations) were performed for the contribution of each soil variable. The importance of individual AM fungal VT was determined using the parameter r ≥ 0.5 (α = 0.05) to better explain the variation among the plantation types. RDA was plotted with two axes in a triplot graphic using functions ordiellipse and envfit (R package vegan) [37], and ggplot and theme (R package ggplot2) [35].
The analyzes were performed in R language, v. 4.0.3 [40] using the interface in InfoStat v. 2020 [41] for the LME, Fisher’s LSD, and Spearman’s tests; the interface in Qeco v. 2018 [42] for the formal ecological analyses; and the interface in RStudio v. 1.3.1093 [43] for the graphical analyses.
3. Results
3.1. Soil Conditions of Rubber Plantations
Soil physicochemical analysis indicated that the soils where the rubber was cultivated had nutritional limitations. The available P was one of the most limited soil nutrients for plant growth, with values lower than 2 mg/kg of soil (Table 1). There was a significant difference in the content of sodium among the plantation types. AS and MP had more sodium than CTCG and CTIG.

Table 1.
Soil physicochemical composition of the rubber tree plantations: clonal trial with Colombian genotypes (CTCG); clonal trial with introduced genotypes (CTIG); agroforestry system (AS); and monoclonal plantation (MP).
3.2. Abundance of Arbuscular Mycorrhizal Spores in Soils
Significant differences in the abundance of AM fungal spores were found among the rubber plantations (p = 0.001). Spore abundance in MP was significantly lower than in AS and CTIG (Figure 2).
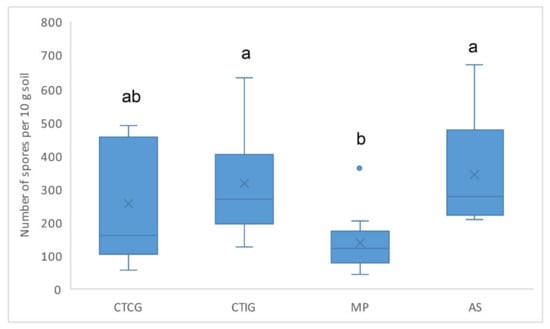
Figure 2.
Spore abundance of arbuscular mycorrhizal fungi in soils on different rubber tree plantations: AS: agroforestry; MP: monoclonal plantation; CTIG: clonal trial with introduced genotypes; and CTCG: clonal trial with Colombian genotypes. Spore abundance expressed as the number of spores per 100 g of dry soil. Letters express significant differences.
3.3. Arbuscular Mycorrhizal Fungal Colonization of Rubber Roots
All rubber root samples were colonized by AM fungi, with percentages of colonization between 1 and 96%. There were significant differences in the percentage of AM root colonization among the different rubber plantations (p < 0.0001). MP and AS had significantly higher percentages of AM root colonization (66 ± 17% and 57 ± 23%, respectively, Figure 3) than the clonal trials. Root colonization on the other rubber plantations was between 36 and 47% on average.
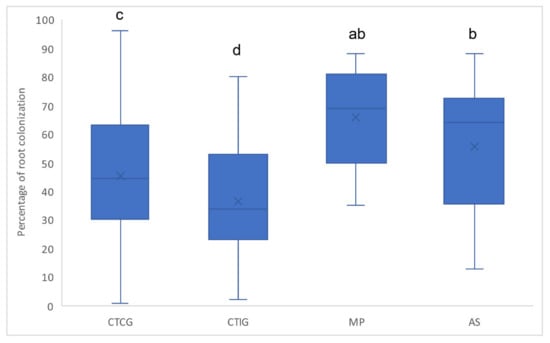
Figure 3.
Percentage of arbuscular mycorrhizal colonization of rubber roots cultivated on different plantation types: clonal trial with Colombian genotypes (CTCG); clonal trial with introduced genotypes (CTIG); monoclonal plantation (MP); and agroforestry system (AS). Letters express significant differences.
There were no significant differences in the percentage of AM root colonization between the Colombian (46%) and introduced (44%) rubber genotypes (p = 0.665), even when Colombian and IAN 873 were growing on the same plantation (p = 0.368; Kruskal-Wallis).
3.4. Arbuscular Mycorrhizal Fungal Community in Rubber Roots
Accumulation curves of VT in rubber roots cultivated on different plantations indicated that AM fungal community composition on CTCG was properly estimated but not on the other rubber tree plantations (Figure 4). According to the Chao index, 95% of the total AM fungal community colonizing rubber tree roots in CTCG was estimated, 85% in CTIG, 84% in MP, and 81% in AS.
