QTL Analysis for Chlorophyll Content in Strawberry (Fragaria × ananassa Duch.) Leaves
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growing Conditions
2.2. Measurement of Chlorophyll Content
2.3. DNA Extraction and Genotyping
2.4. Genetic Map and QTL Detection
2.5. Candidate Gene Analysis
3. Results
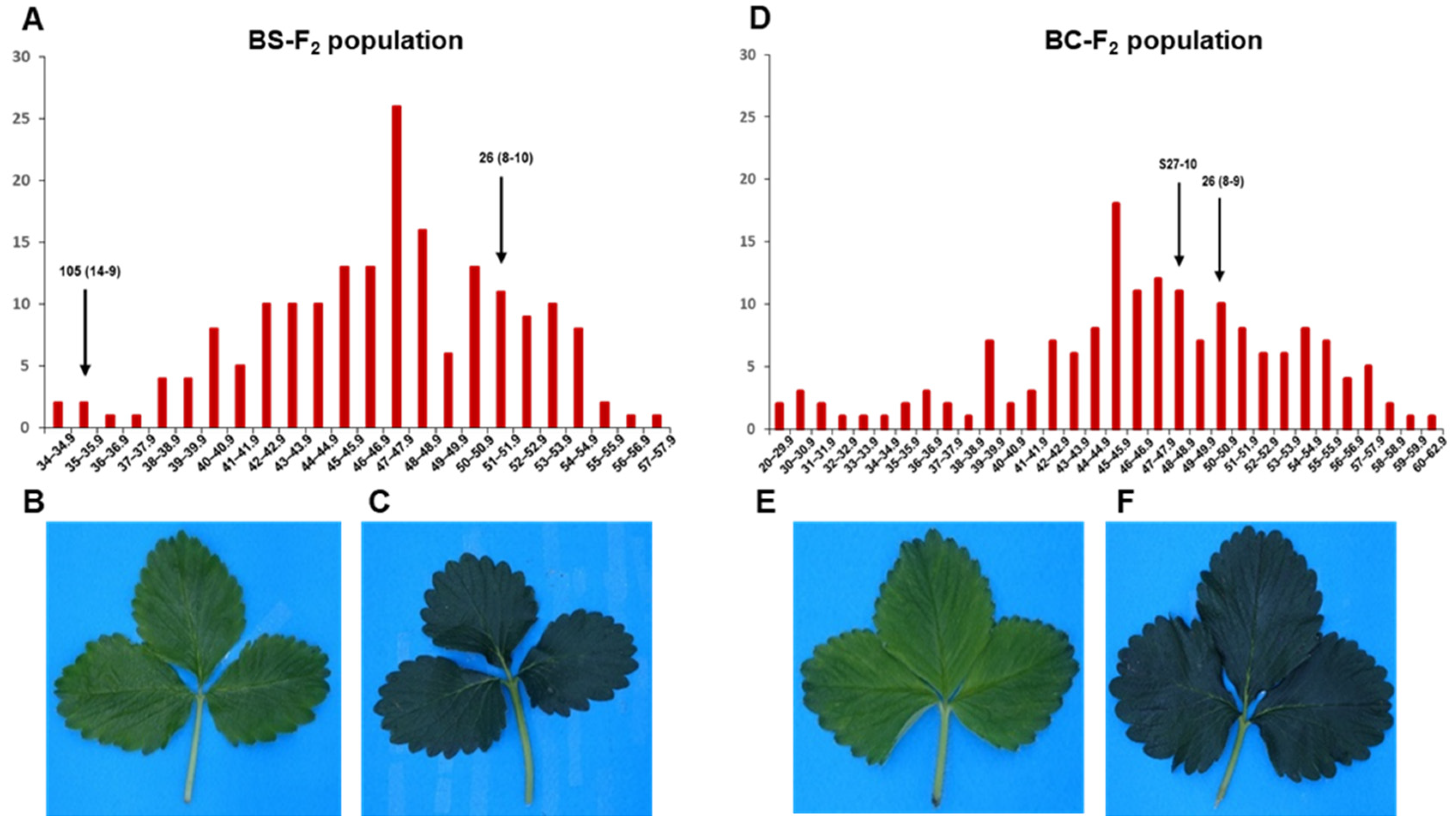
3.1. Phenotypic Analysis of Leaf Chlorophyll Content
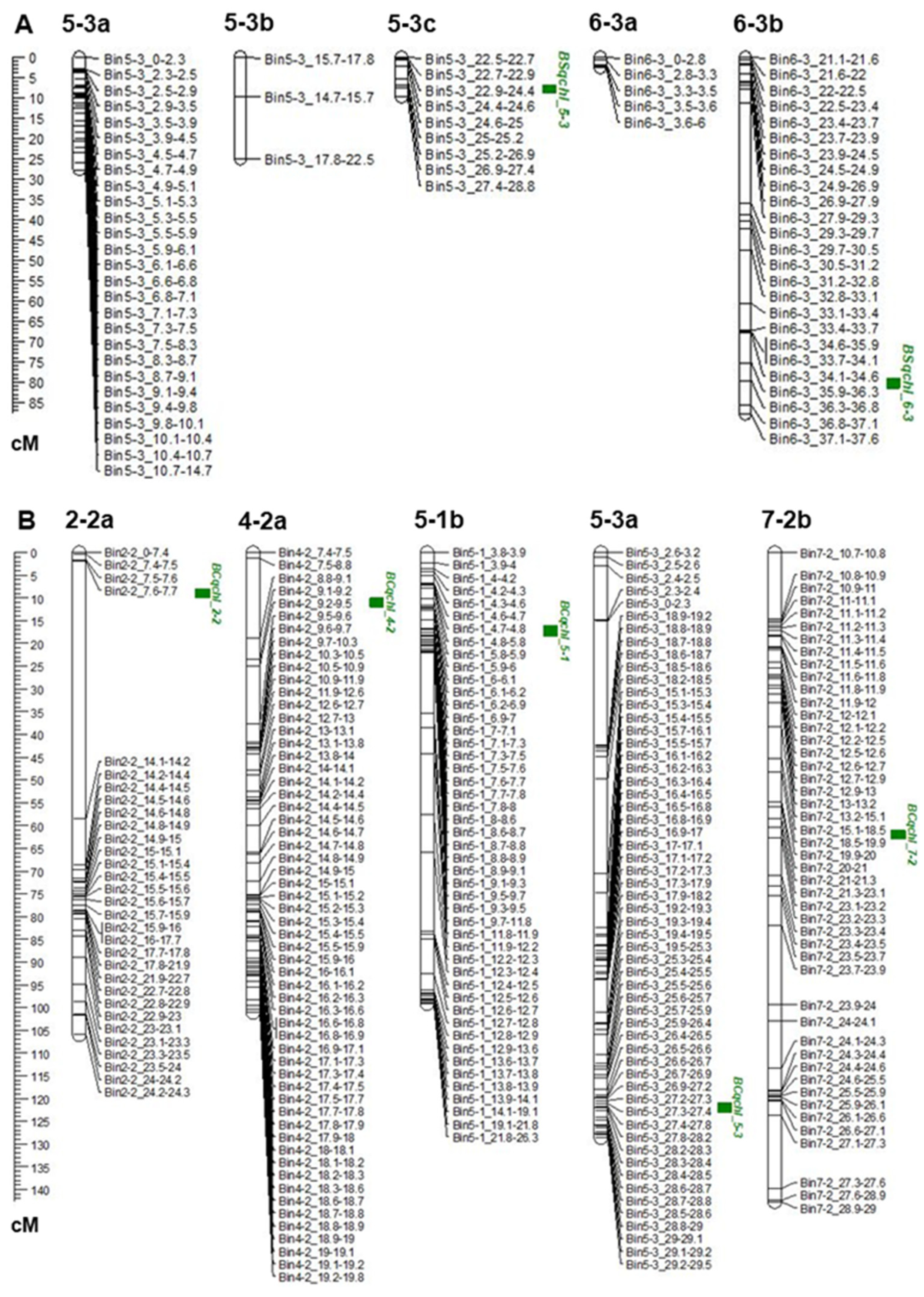
3.2. Construction of Bins and Linkage Map
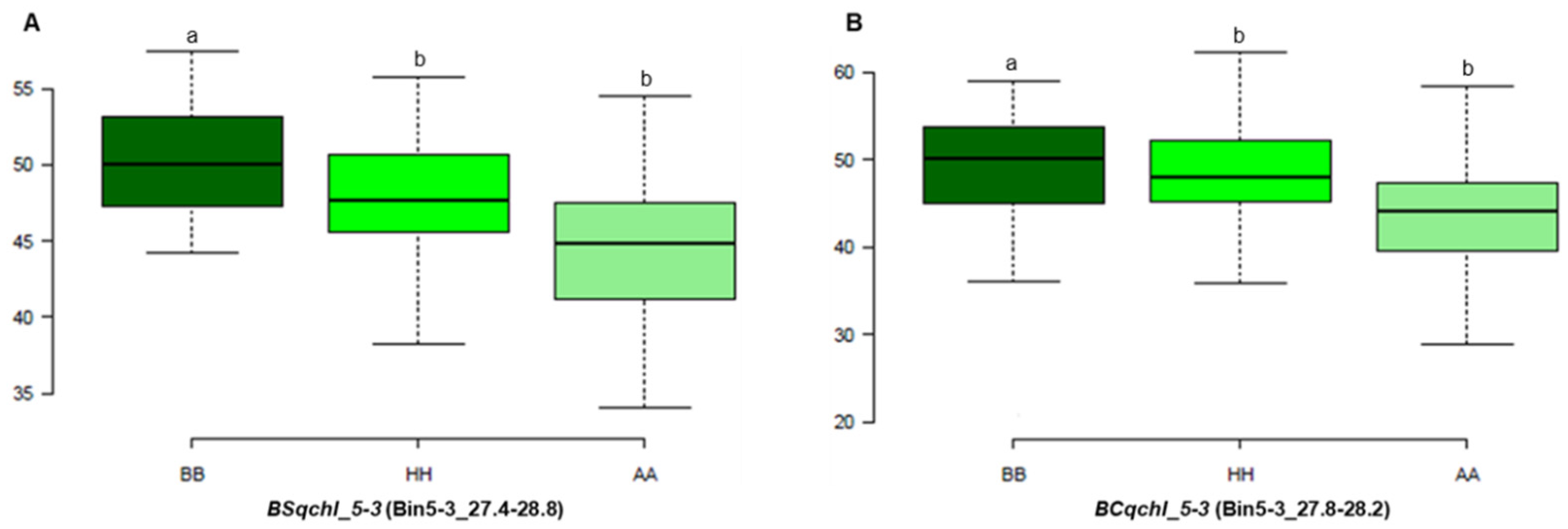
3.3. QTL Analysis and Detection
3.4. Candidate Gene Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Ye, J.; Liu, H.; Zhao, Z.; Xu, L.; Li, K.; Du, D. Fine mapping of the QTL cqSPDA2 for chlorophyll content in Brassica napus L. BMC Plant Biol. 2020, 20, 1–9. [Google Scholar] [CrossRef]
- Raines, C.A. Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: Current and future strategies. Plant Physiol. 2011, 155, 36–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Conteh, B.; Fang, L.; Xia, Q.; Nian, H. QTL mapping for soybean (Glycine max L.) leaf chlorophyll-content traits in a genotyped RIL population by using RAD-seq based high-density linkage map. BMC Genom. 2020, 21, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.A.; Knapp, A.K. Leaf optical properties in higher plants: Linking spectral characteristics to stress and chlorophyll concentration. Am. J. Bot. 2001, 88, 677–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Korff, M.; Grando, S.; Del Greco, A.; This, D.; Baum, M.; Ceccarelli, S. Quantitative trait loci associated with adaptation to Mediterranean dryland conditions in barley. Theor. Appl. Genet. 2008, 117, 653–669. [Google Scholar] [CrossRef] [PubMed]
- Mae, T. Physiological nitrogen efficiency in rice: Nitrogen utilization, photosynthesis, and yield potential. Plant Soil 1997, 196, 201–210. [Google Scholar] [CrossRef]
- Fiorentini, M.; Zenobi, S.; Giorgini, E.; Basili, D.; Conti, C.; Pro, C.; Monaci, E.; Orsini, R. Nitrogen and chlorophyll status determination in durum wheat as influenced by fertilization and soil management: Preliminary results. PLoS ONE 2019, 14, 1–16. [Google Scholar] [CrossRef]
- Meng, L.; Höfte, M.; Van Labeke, M.C. Leaf age and light quality influence the basal resistance against Botrytis cinerea in strawberry leaves. Environ. Exp. Bot. 2019, 157, 35–45. [Google Scholar] [CrossRef]
