Mycorrhizal Fungal Diversity and Its Relationship with Soil Properties in Camellia oleifera
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Setup
2.2. Sampling
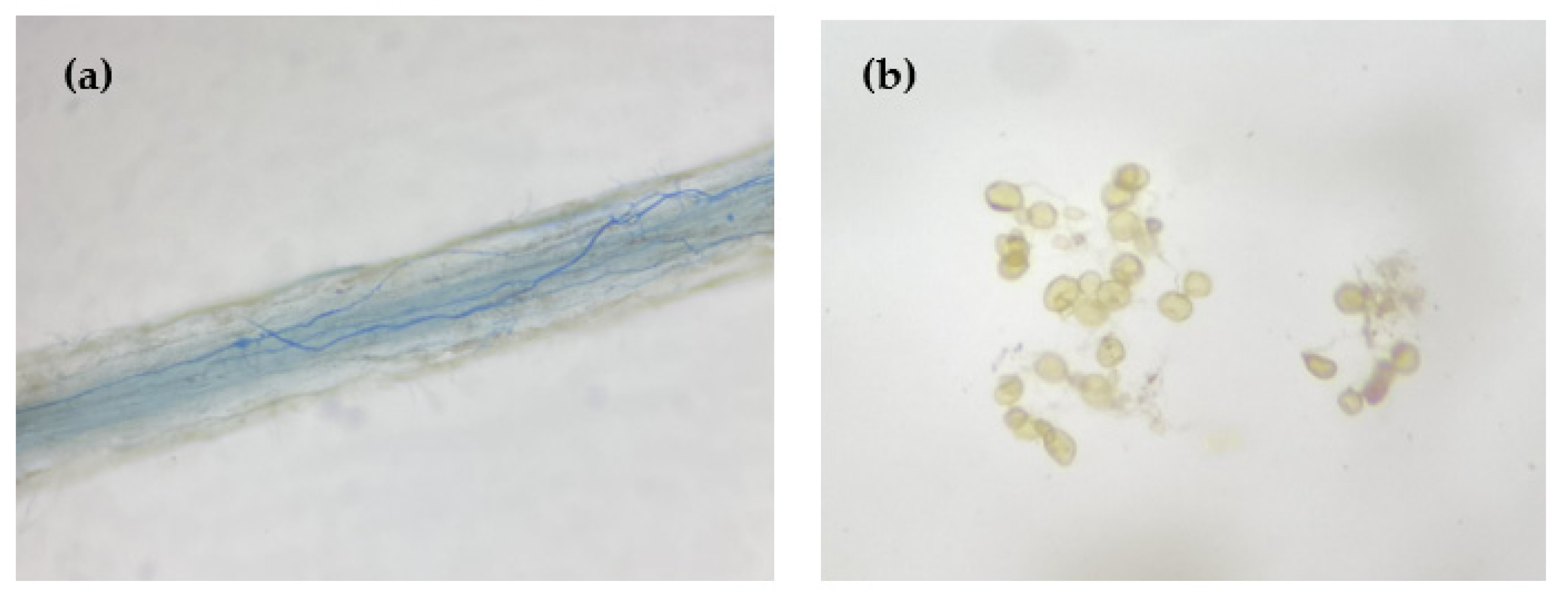
2.3. Determinations of Root Mycorrhizal Colonization
2.4. Determinations of Soil Spore Density and Soil Hyphal Length
2.5. Determinations of Soil Properties
2.6. Analysis of AMF Diversity of Rhizosphere and Endosphere
2.7. Data Analysis
3. Results
3.1. Changes in Soil Chemical Traits
3.2. Changes in Mycorrhizal Status of Rhizosphere and Endosphere
3.3. Correlation between Soil Properties and Mycorrhizal Traits
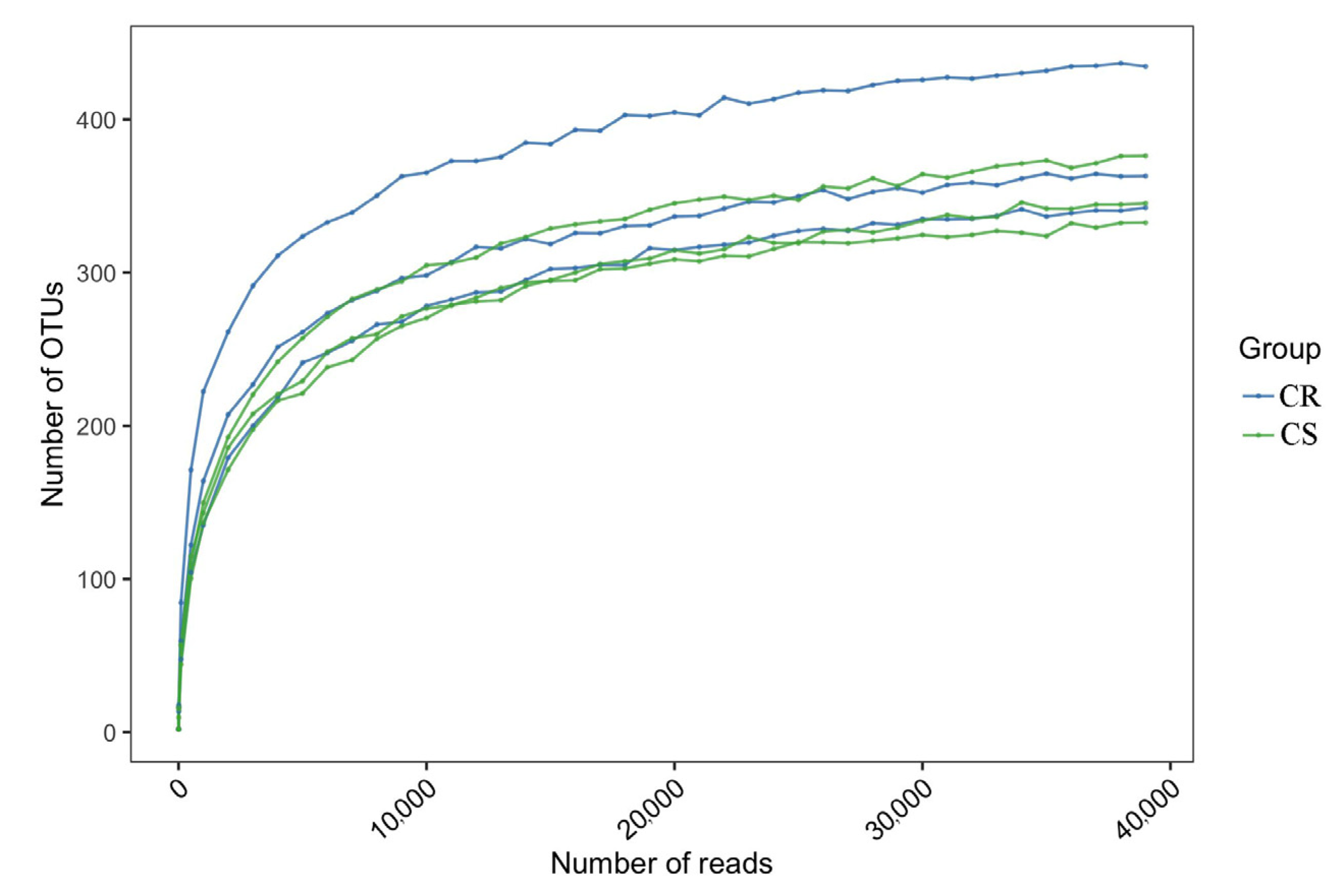
3.4. Sequencing Quality Analysis
3.5. Changes in OTUs in Endosphere and Rhizosphere
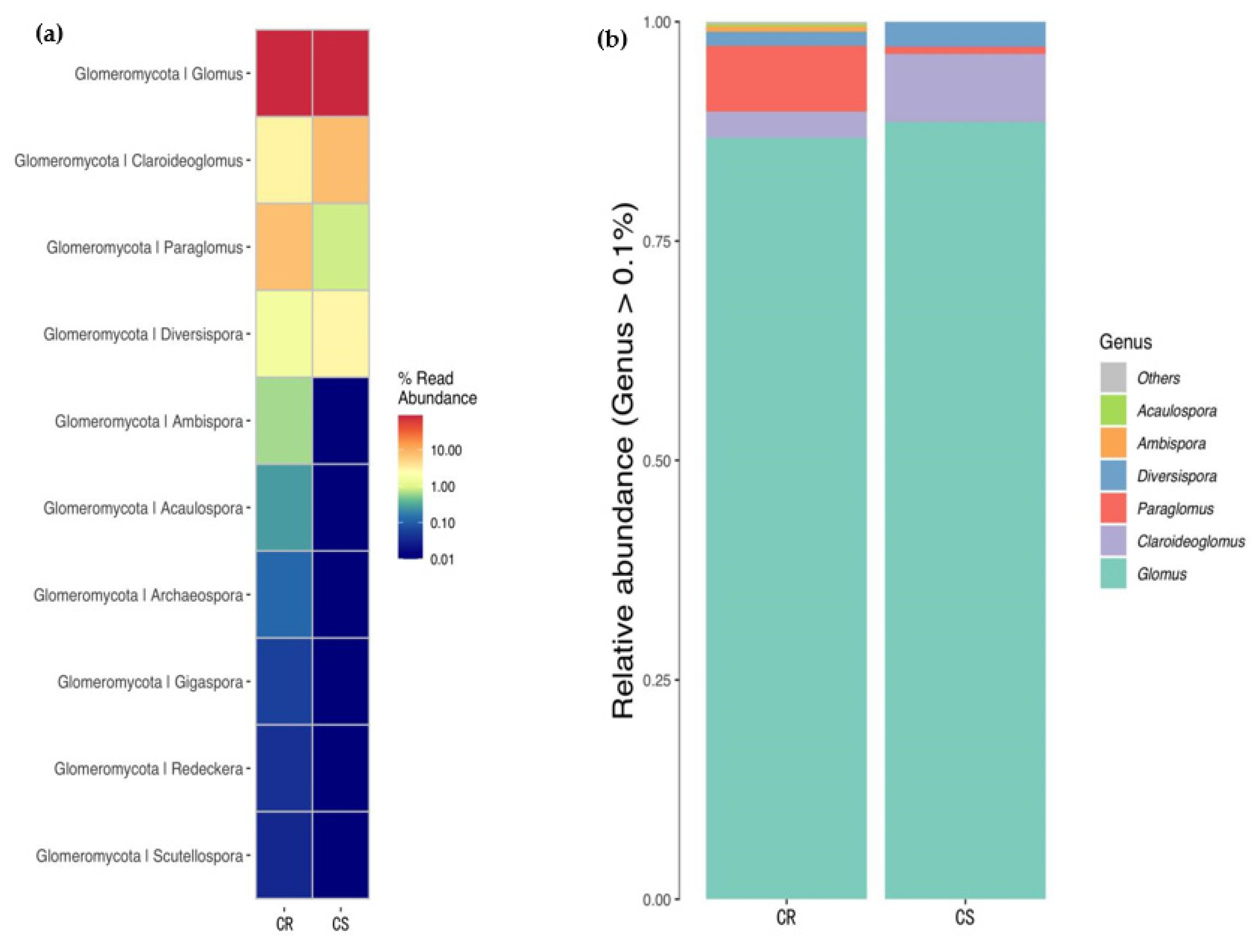
3.6. Changes in Relative Abundance of AMF in the Genus Level
3.7. Changes in Alpha Diversity Index of AMF
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, S.; Li, X.-Z. Inhibition of α-glucosidase by polysaccharides from the fruit hull of Camellia oleifera Abel. Carbohydr. Polym. 2015, 115, 38–43. [Google Scholar] [CrossRef]
- Lee, C.-P.; Shih, P.-H.; Hsu, C.-L.; Yen, G.-C. Hepatoprotection of tea seed oil (Camellia oleifera Abel.) against CCl4-induced oxidative damage in rats. Food Chem. Toxicol. 2007, 45, 888–895. [Google Scholar] [CrossRef]
- Lin, P.; Wang, K.; Zhou, C.; Xie, Y.; Yao, X.; Yin, H. Seed transcriptomics analysis in Camellia oleifera uncovers genes associated with oil content and fatty acid composition. Int. J. Mol. Sci. 2018, 19, 118. [Google Scholar] [CrossRef]
- Li, T.; Zhang, H.; Wu, C.-E. Screening of antioxidant and antitumor activities of major ingredients from defatted Camellia oleifera seeds. Food Sci. Biotechnol. 2014, 23, 873–880. [Google Scholar] [CrossRef]
- Zhang, Z.; Meng, J.; Pan, D.; Yang, C.; Li, Y. Mating system and progeny genetic diversity of Camellia oleifera ‘Ruan Zhi’. J. For. Res. 2018, 30, 1805–1810. [Google Scholar] [CrossRef]