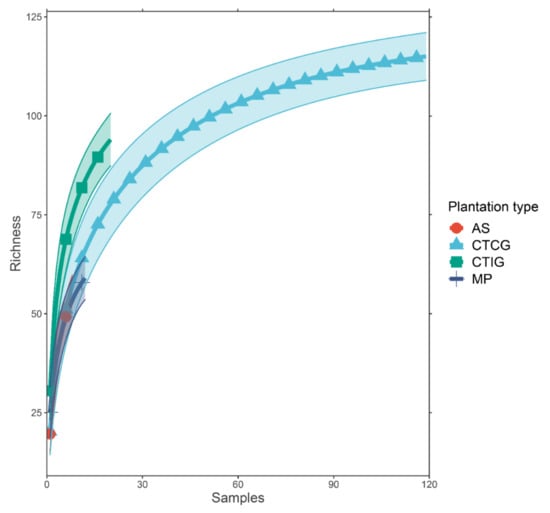
Figure 4.
Accumulation curves of the virtual taxa colonizing rubber root samples collected on different types of plantations: clonal trial with Colombian genotypes (CTCG); clonal trial with introduced genotypes (CTIG); agroforestry system (AS); and monoclonal plantation (MP).
Glomeraceae represented more than 77% of the total AM fungal community associated with rubber tree roots independent of the approach used to estimate the AM fungal community composition. However, the molecular approach increased the number of VT of the families Archaeosporacee and Gigasporaceae, compared to the soil-born spore approach (Figure 5).
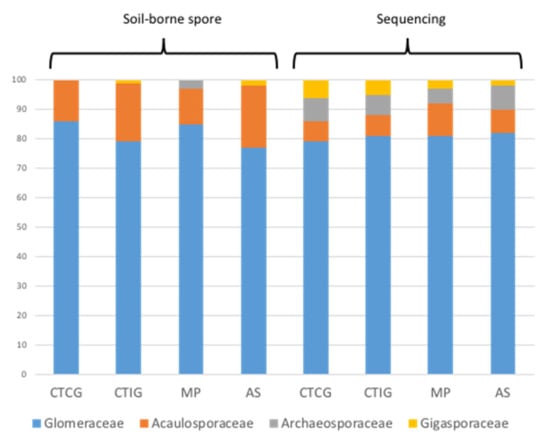
Figure 5.
Community composition of arbuscular mycorrhizal fungi associated with rubber roots cultivated on different types of plantations: clonal trial with Colombian genotypes (CTCG); clonal trial with introduced genotypes (CTIG); monoclonal plantation (MP); and agroforestry system (AS). Community composition was estimated by two different methodologies—soil spore-borne approach and sequencing approach.
The sequencing approach estimated a total of 419,089 Glomeromycotina sequences. Sequences corresponded to 135 VT of 13 genera. Rubber plantations were highly diverse, with more than seven different AM fungal genera per plantation. Glomus with 89 VT was the most diverse and the most frequent genus, representing 66% of the total AM fungal community in rubber roots. Three additional important genera colonizing rubber roots were Acaulospora with 10 VT, Paraglomus with nine VT, and Archaeospora with eight VT, representing 20% of the total AM fungal community. Other AM fungal genera in rubber roots, with less frequency, were Claroideoglomus and Scutellospora with five VT, Rhizophagus with three VT, and Ambispora, Diversispora, Gigaspora, Kuklospora, Redeckera, and Viscospora, with one VT each. On average, each rubber plantation and each rubber root sample hosted 22 and 23 VT, respectively. The complete list of VT obtained in this study is summarized in Table S2, Supplementary Materials.
The PERMANOVA analysis showed significant differences between the AM fungal community composition associated with different rubber plantations (p = 0.027) (Table 2) but did not in those colonizing different rubber genotypes (p = 0.068). The NMDS plot separated the AM fungal communities of MP from AS, CTIC, and CTCG (Figure 6). In addition, the AM fungal communities of AS were separated according to the location of the AS.

Table 2.
Characteristics of rubber plantations and their arbuscular mycorrhizal (AM) fungal composition.
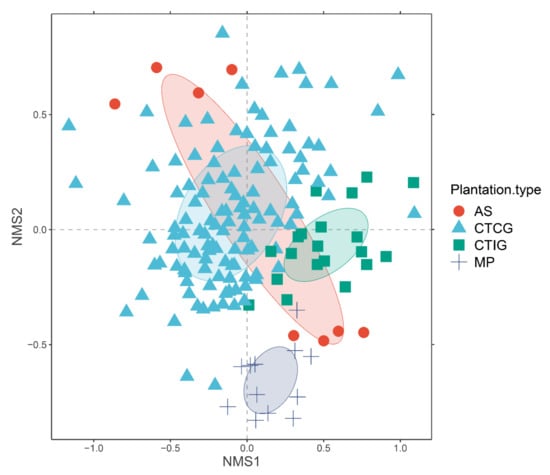
Figure 6.
Non-metric multidimensional scaling (NMDS) plots displaying AM fungal communities colonizing rubber roots. Ellipses indicate one standard deviation around the centroid position of each rubber plantation.
The most abundant VT colonizing rubber roots was Paraglomus VT444, with 50,279 reads. Some VT were identified as indicator species of plantation types or rubber tree genotypes (Table S3, Supplementary Materials). Most of the indicator species identified were Glomus (82% of all indicator species), Acaulospora and Archaeospora (each representing 5% of all indicator species), Gigaspora, and Scutellospora (each representing 2% of all indicator species).
