SLL1-ZH Regulates Spikelets Architecture and Grain Yield in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rice Materials and Plant Growth Conditions
2.2. Phenotyping and Morphological Analysis
2.3. Molecular Cloning of SLL1-ZH
2.4. Genetic Complementation of the SLL1-ZH Mutant
2.5. RNA Extraction and qPCR Analysis
3. Results
3.1. SLL1-ZH Affects Both Plant Architecture and Grain Yields
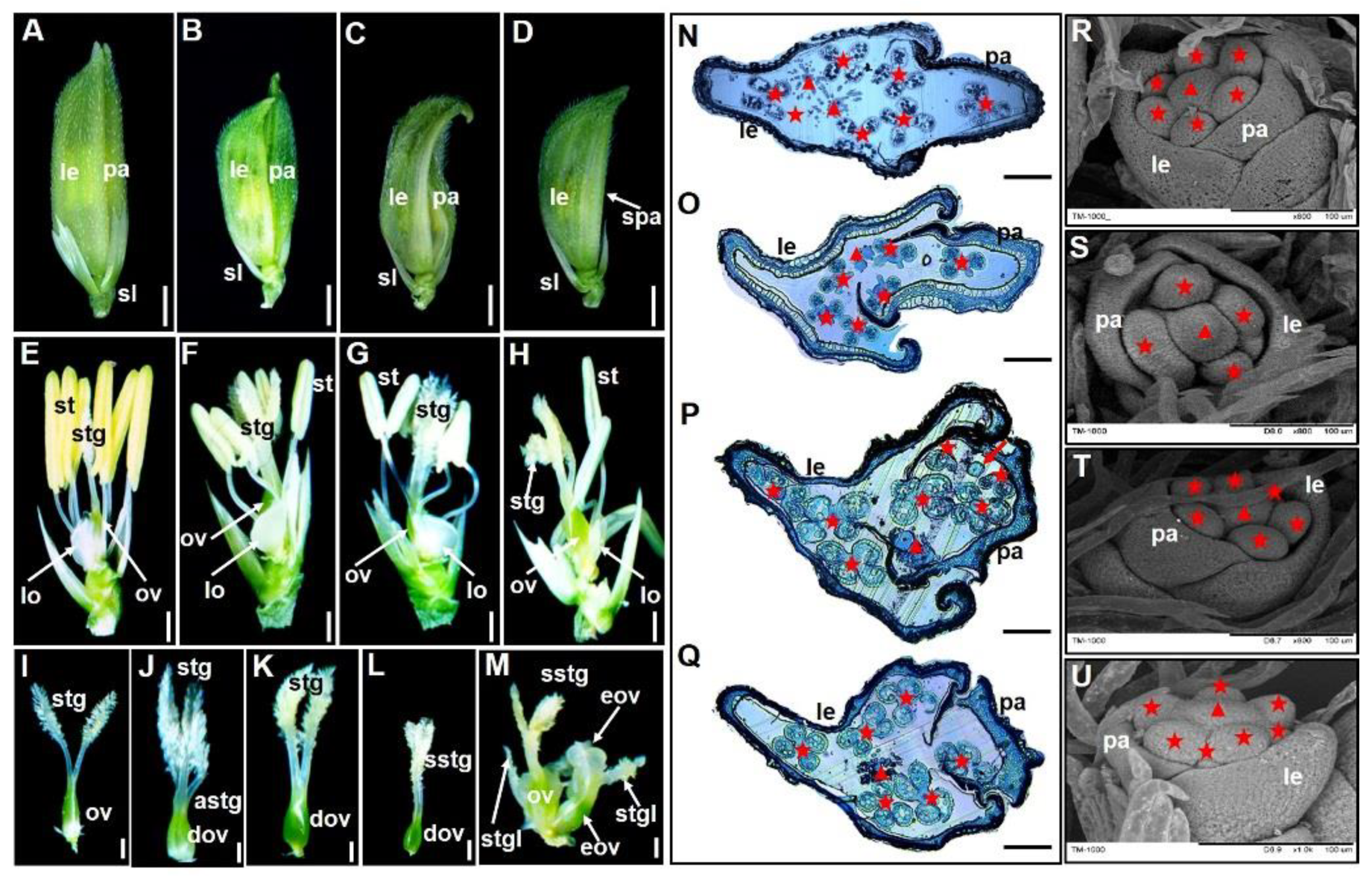
3.2. Abnormal Floral Organ Identity and Development in the SLL1-ZH Mutant
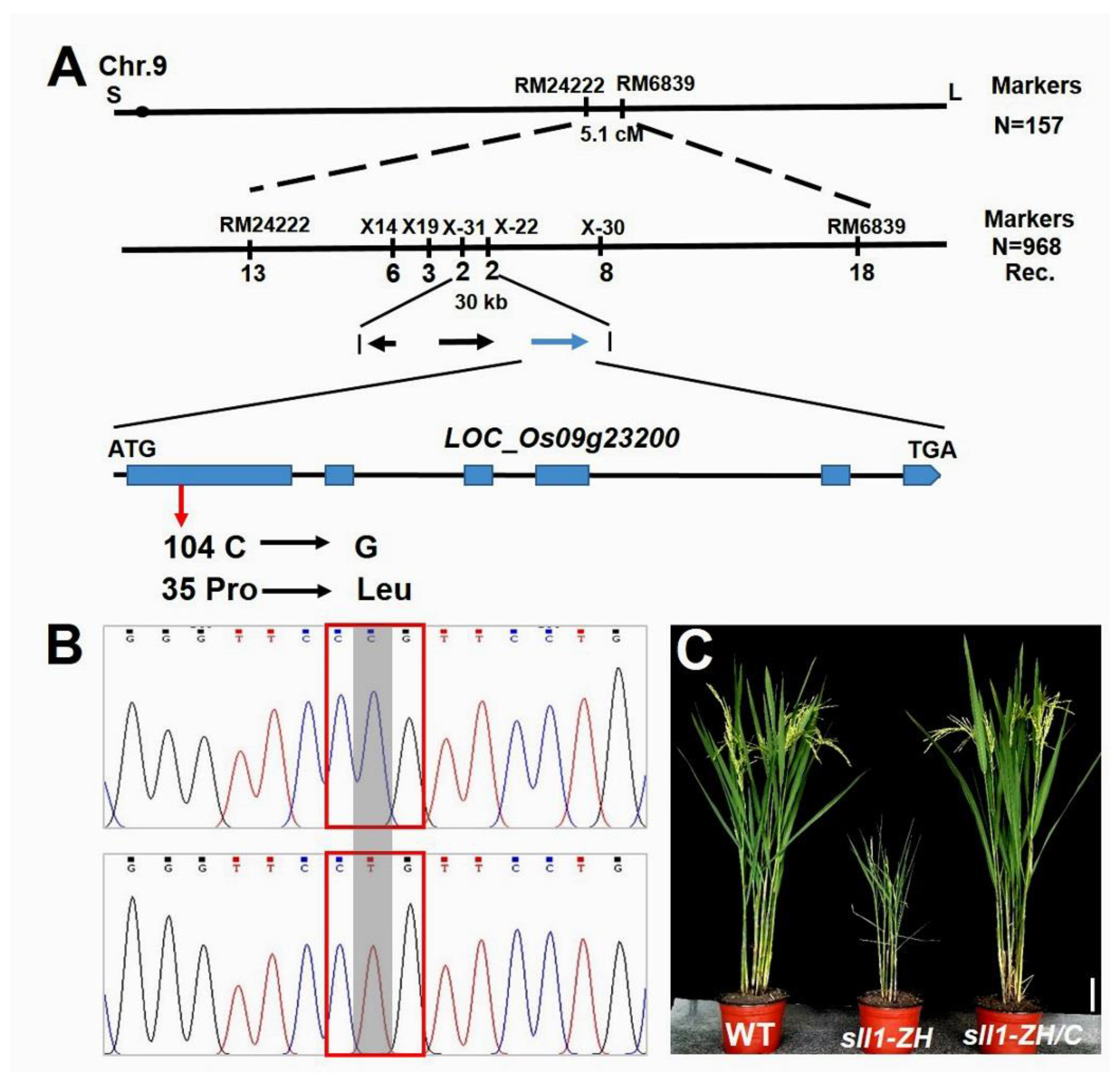
3.3. Map-Based Cloning of the SLL1-ZH Mutant Gene
3.4. Transcription of SLL1-ZH and Floral Organ Development Related Factors in SLL1-ZH
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bommert, P.; Satoh-Nagasawa, N.; Jackson, D.; Hirano, A.H. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 2005, 46, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Itoh, J.; Nonomura, K.; Ikeda, K.; Yamaki, S.; Inukai, Y.; Yamagishi, H.; Kitano, H.; Nagato, A.Y. Rice plant development: From zygote to spikelet. Plant Cell Physiol. 2005, 46, 23–47. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Nagato, A.Y. Flower development in rice. J. Exp. Bot. 2011, 62, 4719–4730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Colombo, L.; Franken, J.; Koetje, E.; van Went, J.; Dons, H.J.; Angenent, G.C.; van Tunen, A.J. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 1995, 7, 1859–1868. [Google Scholar]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrario, S.; Immink, R.G.; Shchennikova, A.; Busscher-Lange, J.; Angenent, G.C. The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 2003, 15, 914–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandel, M.A.; Yanofsky, M.F. The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 1995, 7, 1763–1771. [Google Scholar] [PubMed] [Green Version]
- Lamb, R.S.; Irish, V.F. Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. USA 2003, 100, 6558–6563. [Google Scholar] [CrossRef] [Green Version]