- Peng, J.J.; Zhong, Z.X.; Li, T.T.; Wang, B.M. The effect of bio-fertilizer on the fruit and yield of Camellia oleifera. Anhui Agric. Sci. Bull. 2020, 26, 90–94. [Google Scholar]
- Zhang, F.; Zou, Y.N.; Wu, Q.S.; Kuča, K. Arbuscular mycorrhizas modulate root polyamine metabolism to enhance drought tolerance of trifoliate orange. Environ. Exp. Bot. 2020, 171, 103962. [Google Scholar] [CrossRef]
- He, J.-D.; Chi, G.-G.; Zou, Y.-N.; Shu, B.; Wu, Q.-S.; Srivastava, A.; Kuča, K. Contribution of glomalin-related soil proteins to soil organic carbon in trifoliate orange. Appl. Soil Ecol. 2020, 154, 103592. [Google Scholar] [CrossRef]
- Wu, Q.S.; Gao, W.Q.; Srivastava, A.K.; Zhang, F.; Zou, Y.N. Nutrient acquisition and fruit quality of Ponkan mandarin in response to AMF inoculation. Indian J. Agric. Sci. 2020, 90, 1563–1567. [Google Scholar]
- Cheng, H.-Q.; Giri, B.; Wu, Q.-S.; Zou, Y.-N.; Kuča, K. Arbuscular mycorrhizal fungi mitigate drought stress in citrus by modulating root microenvironment. Arch. Agron. Soil Sci. 2021. [Google Scholar] [CrossRef]
- Yang, L.; Zou, Y.-N.; Tian, Z.-H.; Wu, Q.-S.; Kuča, K. Effects of beneficial endophytic fungal inoculants on plant growth and nutrient absorption of trifoliate orange seedlings. Sci. Hortic. 2021, 277, 109815. [Google Scholar] [CrossRef]
- Zou, Y.-N.; Zhang, F.; Srivastava, A.K.; Wu, Q.-S.; Kuča, K. Arbuscular mycorrhizal fungi regulate polyamine homeostasis in roots of trifoliate orange for improved adaptation to soil moisture deficit stress. Front. Plant Sci. 2021, 11, 600792. [Google Scholar] [CrossRef]
- Wu, Q.S.; Shao, Y.D.; Gao, X.B.; Xia, T.J.; Kuča, K. Characterization of AMF-diversity of endosphere versus rhizosphere of tea (Camellia sinensis) crops. Indian J. Agric. Sci. 2019, 89, 348–352. [Google Scholar]