3.5. Richness and Diversity of Arbuscular Mycorrhizal Fungal in Rubber Roots
Around 27% of the total VT was shared among all rubber tree plantations. Only CTCG showed a high number of exclusive VT (around 19%), while the other plantation types had less than 5% of exclusive VT (Figure 7a). The AM fungal community compositions in roots of Colombian and introduced rubber genotypes were very similar. The Colombian and introduced rubber genotypes shared around 70% of the total AM fungal community (Figure 7b).
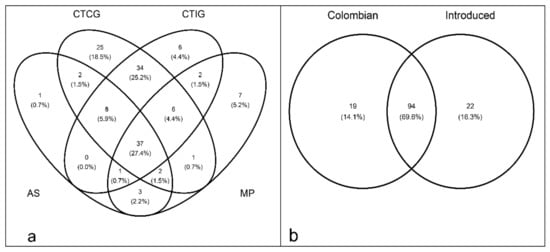
Figure 7.
Arbuscular mycorrhizal fungal community colonizing rubber roots in (a) different plantation types (CTCG: clonal trial with Colombian genotypes; MP: monoclonal plantation; AS: agroforestry system; CTIG: clonal trial with introduced genotypes) and (b) between Colombian and Introduced rubber genotypes.
VT richness colonizing rubber roots was significantly different among the plantation types (p < 0.001) but not between the rubber tree genotypes (p = 0.665), even if they (Colombian vs. IAN 873) were growing on the same plantation (p = 0.429). CTIG and MP hosted significantly more AM fungal VT than the other plantation types (Table 3). Only the exponential Shannon index showed significant differences in the diversity of the different rubber plantations (Table 3).

Table 3.
Richness and diversity indexes of AM fungal colonizing roots of Colombian and introduced rubber genotypes and different plantation types: CTCG (clonal trial with Colombian genotypes); CTIG (clonal trial with introduced genotypes); AS (agroforestry system); and MP (monoclonal plantation).
3.6. Arbuscular Mycorrhizal Fungi and Soil Properties
Three diversity indexes (inverse Simpson (1/D), exponential Shannon (ExpH′), and evenness) were negatively correlated with pH, Ca and Mg (r < −0.64; p < 0.05), two diversity indexes (1/D and ExpH′) were negatively correlated with P (r < −0.65; p < 0.05), and the VT richness was negatively correlated with Ca (r = −0.72; p < 0.05) (Table S4, Supplementary Materials).
Redundancy analysis (RDA) of AM fungal communities associated with soil variables (inertia proportion constrained = 23.77%) showed a clear separation of MP from the other rubber plantations (Figure 8a). The AM fungal communities on MP were strongly associated with a higher soil Na content (Figure 8b), where Glomus VT269 was favored by this condition. RDA plot also showed that the AM fungal communities on CTCG were associated with sandier soils (Figure 8a), where Glomus VT280 was favored by soils with coarse texture. There was a weak relationship between the AM fungal community on CTCG and P (Figure S1, Supplementary Materials).
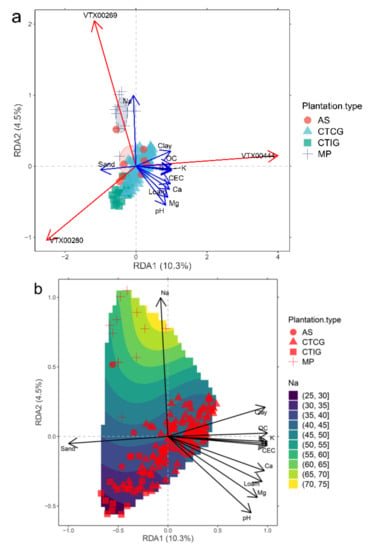
Figure 8.
(a) Redundancy analysis (RDA) of AM fungal communities associating with rubber tree plantations, constrained by soil variables. Blue arrows indicate significant soil variables for the constrained ordination (p < 0.05). Red arrows indicate significant AMF VT species with maximum correlation with ordination scores (r ≥ 0.5; p < 0.05). Ellipses indicate one standard deviation around the centroid position of each rubber tree plantation. (b) Redundancy analysis (RDA) of AM fungal communities associated with rubber tree plantations according to Na soil gradient.
4. Discussion
Rubber planted on degraded soils of the Colombian Amazon harbored highly diverse AM fungal communities. The AM fungal communities in rubber roots differed among the plantation types but not between the rubber genotypes.