- Xu, L.; Liu, H.; Kilian, A.; Bhoite, R.; Liu, G.; Si, P.; Wang, J.; Zhou, W.; Yan, G. QTL Mapping Using a High-Density Genetic Map to Identify Candidate Genes Associated with Metribuzin Tolerance in Hexaploid Wheat (Triticum aestivum L.). Front. Plant Sci. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Yang, D.; Li, M.; Liu, Y.; Chang, L.; Cheng, H.; Chen, J.; Chai, S. Identification of quantitative trait loci and water environmental interactions for developmental behaviors of leaf greenness in wheat. Front. Plant Sci. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.L.; Yu, Q.Y.; Tang, L.L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.F.; Ding, X.D.; Zhu, Y.M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Dai, L.; Wang, L.; Leng, Y.; Yang, Y.; Xu, J.; Hu, J.; Rao, Y.; Zhang, G.; Zhu, L.; et al. Genetic Dissection for Chlorophyll Content of the Top Three Leaves During Grain Filling in Rice (Oryza sativa L.). J. Plant Growth Regul. 2015, 34, 381–391. [Google Scholar] [CrossRef]
- Beale, S.I. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, Y.; Yang, M.; He, J.; Xu, P.; Shao, M.; Chu, P.; Guan, R. Fine mapping of a dominant gene conferring chlorophyll-deficiency in Brassica napus. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Sehgal, S.K.; Kumar, U.; Prasad, P.V.V.; Joshi, A.K.; Gill, B.S. Genomic characterization of drought tolerance-related traits in spring wheat. Euphytica 2012, 186, 265–276. [Google Scholar] [CrossRef]
- Takai, T.; Kondo, M.; Yano, M.; Yamamoto, T. A quantitative trait locus for chlorophyll content and its association with leaf photosynthesis in rice. Rice 2010, 3, 172–180. [Google Scholar] [CrossRef] [Green Version]
- Ye, W.; Hu, S.; Wu, L.; Ge, C.; Cui, Y.; Chen, P.; Xu, J.; Dong, G.; Guo, L.; Qian, Q. Fine mapping a major QTL qFCC7L for chlorophyll content in rice (Oryza sativa L.) cv. PA64s. Plant Growth Regul. 2017, 81, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Wang, T.; Wang, N.; Wang, Z.; Liang, C.; Ramchiary, N.; Choi, S.R.; Lim, Y.P.; Piao, Z.Y. Genetic mapping and localization of quantitative trait loci for chlorophyll content in Chinese cabbage (Brassica rapa ssp. pekinensis). Sci. Hortic. 2012, 147, 42–48. [Google Scholar] [CrossRef]