- Li, X.Q.; Chen, Y.C. Relationship between endophytic mycorrhizal distribution of Camellia oleifera and environmental condi-tion. For. Sci. Technol. 1982, 11, 20–21. [Google Scholar]
- Wang, D.X.; Zhang, N.Y.; Chen, G.C. Effects of AM fungi on the growth and drought-resistance of Camellia oleifera. Guangxi For. Sci. 2011, 40, 259–261, 273. [Google Scholar]
- Lin, Y.L.; Li, Z.Y.; Zhang, L.P.; Wu, F.; Yang, Y.; Tan, M.X.; Hu, D.N. Effects of organic phosphorus and AM fungi on growth, root morphology and photosynthetic characteristics of Camellia oleifera. Non-Wood For. Res. 2021, 39, 121–128. [Google Scholar]
- Jiang, S.; Liu, Y.; Luo, J.; Qin, M.; Johnson, N.C.; Öpik, M.; Vasar, M.; Chai, Y.; Zhou, X.; Mao, L.; et al. Dynamics of arbuscular mycorrhizal fungal community structure and functioning along a nitrogen enrichment gradient in an alpine meadow ecosystem. New Phytol. 2018, 220, 1222–1235. [Google Scholar] [CrossRef]
- Riley, D.; Barber, S.A. Bicarbonate accumulation and ph changes at the soybean (Glycine max (L.) Merr.) root-soil interface. Soil Sci. Soc. Am. J. 1969, 33, 905–908. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, P.; Zou, Y.-N.; Wu, Q.-S.; Kuča, K. Effects of mycorrhizal fungi on root-hair growth and hormone levels of taproot and lateral roots in trifoliate orange under drought stress. Arch. Agron. Soil Sci. 2019, 65, 1316–1330. [Google Scholar] [CrossRef]
- Daniels, B.A.; Skipper, H.D. Methods for the recovery and quantitative estimation of propagules from soil. In Methods and Principles of Mycorrhizal Research; Schenck, N.C., Ed.; American Phytopathological Society: St. Paul, MN, USA, 1982; pp. 29–35. [Google Scholar]
- Bethlenfalvay, G.J.; Ames, R.N. Comparison of two methods for quantifying extraradical mycelium of vesicular-arbuscular mycorrhizal fungi. Soil Sci. Soc. Am. J. 1987, 51, 834–837. [Google Scholar] [CrossRef]