The molecular approach to assess AM fungal community composition was more sensitive than the soil-borne spore approach. The molecular approach recorded a higher number of AM fungi from the Archaeosporaceae and Gigasporaceae families. The Archaeosporaceae and Gigasporaceae families produced low numbers of large-spore size [44] than the Glomeraceae family, which was the best represented and most diverse in the study area (with 77% of the total AM fungal community). Additionally, the molecular approaches provided information for the presence of AM fungi that do not sporulate frequently or do not sporulate at all [45], offering a more accurate picture of the AM fungal community. The AM fungal community in rubber roots was composed of 135 VT in 13 genera. There was a higher number of AM fungi associated with rubber roots than in previous studies (111 VT of 10 genera [9]), even on the rubber plantations planted on areas previously covered by pastures (on average, 22 VT of 7 genera per plot; 16 VT of 5 genera per plot [10]). It is important to indicate that this is the first study evaluating the AM fungal composition of rubber roots using WANDA and AML2 primers, which limits comparisons with results from previous publications. Differences in the number of VT between this study and others might have resulted from the molecular technique used to assess the AM fungi and should not be interpreted as real differences.
Glomus was the dominant genus, representing 66% of the total AM fungal community, followed by Acaulospora, Paraglomus, and Archaeospora. Glomus and Acaulospora were reported previously as frequent genera associated with rubber [9,10], and the main genera composing the AM fungal community of Amazon soils [46,47,48].
In the study area, rubber plantations were planted on degraded pastures after years of cattle ranching. Soils where rubber tree grows presented low fertility, with very low levels of available P and significant differences in the amount of Na among plantations. Pfeiffer and Bloss [49] found that phosphate fertilizers included low concentrations of Cu, Zn, Na, K, and SO4 and were a source of those elements. A significantly higher amount of Na on the older plantations (AS and MP) might have resulted from periodical fertilization in earlier years and later soil accumulation. MP and AS also had significantly higher percentages of root colonization (66% and 57%, respectively). Higher percentages of root colonization in MP and AS might have been the consequence of an accumulative effect from fertilization, through which Na, in combination with P, increased the AM root colonization [49]. On the other plantations, rubber roots presented percentages of AM root colonization around 47%, similar to previous reports [50].
The AM fungal richness and diversity (ExpH′) were significantly higher in CTIG and MP than in the other plantation types. Feldmann et al. [18] found lower spore diversity on commercial rubber plantations than in natural tree stands. They attributed differences to agronomic practices on commercial plantations and mainly to the use of pesticides and extensive weeding. Although there were differences in the agronomic practices used on the different rubber plantations (e.g., MP received less fertilization than the other plantations), our results are best explained by differences in age between rubber plantations. Plantations over nine years old (MP and CTIG) had higher AM fungal richness and diversity than younger plantations. Even with the limitations to compare results obtained from different molecular techniques, our result was similar to the one reported by Herrmann and collaborators. Herrmann et al. [9] also found differences in AM fungal richness related to plant age in a rubber chronosequence of 3, 6, and 16 years old. The NMDS analysis clearly showed how the AM fungal community composition of very old rubber plantations was sufficiently different from the others, forming a separate cluster (Figure 6). Additionally, the low numbers of AM fungal spores in MP resembled the abundance of AM fungal spores in soils of mature Amazon forests [46].
Diversity indexes were negatively correlated with pH, Ca, Mg, and P. Similar results had been reported previously for rubber plantations [9]. The edaphic condition is one of the main factors affecting AM fungal community composition [51,52]. Soil pH has been clearly identified as one of the most important abiotic drivers of AM fungal communities [28,53]. pH directly influences the availability of Ca, Mg, and P in soil. In soils with a low pH, P became particularly less available as it is sorbed in insoluble compounds [54]. Some specific AM fungi are commonly associated with acid soils, such as Acaulosporaceae, and some Glomeraceae, such as Rhizophagus manihotis [28,55]. Particularly, Rhizophagus manihotis is commonly associated with acidic poor-nutrient soils [56,57].
In clonal trials, selected Colombian rubber genotypes performed better (growth, early latex production, and resistance to pathogens) than the IAN 873 (control) genotype [19]. Our results indicated that AM root colonization, richness, and composition of AM fungal communities in roots of different rubber genotypes were similar, even if different rubber genotypes (Colombian vs. IAN 873) were under the same conditions (location, plantation type, and age). The AM fungal community composition in roots of Colombian and introduced genotypes shared around 70% of the total community (Figure 7b). Herrmann et al. [9] identified four VT as indicator species for rubber, namely, Glomus VT399, Acaulospora VT227, Rhizophagus manihotis VT90, and Glomus VT124. From those VT, three were recorded in this study. Rhizophagus manihotis was one of the 20 more abundant VT colonizing rubber tree roots. Glomus VT399 was an indicator species of introduced rubber tree genotypes, and Acaulospora VT227 was an indicator species of MP, but both had a low number of reads. The most abundant VT colonizing rubber roots was Paraglomus VT444, which was not reported before in rubber tree roots. Paraglomus VT444 was an indicator taxon of Colombian rubber genotypes and AS. This VT was isolated before in the study area from pastures, and its abundance increased in soils of secondary forests that originated from the natural regrowth of those pastures [58]. Since AS was similar to natural secondary forests, Paraglomus VT444 persisted in AS soils. The particular association of this VT with Colombian rubber genotypes might be related to the fact that the Colombian genotypes were the youngest rubber trees and were cultivated on recently used pastures.