- Lee, H.E.; Manivannan, A.; Lee, S.Y.; Han, K.; Yeum, J.G.; Jo, J.; Kim, J.; Rho, I.R.; Lee, Y.R.; Lee, E.S.; et al. Chromosome level assembly of homozygous inbred line ‘Wongyo 3115’ facilitates the construction of a high-density linkage map and identification of QTLs associated with fruit firmness in octoploid strawberry (Fragaria × ananassa). Front. Plant Sci. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Alarfaj, R.; El-Soda, M.; Antanaviciute, L.; Vickerstaff, R.; Hand, P.; Harrison, R.J.; Wagstaff, C. Mapping QTL underlying fruit quality traits in an F1 strawberry population. J. Hortic. Sci. Biotechnol. 2021, 96, 634–645. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vining, K.J.; Salinas, N.; Tennessen, J.A.; Zurn, J.D.; Sargent, D.J.; Hancock, J.; Bassil, N.V. Genotyping-by-sequencing enables linkage mapping in three octoploid cultivated strawberry families. Peer J. 2017, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.; Zurn, J.D.; Salinas, N.; Mathey, M.M.; Denoyes, B.; Hancock, J.F.; Finn, C.E.; Bassil, N.V.; Whitaker, V.M. Clarifying sub-genomic positions of QTLs for flowering habit and fruit quality in U.S. strawberry (Fragaria × ananassa) breeding populations using pedigree-based QTL analysis. Hortic. Res. 2017, 4, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sargent, D.J.; Buti, M.; Šurbanovski, N.; Brurberg, M.B.; Alsheikh, M.; Kent, M.P.; Davik, J. Identification of QTLs for powdery mildew (Podosphaera aphanis; syn. Sphaerotheca macularis f. sp. fragariae) susceptibility in cultivated strawberry (Fragaria × ananassa). PLoS ONE 2019, 14, 1–17. [Google Scholar] [CrossRef]
- Hossain, M.R.; Natarajan, S.; Kim, H.T.; Jesse, D.M.I.; Lee, C.G.; Park, J.I.; Nou, I.S. High density linkage map construction and QTL mapping for runner production in allo-octoploid strawberry Fragaria × ananassa based on ddRAD-seq derived SNPs. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Dong, H.; Yang, L.; Li, L.; Zhang, J.; Fan, J.; Song, S.; Yang, L.; Li, H. High Density Genetic Map of Cultivated Strawberry by SLAF-seq. Acta Agric. Boreali-Sin. 2020, 35, 52–57. [Google Scholar] [CrossRef]