- Fornara, F.; Parenicová, L.; Falasca, G.; Pelucchi, N.; Masiero, S.; Ciannamea, S.; Lopez-Dee, Z.; Altamura, M.M.; Colombo, L.; Kater, M.M. Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol. 2004, 135, 2207–2219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.S.; Jang, S.; Lee, S.; Nam, J.; Kim, C.; Lee, S.H.; Chung, Y.Y.; Kim, S.R.; Lee, Y.H.; Cho, Y.G.; et al. Leafy hull sterile1 Is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 2000, 12, 871–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K.; Yasuno, N.; Sato, Y.; Yoda, M.; Yamazaki, R.; Kimizu, M.; Yoshida, H.; Nagamura, Y.; Kyozuka, J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 2012, 24, 1848–1859. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.J.; Wei, H.; Wang, Y.; Wang, H.M.; Yang, R.F.; Zhang, X.B.; Tu, J.M. Overexpression of a transcription factor OsMADS15 modifies plant architecture and flowering time in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 2012, 30, 1461–1469. [Google Scholar] [CrossRef]
- Sentoku, N.; Kato, H.; Kitano, H.; Imai, R. OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy. Mol. Genet. Genom. 2005, 273, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, L.; Cai, Q.; Hu, Y.; Jin, Z.; Zhao, X.; Fan, W.; Huang, Q.; Luo, Z.; Chen, M.; et al. OsMADS32 interacts with PI-like proteins and regulates rice flower development. J. Integr. Plant Biol. 2015, 57, 504–513. [Google Scholar] [CrossRef]
- Nagasawa, N.; MiyOshi, M.; Sano, Y.; Satoh, H.; Hirano, H.; Sakai, H.; Nagato, Y. SUPERWOMAN1 and DROOPING LEAF Genes control floral organ identity in rice. Development 2003, 130, 705–718. [Google Scholar] [CrossRef] [Green Version]
- Xiao, H.; Wang, Y.; Liu, D.F.; Wang, W.M.; Li, X.B.; Zhao, X.F.; Xu, J.C.; Zhai, W.X.; Zhu, L.H. Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol. Biol. 2003, 52, 957–966. [Google Scholar] [CrossRef]
- Li, H.F.; Liang, W.Q.; Hu, Y.; Zhu, L.; Yin, C.S.; Xu, J.; Dreni, L.; Kater, M.M.; Zhang, D.B. Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 2011, 23, 2536–2552. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, T.; Lee, D.Y.; Miyao, A.; Hirochika, H.; An, G.; Hirano, H.Y. Functional diversification of the two C-class MADS box genes OsMADS3 and OsMADS58 in Oryza sativa. Plant Cell 2006, 18, 15–18. [Google Scholar] [CrossRef] [Green Version]
- Dreni, L.; Jacchia, S.; Fornara, F.; Fornari, M.; Ouwerkerk, P.B.; An, G.; Colombo, L.; Kater, M.M. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 2007, 52, 690–699. [Google Scholar] [CrossRef]