Rubber preferences for a particular AM fungal taxon were weak. Strong relationships between particular AM fungi and rubber seem to be context dependent, varying from one site to another. Apparently, plant host-AM fungal preferences occur at the plant level (functional) groups [59] rather than at the level of individual plant species. Even if grifting plants such as rubber tree clones behave as particular individuals [15], the arbuscular mycorrhizal association would not be affected by genotypic differences between genotypes. Rubber tree genotypes apparently behave as natural varieties, even if genotypic variability was induced artificially. The similarity in arbuscular mycorrhization among manioc varieties (Manihot esculenta Crantz), another important commercial Euphorbiaceae species, has been reported [60], supporting the interpretation of our results.
According to Chen et al. [3], agroforestry systems for rubber trees are the best alternative for improving soil properties. Our results indicated that none of the rubber plantations improved the degraded condition of the soils. Significant differences in the soil composition occurred only on rubber plantations older than 30 years. Herrmann et al. [9] also found that the soil fertility of rubber plantations did not change over time despite fertilization. Frequent weed control and other agronomic practices could be responsible for this result. Although soil condition is not improved during the productive cycle of rubber, its roots host significantly more AM fungi (in average 22 VT per plot) than other commercial crops cultivated in Amazon, i.e., mahogany (Swietenia macrophylla; 19 VT [10]), eucalyptus (16 VT [10]), and soybean (17 VT [61]). A higher AM fungal diversity will enhance the external mycelial network, improving the nutrition of the plant community [16]. Additional traits (e.g., soil aggregation, soil organic C, biomass C sequestrated) and ecological services (plant, enthomofauna, and bird diversity) have been evaluated on different rubber plantations and genotypes [3,22,62], corroborating the importance of this plant species for soil restoration.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/agriculture11040361/s1, Table S1: Soil and rubber tree samples collected and analyzed, Table S2: Arbuscular mycorrhizal fungal virtual taxa recovered from different rubber plantations, Table S3: Arbuscular mycorrhizal fungal virtual taxa with significant indicator species value for rubber genotypes and plantation types, Table S4: Spearman’s correlation coefficients between soil parameters, and richness and diversity indexes of arbuscular mycorrhizal fungi, Figure S1: Redundancy analysis (RDA) of arbuscular mycorrhizal fungal communities associated with rubber plantations according to phosphorus soil gradient.
Author Contributions
Conceptualization, C.P.P.-V. and A.S.; methodology, A.S., C.P.P.-V. and T.K.A.-R.; software, A.S.; validation, C.P.P.-V. and A.S.; formal analysis, A.S. and C.P.P.-V.; investigation, C.P.P.-V.; resources, A.S. and C.P.P.-V.; data curation, T.K.A.-R. and A.S.; writing—original draft preparation, C.P.P.-V.; writing—review and editing, C.P.P.-V. and A.S.; visualization, T.K.A.-R. and A.S.; supervision, C.P.P.-V.; project administration, A.S.; funding acquisition, A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by FCTeI-SGR, Contract 59/2013 Instituto Amazónico de Investigaciones Científicas SINCHI–Gobernación del Caquetá–Universidad de la Amazonía–Asociación de Reforestadores y Cultivadores de Caucho del Caquetá; and by the Government of Colombia through the project BPIN 2017011000137 “Investigación en conservación y aprovechamiento sostenible de la diversidad biológica, socioeconómica y cultural de la Amazonia colombiana.”
Acknowledgments
We thank Daniela León and Martti Vasar for their help with bioinformatic analyzes. We also thank all the farmers of the study area for their help and support during the fieldwork.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Verheye, W. Growth and production of rubber. In Land Use, Land Cover and Soil Sciences. Encyclopedia of Life Support Systems (EQLSS); Verheye, W., Ed.; UNESCO-EOLSS Publishers: Oxford, UK, 2010; p. 16. [Google Scholar]