- Siddique, M.I.; Lee, H.-Y.; Ro, N.-Y.; Han, K.; Venkatesh, J.; Solomon, A.M.; Patil, A.S.; Changkwian, A.; Kwon, J.-K.; Kang, B.-C. Identifying candidate genes for Phytophthora capsici resistance in pepper (Capsicum annuum) via genotyping-by-sequencing-based QTL mapping and genome-wide association study. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lee, H.Y.; Ro, N.Y.; Hur, O.S.; Lee, J.H.; Kwon, J.K.; Kang, B.C. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnol. J. 2018, 16, 1546–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solomon, A.M.; Kim, T.G.; Han, K.; Lee, H.Y.; Patil, A.; Siddique, M.I.; Ahn, J.; Kang, B.C. Fine Mapping and Candidate Gene Identification for the CapUp Locus Controlling Fruit Orientation in Pepper (Capsicum spp.). Front. Plant Sci. 2021, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Damerum, A.; Selmes, S.L.; Biggi, G.F.; Clarkson, G.J.J.; Rothwell, S.D.; Truco, M.J.; Michelmore, R.W.; Hancock, R.D.; Shellcock, C.; Chapman, M.A.; et al. Elucidating the genetic basis of antioxidant status in lettuce (Lactuca sativa). Hortic. Res. 2015, 2, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Yu, Z.; Qi, X.; Wang, Z.; Zheng, Y.; Ren, H.; Liang, S.; Zheng, X. Construction of a High-Density Genetic Map and Identification of Leaf Trait-Related QTLs in Chinese Bayberry (Myrica rubra). Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Labadie, M.; Vallin, G.; Petit, A.; Ring, L.; Hoffmann, T.; Gaston, A.; Potier, A.; Schwab, W.; Rothan, C.; Denoyes, B. Metabolite Quantitative Trait Loci for Flavonoids Provide New Insights into the Genetic Architecture of Strawberry (Fragaria × ananassa) Fruit Quality. J. Agric. Food Chem. 2020, 68, 6927–6939. [Google Scholar] [CrossRef] [PubMed]
- Santin, A.; Villa, F.; Paulus, D. Chlorophyll content in plants and fruit yield of strawberry plants grown on mulching. Rev. Ciencias Agroveterinarias 2017, 16, 262–268. [Google Scholar] [CrossRef] [Green Version]
- Güler, S.; Macit, I.; Koç, A.; Ibrikci, H. Estimating leaf nitrogen status of strawberry by using chlorophyll meter reading. J. Biol. Sci. 2006, 6, 1011–1016. [Google Scholar]
- Jiang, C.; Johkan, M.; Hohjo, M.; Tsukagoshi, S.; Maturo, T. A correlation analysis on chlorophyll content and SPAD value in tomato leaves. Hort. Res. 2017, 71, 37–42. [Google Scholar] [CrossRef]
