The World Vegetable Center Okra (Abelmoschus esculentus) Core Collection as a Source for Flooding Stress Tolerance Traits for Breeding
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Genotyping, Core Collection Selection
2.2. Phenotyping Flooding Stress Tolerance
3. Results
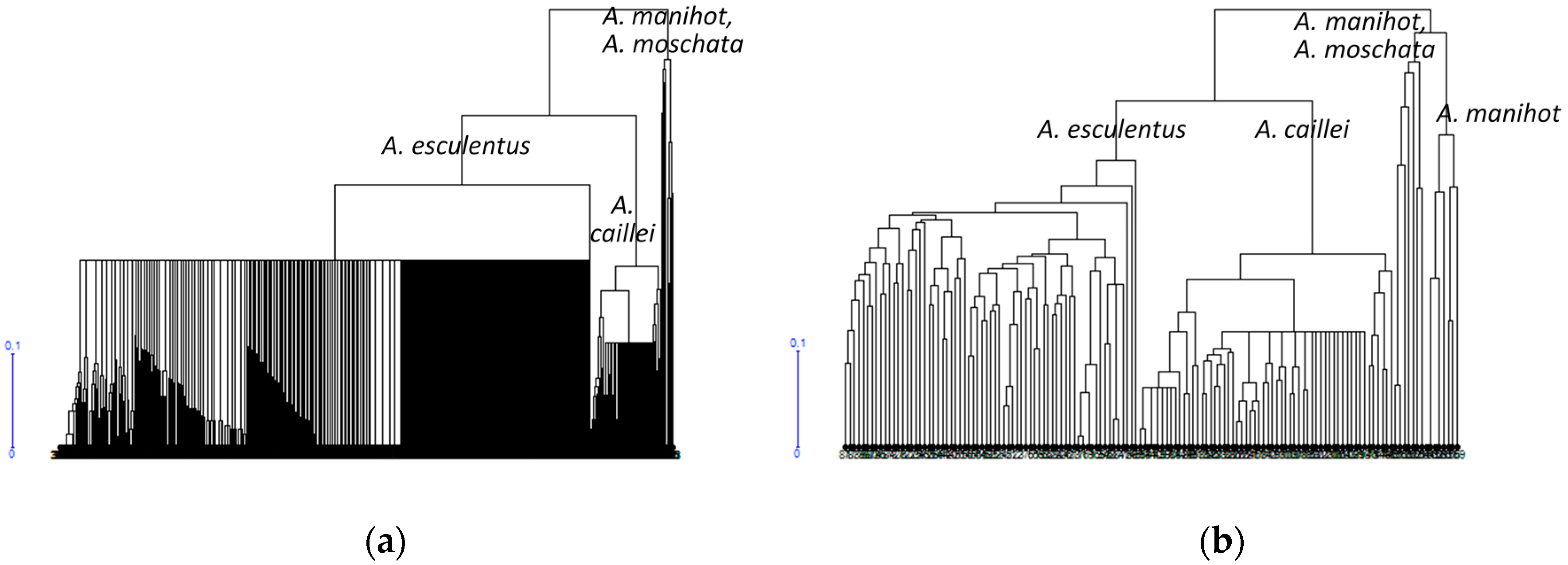
3.1. Diversity of the Okra Genebank Collection
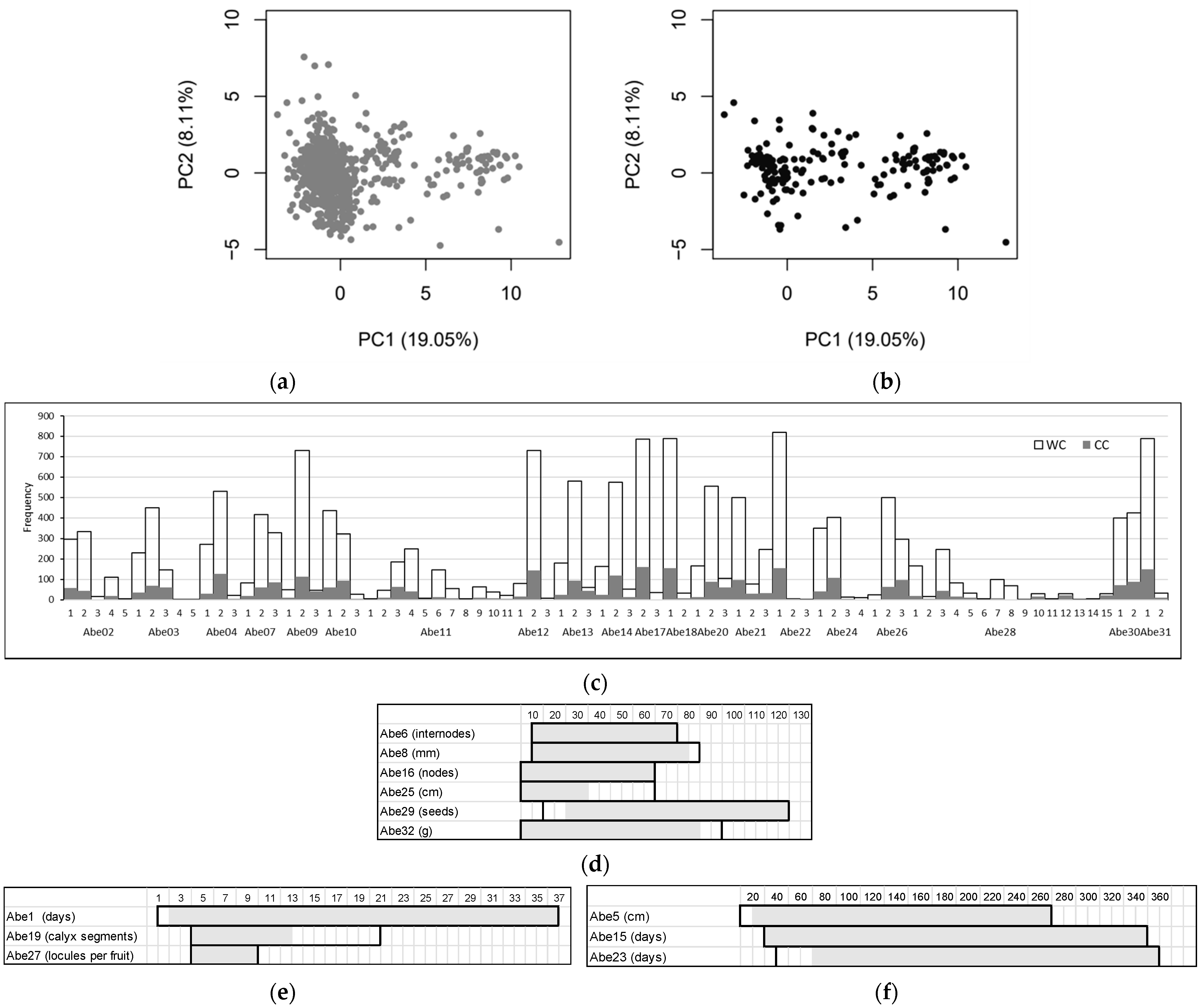
3.2. Okra Core Collection
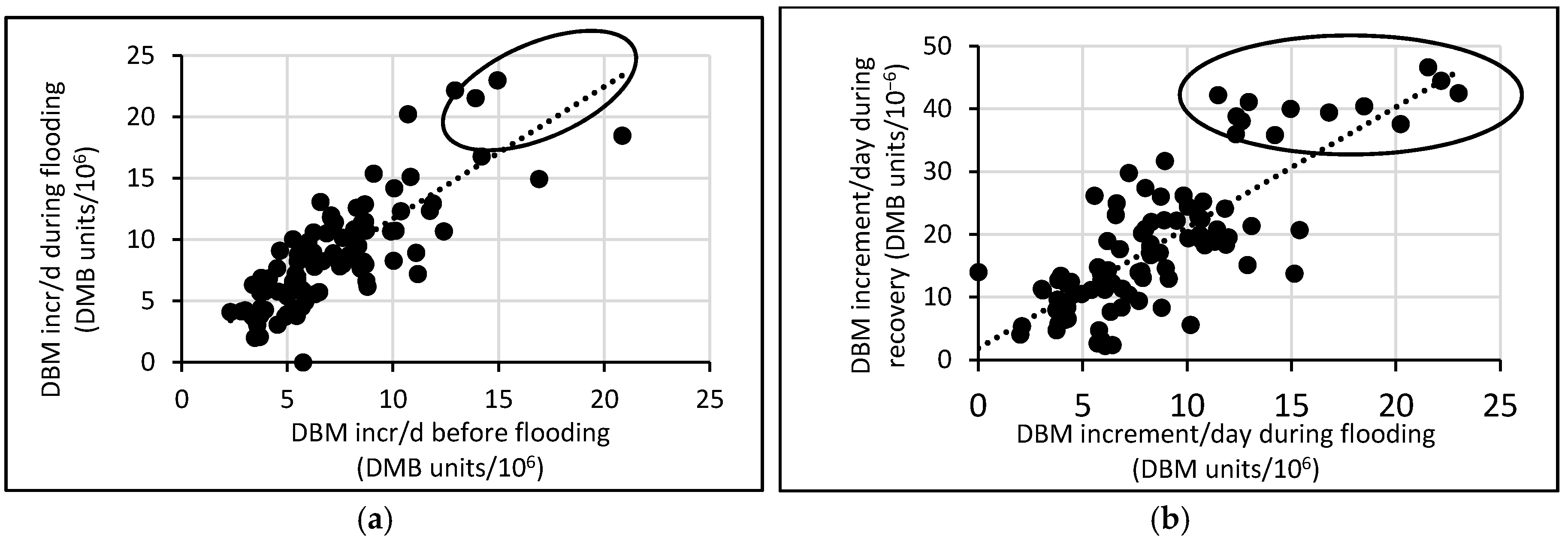
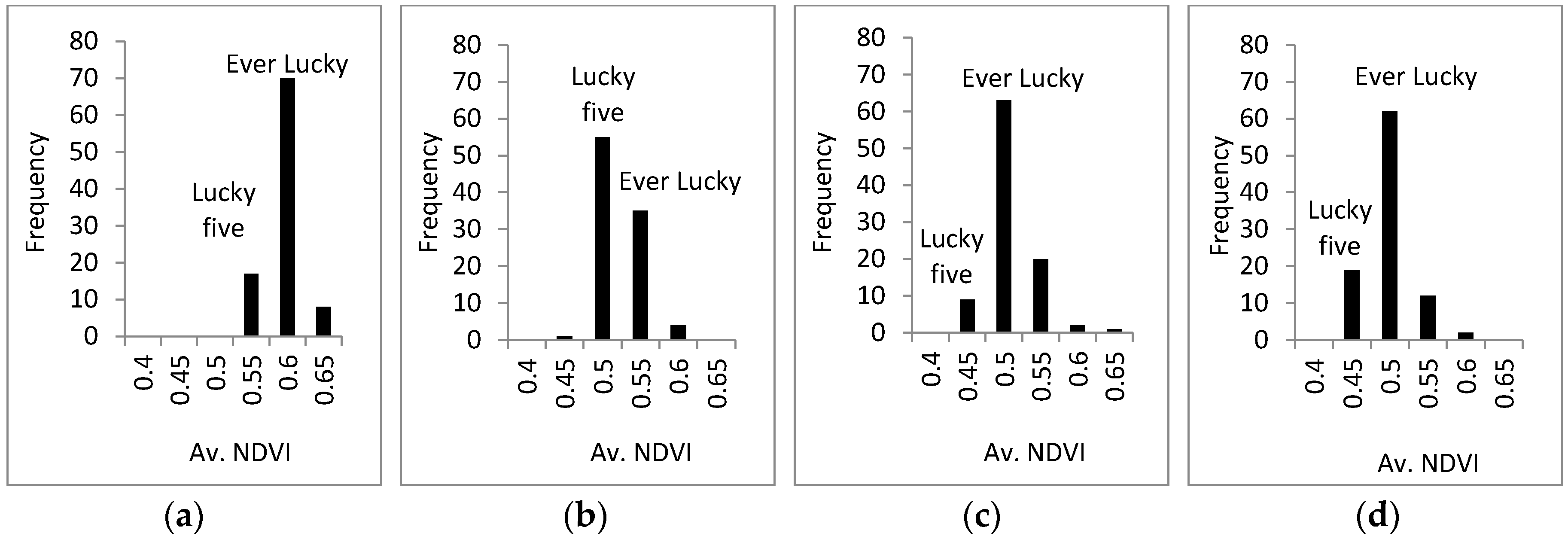
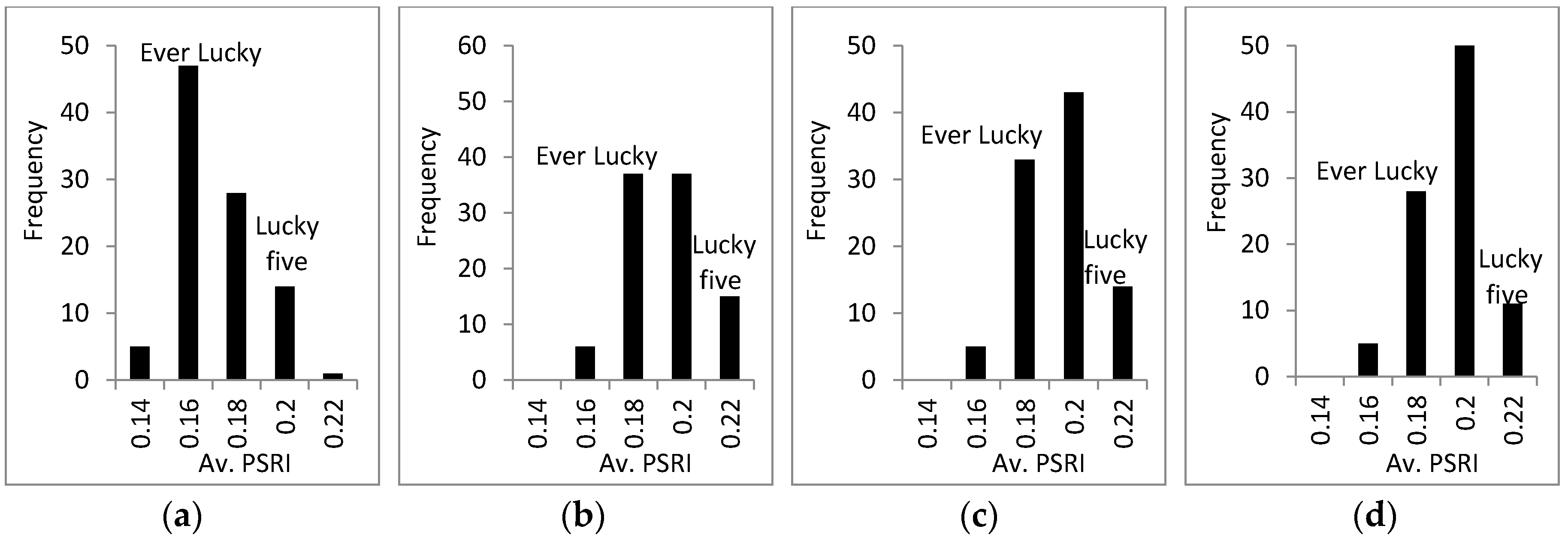
3.3. Response of A. Esculentus to Flooding