- Dreni, L.; Pilatone, A.; Yun, D.; Erreni, S.; Pajoro, A.; Caporali, E.; Zhang, D.B.; Kater, M.M. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 2011, 23, 2850–2863. [Google Scholar] [CrossRef] [Green Version]
- Cui, R.F.; Han, J.K.; Zhao, S.Z.; Su, K.M.; Wu, F.; Du, X.Q.; Xu, Q.J.; Chong, K.; Theissen, G.; Meng, Z. Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 2010, 61, 767–781. [Google Scholar] [CrossRef]
- Jeon, J.S.; Lee, S.; Jung, K.H.; Yang, W.S.; Yi, G.H.; Oh, B.G.; An, G. Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Mol. Breed. 2000, 6, 581–592. [Google Scholar] [CrossRef]
- Khanday, I.; Yadav, S.R.; Vijayraghavan, U. Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol. 2013, 161, 1970–1983. [Google Scholar] [CrossRef] [Green Version]
- Prasad, K.; Parameswaran, S.; Vijayraghavan, U. OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 2005, 43, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.P.; Zhang, Y.X.; Zhang, P.P.; Yang, Z.F.; Zhan, X.D.; Shen, X.H.; Zhang, Z.H.; Hu, X.; Xuan, D.D.; Wu, W.X.; et al. K-domain splicing factor OsMADS1 regulates open hull male sterility in rice. Rice Sci. 2015, 22, 207–216. [Google Scholar]
- Yamaguchi, T.; Nagasawa, N.; Kawasaki, S.; Matsuoka, M.; Nagato, Y.; Hirano, H.Y. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 2004, 16, 500–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, H.W.; Qian, Q.; Liang, W.Q.; Yin, C.S.; Tan, H.X.; Yao, X.; Yuan, Z.; Yang, J.; Huang, H.; Luo, D.; et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006, 142, 1039–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.; Jung, K.H.; Lee, D.E.; Lee, D.Y.; Lee, J.; An, K.; Kang, H.G.; An, G. The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol. Cells 2006, 21, 147–152. [Google Scholar]
- Suzaki, T.; Sato, M.; Ashikari, M.; Miyoshi, M.; Nagato, Y.; Hirano, H.Y. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 2004, 131, 5649–5657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzaki, T.; Toriba, T.; Fujimoto, M.; Tsutsumi, N.; Kitano, H.; Hirano, H.Y. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORALORGAN NUMBER2 gene. Plant Cell Physiol. 2006, 47, 1591–1602. [Google Scholar] [CrossRef] [Green Version]
- Yan, D.W.; Zhang, X.M.; Zhang, L.; Ye, S.H.; Zeng, L.J.; Liu, J.Y.; Li, Q.; He, Z.H. CURVED CHIMERIC PALEA 1 encoding an EMF1-like protein maintains epigenetic repression of OsMADS58 in rice palea development. Plant J. 2015, 82, 12–24. [Google Scholar] [CrossRef]