- Mago, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Oberauner, L.; Zachow, C.; Lackner, S.; Högenauer, C.; Smolle, K.H.; Berg, G. The ignored diversity: Complex bacterial com-munities in intensive care units revealed by 16S pyrosequencing. Sci. Rep. 2013, 3, 1413. [Google Scholar] [CrossRef] [PubMed]
- Tuomisto, H. A diversity of beta diversities: Straightening up a concept gone awry. Part 1. Defining beta diversity as a func-tion of alpha and gamma diversity. Ecography 2010, 33, 2–22. [Google Scholar] [CrossRef]
- Wu, Q.S.; He, X.H.; Zou, Y.N.; He, K.P.; Sun, Y.H.; Cao, M.Q. Spatial distribution of glomalin-related soil protein and its re-lationships with root mycorrhization, soil aggregates, carbohydrates, activity of protease and β-glucosidase in the rhizosphere of Citrus unshiu. Soil Biol. Biochem. 2012, 45, 181–183. [Google Scholar] [CrossRef]
- Brady, N.C.; Wei, R.R. Organisms and ecology of the soil. Nat. Prop. Soil 1996, 11, 328–360. [Google Scholar]
- Wu, Q.S.; Wang, S.; Cao, M.Q.; Zou, Y.N.; Yao, Y.X. Tempo-spatial distribution and related functionings of root glomalin and glomalin-related soil protein in a citrus rhizosphere. J. Anim. Plant Sci. 2014, 24, 245–251. [Google Scholar]
- Guo, X.; Gong, J. Differential effects of abiotic factors and host plant traits on diversity and community composition of root-colonizing arbuscular mycorrhizal fungi in a salt-stressed ecosystem. Mycorrhiza 2014, 24, 79–94. [Google Scholar] [CrossRef]
- Gai, J.P.; Feng, G.; Li, X.L. Diversity of arbuscular mycorrhizal fungi in field soils from North China. Biodivers. Sci. 2004, 12, 435–440. [Google Scholar]
- Liu, Z.K.; Tian, S.; Tang, M. Spatial distribution of arbuscular mycorrhizal fungi and its relationship with soil factors in the rhizosphere of Robinia pseudoacacia at different ages. Sci. Silvae Sin. 2013, 49, 89–95. [Google Scholar]