- Allard, R.W. Principles of Plant Breeding; John Wiley and Sons Inc.: Hoboken, NJ, USA, 1960. [Google Scholar]
- Lee, Y.R.; Kim, J.; Lee, S.Y.; Lee, J. Diallelic SNP marker development and genetic linkage map construction in octoploid strawberry (Fragaria × ananassa) through next-generation resequencing and high-resolution melting analysis. Hortic. Environ. Biotechnol. 2020, 61, 371–383. [Google Scholar] [CrossRef]
- Sonah, H.; Bastien, M.; Iquira, E.; Tardivel, A.; Légaré, G.; Boyle, B.; Normandeau, É.; Laroche, J.; Larose, S.; Jean, M.; et al. An Improved Genotyping by Sequencing (GBS) Approach Offering Increased Versatility and Efficiency of SNP Discovery and Genotyping. PLoS ONE 2013, 8, e54603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, K.; Jeong, H.J.; Yang, H.B.; Kang, S.M.; Kwon, J.K.; Kim, S.; Choi, D.; Kang, B.C. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 2016, 23, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5; Department of Statistics, North CarolinaState University: Raleigh, NC, USA, 2007. [Google Scholar]
- Bojovic, B.; Markovic, A. Correlation between nitrogen and chlorophyll content in wheat (Triticum aestivum L.). Kragujev. J. Sci. 2009, 31, 69–74. [Google Scholar]
- Guo, P.; Baum, M.; Varshney, R.K.; Graner, A.; Grando, S.; Ceccarelli, S. QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica 2008, 163, 203–214. [Google Scholar] [CrossRef]
- Zhang, K.; Fang, Z.; Liang, Y.; Tian, J. Genetic dissection of chlorophyll content at different growth stages in common wheat. J. Genet. 2009, 88, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Sun, Z.L.; Xu, X.L.; Jin, M.; Liu, Y.J.; Zhang, H.; An, L.Z. Influence of low temperatures on photosystem II photochemistry and expression of the NADPH:Protochlorophyllide oxidoreductase in the alpine, subnival perennial, Chorispora bungeana. Photosynthetica 2010, 48, 457–468. [Google Scholar] [CrossRef]
- Chen, M.; Li, J.; Dai, X.; Sun, Y.; Chen, F. Effect of phosphorus and temperature on chlorophyll a contents and cell sizes of Scenedesmus obliquus and Microcystis aeruginosa. Limnology 2011, 12, 187–192. [Google Scholar] [CrossRef]
- Gáborčík, N. Relationship between contents of chlorophyll (a+b) (SPAD values) and nitrogen of some temperate grasses. Photosynthetica 2003, 41, 285–287. [Google Scholar] [CrossRef]