- International Rubber Study Group. Rubber Statistical Bulletin; International Rubber Study Group: Singapore, 2019. [Google Scholar]
- Chen, C.; Liu, W.; Jiang, X.; Wu, J. Effects of rubber-based agroforestry systems on soil aggregation and associated soil organic carbon: Implications for land use. Geoderma 2017, 299, 13–24. [Google Scholar] [CrossRef]
- Sterling, A.; Martínez-Vuiche, E.J.; Suárez-Córdoba, Y.D.; Agudelo-Sánchez, A.A.; Fonseca-Restrepo, J.A.; Andrade-Ramírez, T.K.; Virguez-Díaz, Y.R. Assessing growth, early yielding and resistance in rubber tree clones under low South American Leaf Blight pressure in the Amazon region, Colombia. Ind. Crops Prod. 2020, 158, 112958. [Google Scholar] [CrossRef]
- Ramírez, U.; Charry, A.; Jäger, M.; Huertado, J.; Sterling, A.; Romero, M.; Sierra, L.; Quintero, M. Estrategia Sectorial de la Cadena de Caucho en Caquetá, con Enfoque Agroambiental y cero Deforestación; CIAT: Palmira, Colombia, 2018. [Google Scholar]
- Castellanos, O.; Fonseca, S.; Barón, M. Agenda prospectiva de investigación y desarrollo tecnológico para la cadena productiva de caucho natural y su industria en Colombia. Ing. Investig. 2009, 29, 63–65. [Google Scholar]
- Quesada, C.A.; Lloyd, J.; Schwarz, M.; Patiño, S.; Baker, T.R.; Czimczik, C.; Fyllas, M.N.; Martinelli, L.; Nardoto, G.B.; Schmerler, J.; et al. Variations in chemical and physical properties of Amazon forest soils in relation to their genesis. Biogeosciencies 2010, 7, 1515–1541. [Google Scholar] [CrossRef]
- He, P.; Qin, H.; Wu, M.; Wu, B.; Wei, J.; Wang, D. Identification of genes differentially expressed in the roots of rubber tree (Hevea brasiliensis Muell. Arg.) in response to phosphorus deficiency. Mol. Biol. Reprod. 2013, 40, 1397–1405. [Google Scholar] [CrossRef]
- Herrmann, L.; Lesueur, D.; Bräu, L.; Davison, J.; Jairus, T.; Robain, H.; Robin, A.; Vasar, M.; Wiriyakitnateekul, W.; Öpik, M. Diversity of root-associated arbuscular mycorrhizal fungal communities in a rubber tree plantation chronosequence in Northeast Thailand. Mycorrhiza 2016, 26, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.M.R.; da Silva, D.K.A.; Ferreira, A.C.A.; Goto, B.T.; Maia, L.C. Diversity of arbuscular mycorrhizal fungi in Atlantic forest areas under different land uses. Agric. Ecosyst. Environ. 2014, 185, 245–252. [Google Scholar] [CrossRef]
- Schüβler, A.; Schwarzott, D.; Walker, C. A new fungal phylum, the Glomeromycota: Phylogeny and evolution. Mycol. Res. 2001, 105, 1413–1421. [Google Scholar] [CrossRef]
- Cardoso, I.M.; Kuyper, T.W. Mycorrhizas and tropical soil fertility. Agric. Ecosyst. Environ. 2006, 116, 72–84. [Google Scholar] [CrossRef]
- Baum, C.; El-Tohamy, W.; Gruda, N. Increasing the productivity and product quality of vegetable crops using arbuscular mycorrhizal fungi: A review. Sci. Hortic. 2015, 187, 131–141. [Google Scholar] [CrossRef]
- Goldschmidt, E.E. Plant grafting: New mechanisms, evolutionary implications. Front. Plant Sci. 2014, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Aloni, B.; Cohen, R.; Karni, L.; Aktas, H.; Edelstein, M. Hormonal signaling in rootstock-scion interactions. Sci. Hortic. 2010, 127, 119–126. [Google Scholar] [CrossRef]
- Walder, F.; Niemann, H.; Natarajan, M.; Lehmann, M.F.; Boller, T.; Wiemken, A. Mycorrhizal Networks: Common Goods of Plants Shared under Unequal Terms of Trade. Plant Physiol. 2012, 159, 789–797. [Google Scholar] [CrossRef]
- Soka, G.; Ritchie, M. Arbuscular mycorrhizal symbiosis and ecosystem processes: Prospects for future research in tropical soils. Open J. Ecol. 2014, 4, 11–22. [Google Scholar] [CrossRef]
- Feldmann, F.; da Silva, J.J.P.; Idczak, E.; Lieberei, R. AMF spore community composition of natural and agricultural sites in Central Amazonia—A long term study. In German-Brazilian Workshop on Neotropical Ecosystems—Achievements and Prospects of Cooperative Research; FRG and EMBRAPA: Hamburg, Germany, 2000; pp. 1–31. [Google Scholar]
- Sterling, A.; Rodrìguez, C.H. Valoración de Nuevos Clones de Hevea Brasiliendis con Proyección Para la Amazonia Colombiana: Fases pre y Post-Sangría Temprana en el Caquetá; Imagen Editorial SAS: Bogotá, Colombia, 2020; p. 326. [Google Scholar]
- IGAC. Estudio General de Suelos y Zonificación de Tierras Departamento de Caquetá Escala 1:100.000; Instituto Geográfico Agustin Codazzi—IGAC: Bogotá, Colombia, 2014.