- Singh, S.; Pandey, A.; Chaurasia, B.; Palni, L.M.S. Diversity of arbuscular mycorrhizal fungi associated with the rhizosphere of tea growing in ‘natural’ and ‘cultivated’ ecosites. Biol. Fertil. Soils 2008, 44, 491–500. [Google Scholar] [CrossRef]
- Rodrigues, K.M.; Rodrigues, B.F. Glomus. In Beneficial Microbes in Agro-Ecology: Bateria and Fungi; Amaresan, N., Kumar, S.M., Annapurna, K., Kumar, K., Sankaranarayanan, A., Eds.; Academic Press: London, UK, 2020; pp. 561–569. [Google Scholar]
- Redecker, D.; Schüβler, A.; Stockinger, H.; Stürmer, S.L.; Morton, J.B.; Walker, C. An evidence-based consensus for the classi-fication of arbuscular mycorrhizal fungi (Glomeromycota). Mycorrhiza 2013, 23, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.S.; Sun, P.; Srivastava, A.K. AMF diversity in citrus rhizosphere. Indian J. Agric. Sci. 2017, 87, 653–659. [Google Scholar]

| Soil Samples | pH | NH4+-N Content (mg/kg) | NO3−-N Content (mg/kg) | Available K Content (mg/kg) | Olsen-P Content (mg/kg) |
|---|---|---|---|---|---|
| S1 | 6.13 ± 0.10 b | 78.88 ± 9.31 a | 130.43 ± 31.55 a | 215.68 ± 35.15 a | 104.96 ± 36.03 a |
| S2 | 6.51 ± 0.09 a | 61.68 ± 7.47 b | 74.62 ± 21.64 b | 143.85 ± 38.51 b | 102.50 ± 13.95 a |
| Samples | Root AMF Colonization (%) | Soil Spore Density (#/10 g soil) | Soil Hyphal Length (cm/g) | Soil EE-GRSP Level (mg/g) |
|---|---|---|---|---|
| S1 | 41.68 ± 7.01 a | 111.25 ± 12.56 a | 8.94 ± 0.89 a | 0.64 ± 0.02 a |
| S2 | 30.73 ± 4.27 b | 66.33 ± 8.14 b | 3.99 ± 0.34 b | 0.53 ± 0.06 b |
| Root Colonization | Soil Spore Density | Soil Hyphal Length | EE-GRSP | pH | Soil NH4+-N | Soil NO3−-N | Soil Available K | Soil Olsen-P | |
|---|---|---|---|---|---|---|---|---|---|
| Root colonization | 1.00 | 0.80 * | 0.68 | 0.433 | −0.77 * | 0.30 | 0.56 | 0.74 * | −0.08 |
| Soil spore density | 1.00 | 0.95 ** | 0.682 | −0.92 ** | 0.59 | 0.89 ** | 0.65 | −0.08 | |
| Soil hyphal length | 1.00 | 0.74 * | −0.93 ** | 0.68 | 0.84 ** | 0.61 | −0.05 | ||