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Joshi, A.B.; Gadwal, V.R.; Hardas, M.W. Okra. In Evolutionary Studies in World Crops, Diversity and Change in the Indian Subcontinent; Hutchinson, J.B., Ed.; Cambridge University Press: Cambridge, UK, 1974; pp. 99–105. [Google Scholar]
- Vavilov, N.I. Studies on the Origin of Cultivated Plants; L’Institut de Botanique Appliquee Amélioration Plantes: Leningrad, Russian, 1926. [Google Scholar]
- Van Borssum Waalkes, J. Malesian Malvaceae revised. Blumea 1966, 14, 1–213. [Google Scholar]
- Bates, D.M. Notes on the cultivated Malvaceae 2. Abelmoschus. Baileya 1968, 26, 99–112. [Google Scholar]
- Chheda, H.R.; Fatokun, C.A. Numerical analysis of variation patterns in okra (Abelmoschus esculentus L. Moench). Bot. Gaz. 1982, 143, 253–261. [Google Scholar] [CrossRef]
- Lamont, W.J. Okra—A versatile vegetable crop. Horttechnology 1999, 9, 179–184. [Google Scholar] [CrossRef]
- Reddy, M.T. Crossability behaviour and fertility restoration through colchiploidy in interspecific hybrids of Abelmoschus esculentus × Abelmoschus manihot subsp. Tetraphyllus. Int. J. Plant Sci. Ecol. 2015, 1, 172–181. [Google Scholar]
- Amitaaba, T. Hybidization Studies in Okra (Abelmoschus spp. (L.) Moench). Ph.D. Thesis, University of Ghana, Accra, Ghana, July 2016. [Google Scholar]
- Chevalier, A. L’origin, la culture et les usage de cinq Hibiscus de la section Abelmoschus. Rev. Bot. Appl. Agric. Trop. 1940, 20, 319–328. [Google Scholar] [CrossRef]
- Cornucopia, F.S. A Source Book of Edible Plants; Kampong Publications: Vista, CA, USA, 1990. [Google Scholar]
- Luan, F.; Wu, Q.; Zeng, N.; Lv, H.; Liu, D.; Gan, Z. Traditional uses, chemical constituents, and biological properties, clinical settings, and toxicities of Abelmoschus manihot L.: A comprehensive review. Front. Pharmacol. 2020, 11, 1068. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. 2018. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 18 November 2020).