- Zheng, M.; Wang, Y.H.; Wang, Y.L.; Wang, C.M.; Ren, Y.L.; Lv, J.; Peng, C.; Wu, T.; Liu, K.; Zhao, S.L.; et al. DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa). New Phytol. 2015, 206, 1476–1490. [Google Scholar] [CrossRef]
- Tao, J.H.; Liang, W.Q.; An, G.; Zhang, D.B. OsMADS6 Controls Flower Development by Activating Rice FACTOR OF DNA METHYLATION LIKE1. Plant Physiol. 2018, 177, 713–727. [Google Scholar] [CrossRef] [Green Version]
- You, X.M.; Zhu, S.S.; Zhang, W.W.; Zhang, J.; Wang, C.M.; Jing, R.N.; Chen, W.W.; Wu, H.M.; Cai, Y.; Feng, Z.M.; et al. OsPEX5 regulates rice spikelet development through modulating jasmonic acid biosynthesis. New Phytol. 2019, 224, 712–724. [Google Scholar] [CrossRef]
- Zhuang, H.; Wang, H.L.; Zhang, T.; Zeng, X.Q.; Chen, H.; Wang, Z.W.; Zhang, J.; Zheng, H.; Tang, J.; Ling, Y.H.; et al. NONSTOP GLUMES1 Encodes a C2H2 zinc finger protein that regulates spikelet development in rice. Plant Cell 2020, 32, 392–413. [Google Scholar] [CrossRef] [Green Version]
- He, Y.B.; Yan, L.; Ge, C.N.; Yao, X.F.; Han, X.; Wang, R.C.; Xiong, L.Z.; Jiang, L.W.; Liu, C.M.; Zhao, Y.D. PINOID is required for formation of the stigma and style in rice. Plant Physiol. 2019, 180, 926–936. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Tang, D.; Cheng, X.J.; Zhang, J.X.; Tang, Y.J.; Tao, Q.D.; Shi, W.Q.; You, A.Q.; Gu, M.H.; Cheng, Z.K.; et al. OsPINOID regulates stigma and ovule initiation through maintenance of the floral meristem by auxin signaling. Plant Physiol. 2019, 180, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.H.; Xie, D.J.; Tang, Z.S.; Shi, D.Q.; Yang, W.C. PINOID regulates floral organ development by modulating auxin transport and interacts with MADS16 in rice. Plant Biotechnol. J. 2020, 18, 1778–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.P.; Li, H.; Liu, Z.J.; Zhuang, Y.; Wei, M.; Gu, Y.Y.; Liu, Y.X.; Sun, X.Q.; Tang, Y.Y.; Yue, L.; et al. Characterization of the ‘Oat-Like Rice’ caused by a novel allele OsMADS1Olr reveals vital importance of OsMADS1 in regulating grain shape in Oryza sativa L. Rice 2020, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, H.P.; Liu, J.; Wang, W.; Sun, J.; Gao, Q.; Zhang, Y.H.; Ma, D.R.; Wang, J.Y.; Xu, Z.J.; et al. Loss of function of OsMADS34 leads to large sterile lemma and low grain yield in rice (Oryza sativa L.). Mol. Breed. 2016, 36, 147. [Google Scholar] [CrossRef]