- Sterling, A.; Rodríguez, C.H. Agroforestería en el Caquetá: Clones Promisorios de Caucho en Asocio con Copoazú y Plátano Hartón con Potencial para la Amazonia Colombiana; Instituto Amazonico de Investigaciones Científicas Sinchi: Bogotá, Colombia, 2014. [Google Scholar]
- Sterling, A.; Rodríguez, C.H. Valoración y Análisisde la Biodiversidad y Servicios Ecosistémicos Asociados a Campos Clonales de Caucho en Caquetá, Amazonia Colombiana; Instituto Amazonico de Investigaciones Científicas Sinchi: Bogotá, Colombia, 2019. [Google Scholar]
- Sieverding, E. Vesicular-arbuscular mycorrhiza management in tropical ecosystems. In Deutche Gesellschaft für Technische Zusammenarbeit 224; Verlag, H.B., Ed.; Technical Cooperation Federal Republic of Germany: Eschbom, Germany, 1991. [Google Scholar]
- Phillips, J.M.; Hayman, D.S. Improved procedures for cleaning roots and staining parasitic and vesicular-arbuscular fungi for rapid assessment of infection. Transcr. Br. Soc. Mycol. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- Giovannetti, M.; Mosse, B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 1980, 84, 489–500. [Google Scholar] [CrossRef]
- Dumbrell, A.J.; Ashton, P.D.; Aziz, N.; Feng, G.; Nelson, M.; Dytham, C.; Fitter, A.H.; Helgason, T. Distinct seasonal assemblages of arbuscular mycorrhizal fungi revealed by massively parallel pyrosequencing. New Phytol. 2011, 190, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, S.; Young, J.P.W. Improved PCR primers for the detection and identi¢cation of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 2008, 65, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Davison, J.; Moora, M.; Semchenko, M.; Adenan, S.B.; Ahmed, T.; Akhmetzhanova, A.A.; Alatalo, J.M.; Al-Quraishy, S.; Adriyanova, E.; Anslan, S.; et al. Temperature and pH define the realized niche space of arbuscular mycorrhizal fungi. New Phytol. 2021. [Google Scholar] [CrossRef]
- Vasar, M.; Andreson, R.; Davison, J.; Jairus, T.; Moora, M.; Remm, M.; Young, J.P.W.; Zobel, M.; Öpik, M. Increased sequencing depth does not increase captured diversity of arbuscular mycorrhizal fungi. Mycorrhiza 2017, 27, 761–773. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Milne, I.; Lindner, D.; Bayer, M.; Husmeier, D.; McGuire, G.; Marshall, D.F.; Wright, F. TOPALi v2: A rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 2009, 25, 126–127. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Kindt, R. Package ‘BiodiversityR’: Community Ecology and Suitability Analysis; World Agroforestry Centre (ICRAF): Nairobi, Kenya, 2020. [Google Scholar]
- Dunnington, D.; Wickham, H.; Chang, W.; Henry, L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H. Package ‘ggplot2’: Create Elegant Data Visualisations Unisg the Grammar of Graphics. R Package Version 2020. Available online: https://ggplot2.tidyverse.org/reference/ggplot2-package.html (accessed on 9 January 2021).
- Yan, L. Package ‘Ggvenn’. 2021. Available online: https://cren.r-project.org/web/packages/ggvenn/ggvenn.pdf (accessed on 11 January 2021).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Package ‘Vegan’: Community Ecology Package. 2018. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 13 January 2021).
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. R-Core. Nlme: Linear and Nonlinear Mixed Effects Models. 2013. Available online: https://cran.r-project.org/web/packages/nlme/index.html (accessed on 12 January 2021).
- Robert, D.W. Labdsv: Ordination and Multivariate Analysis for Ecology, in R Package. 2012. Available online: https://cran.r-project.org/web/packages/labdsv/index.html (accessed on 13 January 2021).
- R Core Team. Core Team R: A Language and Environment for Statistical Computing. 2020. Available online: http://www.r-project.org/index.html (accessed on 12 January 2021).
- Di Rienzo, J.A.; Casanoves, F.; Pla, L.; Vilchez, S.; Di Renzo, M.J. InfoStat. 2020. Available online: http://www.infostat.com/ (accessed on 15 January 2021).
- Di Rienzo, J.A.; Casanoves, F.; Pla, L.; Vilchez, S.; Di Renzo, M.J. Qeco. 2018. Available online: https://sites.google.com/site/qecosite/home (accessed on 15 January 2021).
- R Studio. 2020. Available online: https://www.rstudio.com/products/rstudio/older-versions/ (accessed on 15 January 2021).