- Fritschi, F.B.; Ray, J.D. Soybean leaf nitrogen, chlorophyll content, and chlorophyll a/b ratio. Photosynthetica 2007, 45, 92–98. [Google Scholar] [CrossRef]
- Suharja, S.; Sutarno, S. Biomass, chlorophyll and nitrogen content of leaves of two chili pepper varieties (Capsicum annum) in different fertilization treatments. Bioscience 2009, 1, 9–16. [Google Scholar]
- Lee, Y.J.; Yang, C.M.; Chang, K.W.; Shen, Y. Effects of nitrogen status on leaf anatomy, chlorophyll content and canopy reflectance of paddy rice. Bot. Stud. 2011, 52, 295–303. [Google Scholar]
- Zucchi, M.R.; Necchi, O. Effects of temperature, irradiance and photoperiod on growth and pigment content in some freshwater red algae in culture. Phycol. Res. 2001, 49, 103–114. [Google Scholar] [CrossRef]
- Seyfabadi, J.; Ramezanpour, Z.; Khoeyi, Z.A. Protein, fatty acid, and pigment content of Chlorella vulgaris under different light regimes. J. Appl. Phycol. 2011, 23, 721–726. [Google Scholar] [CrossRef]
- De Oliveira, A.H.G.; Maciel, G.M.; Siquieroli, A.C.S.; Luz, J.M.Q.; Da Silva, E.C. Dynamics of heritability in different characters of lettuce. Rev. Caatinga. 2021, 34, 514–526. [Google Scholar] [CrossRef]
- Yu, K.; Wang, J.; Sun, C.; Liu, X.; Xu, H.; Yang, Y.; Dong, L.; Zhang, D. High-density QTL mapping of leaf-related traits and chlorophyll content in three soybean RIL populations. BMC Plant Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Si, L.T.; Zhang, G.Y.; Zhang, S.J.; Tian, L.B. Quantitative trait loci analysis for chlorophyll content of cucumber (Cucumis sativus L.) seedlings under low-light stress. African J. Biotechnol. 2010, 9, 4692–4699. [Google Scholar] [CrossRef]
- Ji, S.; Siegel, A.; Shan, S.-O.; Grimm, B.; Wang, P. Chloroplast SRP43 autonomously protects chlorophyll biosynthesis proteins against heat shock. Nat. Plants 2021. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Islam, S.; Juhasz, A.; Yang, R.; She, M.; Alhabbar, Z.; Zhang, J.; Ma, W. Transcriptomic study for identification of major nitrogen stress responsive genes in Australian bread wheat cultivars. Front. Genet. 2020, 11, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Song, J.T.; Lu, H.; Greenberg, J.T. Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant arowth and death2 And AGD2-Like Defense Response Protein 1, Encoding Novel Aminotransferases. Plant Cell 2004, 16, 353–366. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Park, S.J.; Han, J.H.; Jeon, Y.; Pai, H.S.; Kang, H. A chloroplast-targeted pentatricopeptide repeat protein PPR287 is crucial for chloroplast function and Arabidopsis development. BMC Plant Biol. 2019, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Richter, A.S.; Kleeberg, J.R.W.; Geimer, S.; Grimm, B. Post-translational coordination of chlorophyll biosynthesis and breakdown by BCMs maintains chlorophyll homeostasis during leaf development. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, M.K.; Shih, T.H.; Lin, S.H.; Lin, J.W.; Nguyen, H.C.; Yang, Z.W.; Yang, C.M. Transcription profile analysis of chlorophyll biosynthesis in leaves of wild-type and chlorophyll b-deficient rice (Oryza sativa L.). Agriculture 2021, 11, 401. [Google Scholar] [CrossRef]
- Inoue, H.; Li, M.; Schnell, D.J. An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, 3173–3178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.S.; Choi, I.; Jeon, Y.; Ahn, H.K.; Cho, H.; Kim, J.W.; Kim, J.H.; Lee, J.M.; Lee, S.H.; Bünting, J.; et al. Chaperone-like protein DAY plays critical roles in photomorphogenesis. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
| Summary of Illumina Sequencing | Data |
|---|---|
| Number of BC-F2 plants subjected sequencing | 158 |
| Total base number of raw reads (bp) | 124,093,242,956 |
| Average base number of raw reads (bp) | 775,582,768 |
| Average number of trimmed reads | 3,000,099 |
| Average mapping rate (%) | 97.5 |
| Average depth of mapped region | 9.9 |
| Total number of SNPs detected | 17,416 |
| Population | Trait | QTLs | Linkage Group | Chromosome | Position 1 (cM) | Position 2 (Mb) | LOD 3 | R2 (%) 4 | Additive Effect |
|---|---|---|---|---|---|---|---|---|---|
| BS-F2 | Chl | BSqchl_5-3 | 5-3c | 5-3 | 7.01 | 26.5–28 | 8.1 | 17.3 | −2.5 |
| BS-F2 | Chl | BSqchl_6-3 | 6-3b | 6-3 | 79.61 | 36.3–37 | 4.2 | 5.9 | −1.7 |
| BC-F2 | Chl | BCqchl_2-2 | 2-2a | 2-2 | 9.91 | 8–14 | 3.4 | 6.3 | 9 |
| BC-F2 | Chl | BCqchl_4-2 | 4-2a | 4-2 | 10.31 | 8–9 | 2.9 | 26.8 | −3.1 |
| BC-F2 | Chl | BCqchl_5-1 | 5-1b | 5-1 | 17.61 | 7–8 | 3 | 4.1 | 1.8 |