- Moosavi, S.A.; Aghaalikhani, M.; Ghobadian, B.; Fayyazi, E. Okra: A potential future bioenergy crop in Iran. Renew. Sust. Energy Rev. 2018, 93, 517–524. [Google Scholar] [CrossRef]
- Anwar, F.; Qadir, R.; Ahmad, N. Cold pressed okra (Abelmoschus esculentus) seed oil. In Cold Pressed Oils; Ramadan, M.F., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 309–314. [Google Scholar] [CrossRef]
- Das, S.; Nandi, G.; Ghosh, L.K. Okra and its various applications in drug delivery, food technology, health care and pharmacological aspects-a review. Int. J. Pharm. Sci. Res. 2019, 11, 2139–2147. [Google Scholar]
- Chowdhury, S.; Kumar, S. Okra breeding: Recent approaches and constraints. Ann. Biol. 2019, 35, 55–60. [Google Scholar]
- Van Hintum, T.J.; Brown, A.H.; Spillane, C. Core Collections of Plant Genetic Resources; IPGRI Technical Bulletin No. 3; International Plant Genetic Resources Institute: Rome, Italy, 2000. [Google Scholar]
- Ahmad, J.; Balal, R.M.; Shahid, M.A.; Akhtar, G.; Akram, A.; Khan, M.W.; Zubair, M. Characterization of okra genotypes at reproductive stage under high temperature stress. Int. J. Chem. Biochem. Sci. 2016, 9, 44–48. [Google Scholar]
- Zhao, Y.; Wang, C.; Wang, S.; Tibig, L.V. Impacts of present and future climate variability on agriculture and forestry in the humid and sub-humid tropics. Clim. Chang. 2005, 70, 73–116. [Google Scholar] [CrossRef]
- Zhou, W.; Chen, F.; Meng, Y.; Chandrasekaran, U.; Luo, X.; Yang, W.; Shu, K. Plant waterlogging/flooding stress responses: From seed germination to maturation. Plant Physiol. Biochem. 2020, 148, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Polanco, M.; Senorans, J.; Zwiazek, J.J. Role of adventitious roots in water relations of tamarack (Larix laricina) seedlings exposed to flooding. BMC Plant Biol. 2012, 12, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Arfan, M. Gas exchange characteristics and water relations in two cultivars of Hibiscus esculentus under waterlogging. Biol. Plant. 2005, 49, 459–462. [Google Scholar] [CrossRef]
- Vwioko, E.D.; El-Esawi, M.A.; Imoni, M.E.; Al-Ghamdi, A.A.; Ali, H.M.; El-Sheekh, M.M.; Abdeldaym, E.A.; Al-Dosary, M.A. Sodium azide priming enhances waterlogging stress tolerance in okra (Abelmoschus esculentus L.). Agronomy 2019, 9, 679. [Google Scholar] [CrossRef]
- Pratap, A.; Gupta, S.; Nair, R.M.; Gupta, S.K.; Schafleitner, R.; Basu, P.S.; Singh, C.M.; Prajapati, U.; Gupta, A.K.; Nayyar, H.; et al. Using plant phenomics to exploit the gains of genomics. Agronomy 2019, 9, 126. [Google Scholar] [CrossRef]
- Schafleitner, R.; Kumar, S.; Lin, C.Y.; Hegde, S.G.; Ebert, A. The okra (Abelmoschus esculentus) transcriptome as a source for gene sequence information and molecular markers for diversity analysis. Gene 2013, 517, 27–36. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr/darwin (accessed on 2 November 2020).
