Evaluating Multiple Allelic Combination to Determine Tiller Angle Variation in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Measurement of Tiller Angle
2.2. Development of Single-Nucleotide Polymorphism (SNP) Markers and PCR Condition
2.3. Development of Linear Regression Equation Model and Statistical Analysis
3. Results
3.1. Variation of Tiller Angle in Rice Germplasms
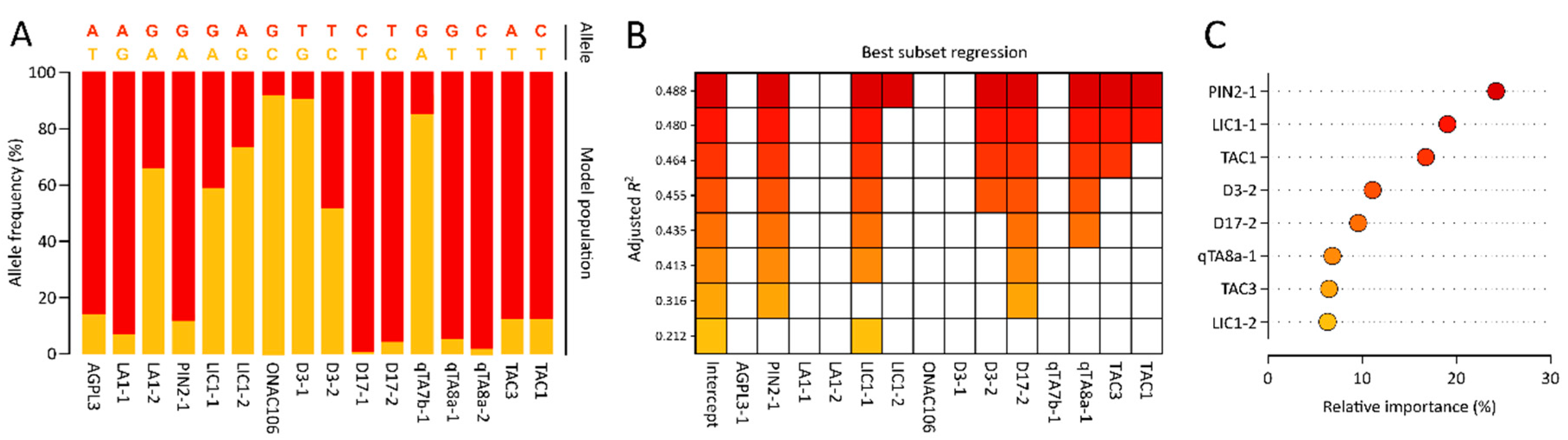
3.2. Development of SNP Markers Associated with Tiller Angle
3.3. Development of a Multiple Linear Regression Model for Tiller Angle Using a Model Rice Population
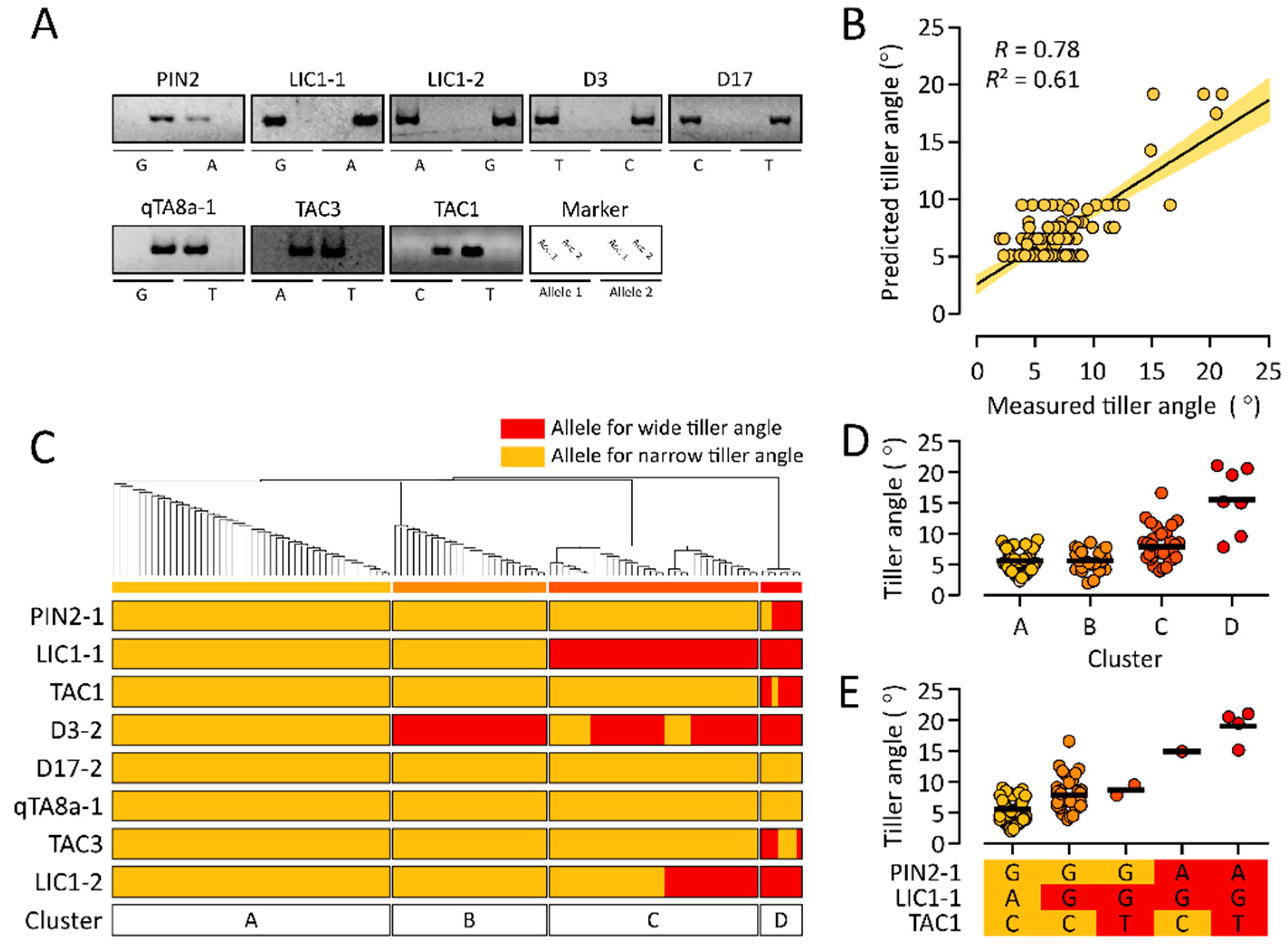
3.4. Test of a Multiple Linear Regression Model for Rice Tiller Angle Using a Test Population
3.5. Allelic Combinations of Rice Tiller Angle-Related Markers in a Test Population
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dong, H.; Zhao, H.; Xie, W.; Han, Z.; Li, G.; Yao, W.; Bai, X.; Hu, Y.; Guo, Z.; Lu, K.; et al. A Novel Tiller Angle Gene, TAC3, together with TAC1 and D2 Largely Determine the Natural Variation of Tiller Angle in Rice Cultivars. PLoS Genet. 2016, 12, e1006412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; McCouch, S.R.; Shen, Z. Transgressive Segregation of Tiller Angle in Rice Caused by Complementary Gene Action. Crop Sci. 1998, 38. [Google Scholar] [CrossRef]
- Yu, B.; Lin, Z.; Li, H.; Li, X.; Li, J.; Wang, Y.; Zhang, X.; Zhu, Z.; Zhai, W.; Wang, X.; et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007, 52, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Zhang, Y.; Chen, Z.; Jiang, W.; Feng, M.; Pan, X. Improvement of Rice Resistance to Sheath Blight by Pyramiding QTLs Conditioning Disease Resistance and Tiller Angle. Rice Sci. 2014, 21, 318–326. [Google Scholar] [CrossRef]