| BC-F2 | Chl | BCqchl_5-3 | 5-3a | 5-3 | 122.01 | 27–28 | 3.4 | 1.4 | 2.4 |
| BC-F2 | Chl | BCqchl_7-2 | 7-2b | 7-2 | 61.41 | 23–24 | 2.9 | 5.4 | 0 |
| QTLs | Chromosome | Location | Gene ID | Functional Annotation |
|---|---|---|---|---|
| BSqchl_5-3 BCqchl_5-3 | Chr5-3 | 26833856–26835283 | ID = g00061270 | Glutamate-tRNA ligase |
| Chr5-3 | 26902815–26905722 | ID = g00061283 | Aminotransferase ALD1 | |
| Chr5-3 | 26938249–26940105 | ID = g00061289 | Pentatricopeptide repeat-containing protein | |
| Chr5-3 | 27008147–27010431 | ID = g00061300 | Chaperone protein dnaJ | |
| Chr5-3 | 27323239–27328734 | ID = g00061382 | Glutamyl endopeptidase | |
| Chr5-3 | 27339049–27339435 | ID = g00061384 | Calvin cycle protein CP12-2 | |
| Chr5-3 | 27351854–27352555 | ID = g00061388 | Glutamate--tRNA ligase | |
| Chr5-3 | 27536789–27540838 | ID = g00061428 | Phospholipase A1 PLIP2 | |
| Chr5-3 | 27596348–27596541 | ID = g00061448 | Senescence-associated protein | |
| Chr5-3 | 27630517–27630918 | ID = g00061457 | Peroxiredoxin | |
| Chr5-3 | 27652144–27655367 | ID = g00061460 | Heat shock protein 90-5 | |
| Chr5-3 | 27706473–27712240 | ID = g00061470 | WAT1-related protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddique, M.I.; Han, K.; Lee, J.; Lee, E.S.; Lee, Y.-R.; Lee, H.-E.; Lee, S.Y.; Kim, D.-S. QTL Analysis for Chlorophyll Content in Strawberry (Fragaria × ananassa Duch.) Leaves. Agriculture 2021, 11, 1163. https://doi.org/10.3390/agriculture11111163
Siddique MI, Han K, Lee J, Lee ES, Lee Y-R, Lee H-E, Lee SY, Kim D-S. QTL Analysis for Chlorophyll Content in Strawberry (Fragaria × ananassa Duch.) Leaves. Agriculture. 2021; 11(11):1163. https://doi.org/10.3390/agriculture11111163
Chicago/Turabian StyleSiddique, Muhammad Irfan, Koeun Han, Jieun Lee, Eun Su Lee, Ye-Rin Lee, Hye-Eun Lee, Sun Yi Lee, and Do-Sun Kim. 2021. "QTL Analysis for Chlorophyll Content in Strawberry (Fragaria × ananassa Duch.) Leaves" Agriculture 11, no. 11: 1163. https://doi.org/10.3390/agriculture11111163
APA StyleSiddique, M. I., Han, K., Lee, J., Lee, E. S., Lee, Y.-R., Lee, H.-E., Lee, S. Y., & Kim, D.-S. (2021). QTL Analysis for Chlorophyll Content in Strawberry (Fragaria × ananassa Duch.) Leaves. Agriculture, 11(11), 1163. https://doi.org/10.3390/agriculture11111163