- De Beukelaer, H.; Smýkal, P.; Davenport, G.F.; Fack, V. Core Hunter II: Fast core subset selection based on multiple genetic diversity measures using Mixed Replica search. BMC Bioinform. 2012, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Chavent, M.; Kuentz-Simonet, V.; Labenne, A.; Saracco, J. Multivariate analysis of mixed data: The PCAmixdata R package. arXiv 2014, arXiv:1411.4911, 132. [Google Scholar]
- Mahajan, R.K.; Bisht, I.S.; Agrawal, R.C.; Rana, R.S. Studies on South Asian okra collection: Methodology for establishing a representative core set using characterization data. Genet. Resour. Crop. Evol. 1996, 43, 249–255. [Google Scholar] [CrossRef]
- Setter, T.L.H.; Khabaz-Saberi, W.I.; Singh, K.N.; Kulshreshtha, N.; Sharma, S.K. Review of waterlogging tolerance in wheat in India: Involvement of element/microelement toxicities, relevant to yield plateau and opportunities for crop management. In International Symposium on Balanced Fertilization; Ludhiana Benbi, D.K., Brar, M.S., Bansal, S.K., Eds.; Punjab Agricultural University: Ludhiana, India, 2006; pp. 513–520. [Google Scholar]
- Chittora, A.; Singh, N. Production technology of okra. Marumegh 2016, 1, 48–51. [Google Scholar]
- Caudle, K.L.; Maricle, B.R. Effects of flooding on photosynthesis, chlorophyll fluorescence, and oxygen stress in plants of varying flooding tolerance. Trans. Kans. Acad. Sci. 2012, 115, 5–18. [Google Scholar] [CrossRef]
- Flood, P.J.; Harbinson, J.; Aarts, M.G. Natural genetic variation in plant photosynthesis. Trends Plant Sci. 2011, 16, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Ehlert, D.; Horn, H.J.; Adamek, R. Measuring crop biomass density by laser triangulation. Comput. Electron. Agric. 2008, 61, 117–125. [Google Scholar] [CrossRef]
- Maphosa, L.; Thoday-Kennedy, E.; Vakani, J.; Phelan, A.; Badenhorst, P.; Slater, A.; Spangenberg, G.; Kant, S. Phenotyping wheat under salt stress conditions using a 3D laser scanner. Isr. J. Plant Sci. 2017, 64, 55–62. [Google Scholar] [CrossRef]