| EE-GRSP | 1.00 | −0.84 ** | 0.68 | 0.71 * | 0.73 * | −0.11 |
| Samples | Sample Number | Raw PE | Clean Tags | Effective Tags | Effective Rates (%) | Average Length (nt) |
|---|---|---|---|---|---|---|
| Root | CR1 | 47,469 | 44,188 | 43,030 | 90.65 | 263 |
| CR2 | 45,960 | 41,329 | 40,105 | 87.26 | 262 | |
| CR3 | 44,237 | 39,958 | 35,431 | 86.88 | 261 | |
| Soil | CS1 | 43,739 | 40,814 | 39,320 | 89.90 | 262 |
| CS2 | 42,021 | 39,852 | 38,437 | 91.47 | 261 | |
| CS3 | 47,477 | 43,361 | 42,042 | 88.55 | 262 |
| Observed | Chao1 | ACE Estimator | Shannon | Simpson | Phylogenetic Diversity | |
|---|---|---|---|---|---|---|
| CR | 257.67 ± 14.57 a | 286.81 ± 10.99 a | 292.10 ± 5.16 a | 3.39 ± 0.27 a | 0.93 ± 0.03 a | 12.70 ± 1.14 a |
| CS | 234.67 ± 12.50 a | 286.73 ± 9.65 a | 279.92 ± 8.87 a | 2.85 ± 0.13 b | 0.83 ± 0.05 b | 10.56 ± 0.53 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.-C.; Xiao, Z.-Y.; Hashem, A.; Abd_Allah, E.F.; Wu, Q.-S. Mycorrhizal Fungal Diversity and Its Relationship with Soil Properties in Camellia oleifera. Agriculture 2021, 11, 470. https://doi.org/10.3390/agriculture11060470
Liu R-C, Xiao Z-Y, Hashem A, Abd_Allah EF, Wu Q-S. Mycorrhizal Fungal Diversity and Its Relationship with Soil Properties in Camellia oleifera. Agriculture. 2021; 11(6):470. https://doi.org/10.3390/agriculture11060470
Chicago/Turabian StyleLiu, Rui-Cheng, Zhi-Yan Xiao, Abeer Hashem, Elsayed Fathi Abd_Allah, and Qiang-Sheng Wu. 2021. "Mycorrhizal Fungal Diversity and Its Relationship with Soil Properties in Camellia oleifera" Agriculture 11, no. 6: 470. https://doi.org/10.3390/agriculture11060470
APA StyleLiu, R.-C., Xiao, Z.-Y., Hashem, A., Abd_Allah, E. F., & Wu, Q.-S. (2021). Mycorrhizal Fungal Diversity and Its Relationship with Soil Properties in Camellia oleifera. Agriculture, 11(6), 470. https://doi.org/10.3390/agriculture11060470