- Aguilar-Trigueros, C.A.; Hempel, S.; Powell, J.R.; Cornwell, W.K.; Rilling, M.C. Bridging reproductive and microbial ecology: A case study in arbuscular mycorrhizal fungi. ISME J. 2019, 13, 873–884. [Google Scholar] [CrossRef]
- Sanders, I.R. Plant and arbuscular mycorrhizal fungal diversity: Are we looking at the relevant levels of diversity and are we using the right techniques? New Phytol. 2004, 164, 415–418. [Google Scholar] [CrossRef]
- Leal, P.L.; Siqueira, J.O.; Stürmer, S.L. Switch of tropical Amazon forest to pasture affects taxonomic composition but not species abundance and diversity of arbuscular mycorrhizal fungal community. Appl. Soil Ecol. 2013, 71, 72–80. [Google Scholar] [CrossRef]
- Reyes, H.A.; Ferreira, P.F.A.; Silva, L.C.; da Costa, M.G.; Nobre, C.P.; Gehring, C. Arbuscular mycorrhizal fungi along secondary forest succession at the eastern periphery of Amazonia: Seasonal variability and impacts of soil fertility. Appl. Soil Ecol. 2019, 136, 1–10. [Google Scholar] [CrossRef]
- Stürmer, S.L.; Siqueira, J.O. Species richness and spore abundance of arbuscular mycorrhizal fungi across distinct land uses in Western Brazilian Amazon. Mycorrhiza 2011, 21, 255–267. [Google Scholar] [CrossRef]
- Pfeiffer, C.M.; Bloss, H.E. Growth and nutrition of guayule (Parthenium argentatum) in a saline soil as influenced by vesicular-arbuscular mycorrhiza and phosphorus fertilization. New Phytol. 1988, 108, 315–321. [Google Scholar] [CrossRef]
- Tawaraya, K.; Tacaya, Y.; Turjaman, M.; Tuah, S.J.; Limin, S.H.; Tamai, Y.; Cha, J.Y.; Wagatsuma, T.; Osaki, M. Arbuscular mycorrhizal colonization of tree species grown in peat swamp forest of Central Kalimantan, Indonesia. Forest Ecol. Manag. 2003, 182, 381–386. [Google Scholar] [CrossRef]
- Dumbrell, A.J.; Nelson, M.; Helgason, T.; Dytham, C.; Fitter, A.H. Idiosyncrasy and overdominance in the structure of natural communities of arbuscular mycorrhizal fungi: Is there a role for stochastic processes? J. Ecol. 2010, 98, 419–428. [Google Scholar] [CrossRef]
- Entry, J.A.; Rygiewicz, P.T.; Watrud, L.S.; Donelly, P.K. Influence of adverse soil conditions on the formation and function of Arbuscular mycorrhizas. Adv. Environ. Res. 2002, 7, 123–138. [Google Scholar] [CrossRef]
- Moora, M.; Davison, J.; Öpik, M.; Metsis, M.; Saks, Ü.; Jairus, T.; Vasar, M.; Zobel, M. Anthropogenic land use shapes the composition and phylogenetic structure of soil arbuscular mycorrhizal fungal communities. FEMS Microbiol. Ecol. 2014, 90, 609–621. [Google Scholar] [CrossRef]
- García-Montiel, D.C.; Neill, C.; Melillo, J.; Thomas, S.; Steudler, P.A.; Cerri, C.C. Soil phosphorus transformations following forest clearing for pasture in the Brazilian Amazon. Soil Sci. Soc. Am. J. 2000, 64, 1792–1804. [Google Scholar] [CrossRef]
- Clark, R.B. Arbuscular mycorrhizal adaptation, spore germination, root colonization, and host plant growth and mineral adquisition at low pH. Plant Soil 1997, 192, 15–22. [Google Scholar] [CrossRef]
- Cuenca, G.; Cáceres, A.; González, M.G. AM inoculation in tropical agroculture: Field results. In Mycorrhiza; Varma, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 403–417. [Google Scholar]
- Izaguirre-Mayoral, M.L.; Flores, S.; Pieters, A.; Olivares, E.; Cuenca, C. Rhizophagus manihotis promotes the growth of rhizobia-nodulated Vigna luteola L. in phosphorus deficient acid montane soils devoid of ground cover vegetation. Symbiosis 2011, 55, 1–9. [Google Scholar] [CrossRef]
- Peña-Venegas, C.P.; Castro, D.; Fausto, M.; Silva, A.; Rodríguez, C.H. Comportamiento de Variables del Suelo en la Sucesión de Bosques Secundarios, in Sucesión Ecológica y Restauración en Paisajes Fragmentados de la Amazonia Colombiana; Rodríguez, C.H., Sterling, A., Eds.; Instituto Amazónico de Investigaciones Científicas Sinchi: Bogotá, Colombia, 2020; pp. 141–170. [Google Scholar]
- Sepp, S.-K.; Davison, J.; Jairus, T.; Vasar, M.; Moora, M.; Zobel, M.; Öpik, M. Non-random association patterns in a plant-mycorrhizal fungal network reveal host-symbiont specificity. Mol. Ecol. 2019, 28, 365–378. [Google Scholar] [CrossRef]
- Peña-Venegas, C.P.; Kuyper, T.W.; Davison, J.; Jairus, T.; Vasar, M.; Stomph, T.J.; Struik, P.C.; Öpik, M. Distinct arbuscular mycorrhizal fungal communities associate with different manioc landraces and Amazonian soils. Mycorrhiza 2019, 29, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Faggioli, V.S.; Cabello, M.N.; Grilli, G.; Vasar, M.; Covacevich, F.; Öpik, M. Root colonizing and soil borne communities of arbuscular mycorrhizal fungi differ among soybean fields with contrasting historical land use. Agric. Ecosyst. Environ. 2019, 269, 174–182. [Google Scholar] [CrossRef]
- Beukema, H.; Danielsen, F.; Vincent, G.; Hardiwinoto, S.; van Andel, J. Plant and bird diversity in rubber agroforests in the lowlands of Sumatra, Indonesia. Agrofor. Syst. 2007, 70, 217–242. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).