- Berger, A.; Ettlin, G.; Quincke, C.; Rodríguez-Bocca, P. Predicting the Normalized Difference Vegetation Index (NDVI) by training a crop growth model with historical data. Comput. Electron. Agric. 2019, 161, 305–311. [Google Scholar] [CrossRef]
- Holtgrave, A.K.; Röder, N.; Ackermann, A.; Erasmi, S.; Kleinschmit, B. Comparing Sentinel-1 and-2 Data and indices for agricultural land use monitoring. Remote Sens. 2020, 12, 2919. [Google Scholar] [CrossRef]
- Shrestha, R.; Di, L.; Eugene, G.Y.; Kang, L.; Shao, Y.Z.; Bai, Y.Q. Regression model to estimate flood impact on corn yield using MODIS NDVI and USDA cropland data layer. J. Integr. Agric. 2017, 16, 398–407. [Google Scholar] [CrossRef]
- Phyu, P.; Islam, M.R.; Sta Cruz, P.C.; Collard, B.C.Y.; Kato, Y. Use of NDVI for indirect selection of high yield in tropical rice breeding. Euphytica 2020, 216, 74. [Google Scholar] [CrossRef]
- Rizk, H.; Habib, M.K. Robotized early plant health monitoring system. In Proceedings of the IECON 2018-44th Annual Conference of the IEEE Industrial Electronics Society, Washington, DC, USA, 21 October 2018; pp. 3795–3800. [Google Scholar]
- Beisel, N.S.; Callaham, J.B.; Sng, N.J.; Taylor, D.J.; Paul, A.L.; Ferl, R.J. Utilization of single-image normalized difference vegetation index (SI-NDVI) for early plant stress detection. Appl. Plant Sci. 2018, 6, e01186. [Google Scholar] [CrossRef] [PubMed]
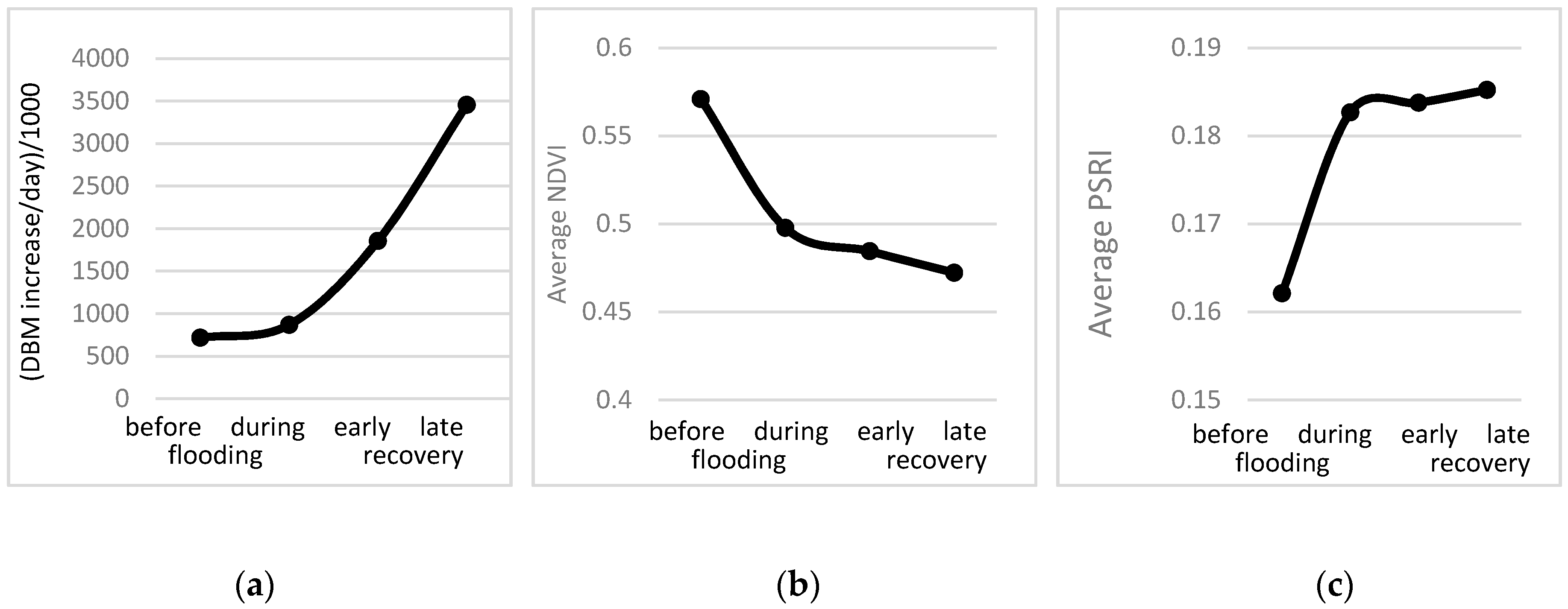
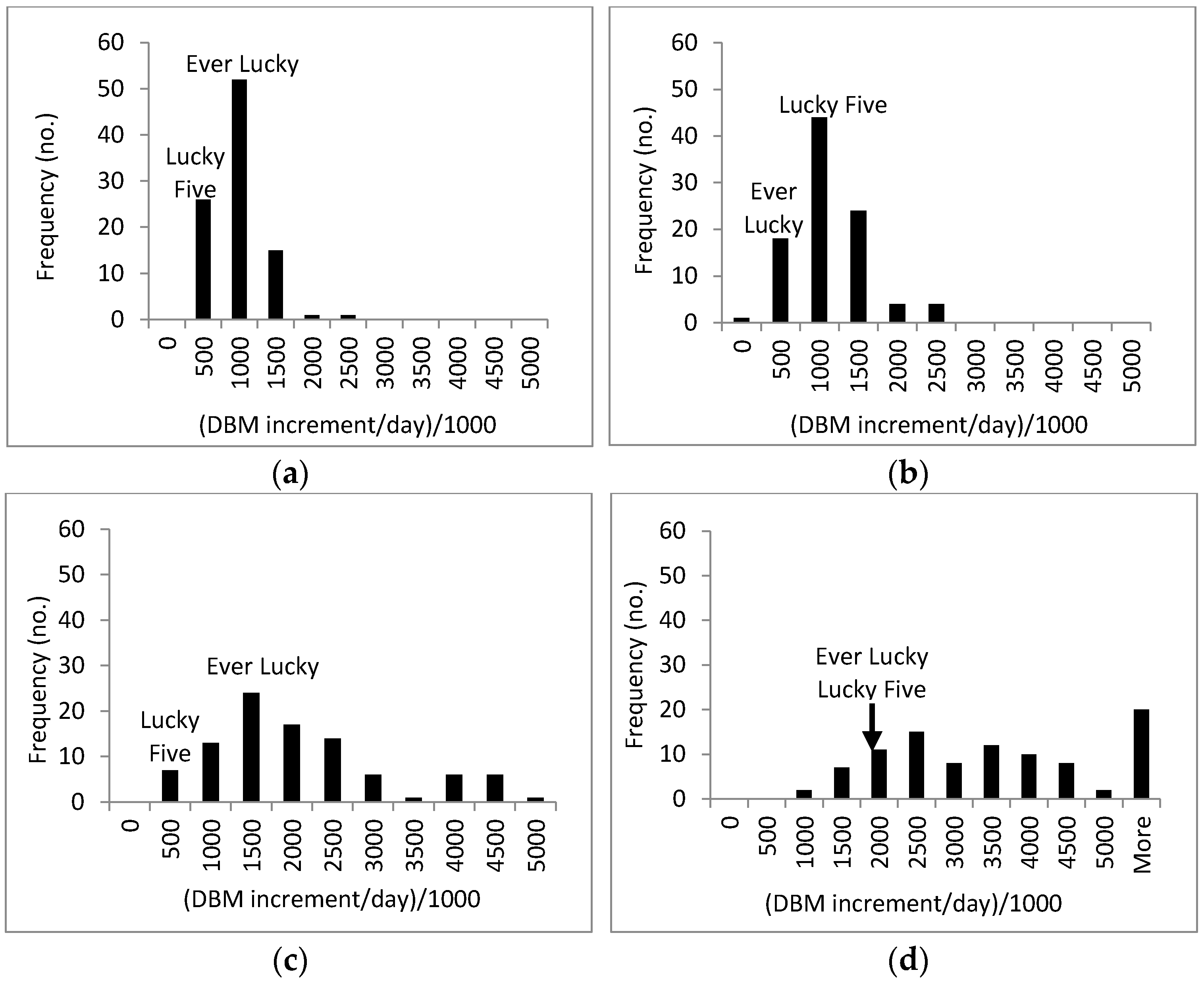
| Period (before, during and after Flooding) | Genotype | Replication | |
|---|---|---|---|
| DBM (AUC) | <0.001 * | <0.001 * | 0.6330 |
| NDVI average slope | <0.001 * | 0.001 * | 0.6241 |
| PSRI average slope | 0.1294 | 0.0353 * | 0.1453 |
| Accession | Origin | Flowering and Fruit Setting before 65 Days after Sowing | Maintained Growth under Flooding | Good Recovery after Flooding | Maintained NDVI under Flooding | Low Increase of PSRI under Flooding |
|---|---|---|---|---|---|---|
| VI033791 | Malaysia | √ | √ | |||
| VI047518 | Bangladesh | √ | √ | |||
| VI050170 | Taiwan | √ | √ | |||
| VI055884 | Laos | √ | ||||