- Ma, X.D.; Zhang, J.N.; Han, B.; Tang, J.H.; Cui, D.; Han, L.Z. FLA, which encodes a homolog of UBP, is required for chlorophyll accumulation and development of lemma and palea in rice. Plant Cell Rep. 2019, 38, 321–331. [Google Scholar] [CrossRef]
- Zhang, G.H.; Xu, Q.; Zhu, X.D.; Qian, Q.; Xue, H.W. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 2009, 21, 719–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, D.Y.; Cui, Y.J.; Hu, H.T.; Xu, Q.K.; Rao, Y.C.; Yu, X.Q.; Zhang, Y.; Wang, Y.X.; Peng, Y.L.; Zeng, D.L.; et al. AH2 encodes a MYB domain protein that determines hull fate and affects grain yield and quality in rice. Plant J. 2019, 100, 813–824. [Google Scholar] [CrossRef]
- Yan, D.W.; Zhou, Y.; Ye, S.H.; Zeng, L.J.; Zhang, X.M.; He, Z.H. BEAK-SHAPED GRAIN 1/TRIANGULAR HULL 1, a DUF640 gene, is associated with grain shape, size and weight in rice. Sci. China Life Sci. 2013, 56, 275–283. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.J.; Zhang, X.W.; Shao, G.N.; He, J.W.; Jiao, G.A.; Xie, L.H.; Sheng, Z.H.; Tang, S.Q.; Hu, P.S. Fine mapping of BH1, a gene controlling lemma and palea development in rice. Plant Cell Rep. 2013, 32, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.Y.; Rao, Y.C.; Wu, L.W.; Xu, Q.K.; Li, Z.Z.; Yu, H.P.; Zhang, Y.; Leng, Y.J.; Hu, J.; Zhu, L.; et al. The pleiotropic ABNORMAL FLOWER AND DWARF1 affects plant height, floral development and grain yield in rice. J. Integr. Plant Biol. 2016, 58, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Li, Y.F.; Ma, L.; Sang, X.C.; Ling, Y.H.; Wang, Y.T.; Yu, P.; Zhuang, H.; Huang, J.Y.; Wang, N.; et al. LATERAL FLORET 1 induced the three-florets spikelet in rice. Proc. Natl. Acad. Sci. USA 2017, 114, 9984–9989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.G.; Xue, D.W.; Gao, Z.Y.; Yan, M.X.; Xu, W.Y.; Xing, Z.; Huang, D.N.; Qian, Q.; Xue, Y.B. A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J. 2009, 57, 593–605. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.Y.; Yu, H.P.; Rao, Y.C.; Xu, Q.K.; Zhou, T.T.; Hu, J.; Zhang, Y.; Zhang, G.H.; Zhu, L.; Gao, Z.Y.; et al. ‘Two-floret spikelet’ as a novel resource has the potential to increase rice yield. Plant Biot. J. 2018, 16, 351–353. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.P.; Xiang, X.J.; Yang, Z.F.; Yu, P.; Wen, X.X.; Wang, H.; Adil, A.; Riaz, M.K.; Zhang, Y.X.; Cheng, S.H.; et al. OsGPAT3 plays a critical role in anther wall programmed cell death and pollen development in rice. Int. J. Mol. Sci. 2018, 19, 4017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.P.; Zhang, Y.X.; Zhang, P.P.; Yang, Z.F.; Zhou, X.X.; Xuan, D.D.; Li, Z.H.; Wu, W.X.; Zhan, X.D.; Shen, X.H.; et al. Morphogenesis and Gene Mapping of deformed interior floral organ 1 (difo1), a novel mutant associated with floral organ development in rice. Plant Mol. Biol. Rep. 2017, 35, 330–344. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C (T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]


| Genotype | Glumes | Lodicules | Lemmas/Paleas | Stamens | Stigmas | Ovaries | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | ≤3 | 4 | 5 | 6 | ≥7 | 1 | 2 | 3 | 1 | ≥2 | |||
| Wild type (200) | 2 (100%) | 2 (100%) | 200 | 0 | 0 | 0 | 0 | 200 | 0 | 0 | 200 | 0 | 200 | 0 |
| SLL1-ZH (400) | 2 (100%) | 2 (100%) | 379 | 21 | 2 | 11 | 21 | 357 | 9 | 9 | 256 | 135 | 393 | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Wang, J.; Wen, X.; Peng, Z.; Chen, D.; Zhang, Y.; Cheng, S.; Cao, L.; Zhan, X. SLL1-ZH Regulates Spikelets Architecture and Grain Yield in Rice. Agriculture 2021, 11, 1162. https://doi.org/10.3390/agriculture11111162
Sun L, Wang J, Wen X, Peng Z, Chen D, Zhang Y, Cheng S, Cao L, Zhan X. SLL1-ZH Regulates Spikelets Architecture and Grain Yield in Rice. Agriculture. 2021; 11(11):1162. https://doi.org/10.3390/agriculture11111162
Chicago/Turabian StyleSun, Lianping, Jingxin Wang, Xiaoxia Wen, Zequn Peng, Daibo Chen, Yingxin Zhang, Shihua Cheng, Liyong Cao, and Xiaodeng Zhan. 2021. "SLL1-ZH Regulates Spikelets Architecture and Grain Yield in Rice" Agriculture 11, no. 11: 1162. https://doi.org/10.3390/agriculture11111162
APA StyleSun, L., Wang, J., Wen, X., Peng, Z., Chen, D., Zhang, Y., Cheng, S., Cao, L., & Zhan, X. (2021). SLL1-ZH Regulates Spikelets Architecture and Grain Yield in Rice. Agriculture, 11(11), 1162. https://doi.org/10.3390/agriculture11111162