| VI056451 | USA | √ | ||||
| VI059479 | Malawi | √ | √ | |||
| VI060132 | Mali | √ | √ | |||
| VI060690B | Benin | √ | √ | |||
| VI060739A | Thailand | √ | √ | |||
| VI060748A | Philippines | √ | ||||
| VI060784 | USA (heirloom variety “Burgundy”) | √ | √ | |||
| VI060801 | Turkey | √ | √ | |||
| VI060806 | Turkey (local variety “Amasya”) | √ | √ | |||
| VI060822 | Nigeria | √ | ||||
| VI060837B | Mali | √ | ||||
| VI060838B | Mali | √ | √ | |||
| VI060850 | Mali | √ | √ | |||
| VI061719 | Guinea | √ | √ | √ | ||
| VI061723 | Senegal | √ | √ | |||
| VI061750 | Senegal | √ | √ | |||
| VI061803 | Senegal | √ | ||||
| VI062547 | Niger (local variety “Gaya”) | √ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schafleitner, R.; Lin, C.-Y.; Lin, Y.-P.; Wu, T.-H.; Hung, C.-H.; Phooi, C.-L.; Chu, S.-H.; Jhong, Y.-C.; Hsiao, Y.-Y. The World Vegetable Center Okra (Abelmoschus esculentus) Core Collection as a Source for Flooding Stress Tolerance Traits for Breeding. Agriculture 2021, 11, 165. https://doi.org/10.3390/agriculture11020165
Schafleitner R, Lin C-Y, Lin Y-P, Wu T-H, Hung C-H, Phooi C-L, Chu S-H, Jhong Y-C, Hsiao Y-Y. The World Vegetable Center Okra (Abelmoschus esculentus) Core Collection as a Source for Flooding Stress Tolerance Traits for Breeding. Agriculture. 2021; 11(2):165. https://doi.org/10.3390/agriculture11020165
Chicago/Turabian StyleSchafleitner, Roland, Chen-Yu Lin, Ya-Ping Lin, Tien-Hor Wu, Cian-Huei Hung, Chooi-Lin Phooi, Shu-Hui Chu, Yu-Cen Jhong, and Yun-Yin Hsiao. 2021. "The World Vegetable Center Okra (Abelmoschus esculentus) Core Collection as a Source for Flooding Stress Tolerance Traits for Breeding" Agriculture 11, no. 2: 165. https://doi.org/10.3390/agriculture11020165
APA StyleSchafleitner, R., Lin, C.-Y., Lin, Y.-P., Wu, T.-H., Hung, C.-H., Phooi, C.-L., Chu, S.-H., Jhong, Y.-C., & Hsiao, Y.-Y. (2021). The World Vegetable Center Okra (Abelmoschus esculentus) Core Collection as a Source for Flooding Stress Tolerance Traits for Breeding. Agriculture, 11(2), 165. https://doi.org/10.3390/agriculture11020165