- Dong, Y.; Lin, D.; Kamiuntent, H.; Cai, Q.; Tsuzuki, E. Molecular Genetic Mapping of QTLs for Tiller Angle in Rice (Oryza sativa L.). Jpn. J. Trop. Agric. 2004, 48, 228–234. [Google Scholar]
- Wu, Y.; Zhao, S.; Li, X.; Zhang, B.; Jiang, L.; Tang, Y.; Zhao, J.; Ma, X.; Cai, H.; Sun, C.; et al. Deletions linked to PROG1 gene participate in plant architecture domestication in Asian and African rice. Nat. Commun. 2018, 9, 4157. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Tan, L.; Sun, H.; Zhao, X.; Liu, F.; Cai, H.; Fu, Y.; Sun, X.; Gu, P.; Zhu, Z.; et al. Natural Variations at TIG1 Encoding a TCP Transcription Factor Contribute to Plant Architecture Domestication in Rice. Mol. Plant 2019, 12, 1075–1089. [Google Scholar] [CrossRef]
- Zhang, N.; Yu, H.; Yu, H.; Cai, Y.; Huang, L.; Xu, C.; Xiong, G.; Meng, X.; Wang, J.; Chen, H.; et al. A Core Regulatory Pathway Controlling Rice Tiller Angle Mediated by the LAZY1—Dependent Asymmetric Distribution of Auxin. Plant Cell 2018, 30, 1461–1475. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Zhang, M.; Niu, X.; Wang, S.; Xu, Q.; Feng, Y.; Wang, C.; Deng, H.; Yuan, X.; Yu, H.; et al. Genetic variation and association mapping for 12 agronomic traits in indica rice. BMC Genom. 2015, 16, 1067. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wang, Y.; Qian, Q.; Fu, Z.; Wang, M.; Zeng, D.; Li, B.; Wang, X.; Li, J. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007, 17, 402–410. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Xu, Y.; Zhang, C.; Ma, Q.; Joo, S.-H.; Kim, S.-K.; Xu, Z.; Chong, K. OsLIC, a Novel CCCH-Type Zinc Finger Protein with Transcription Activation, Mediates Rice Architecture via Brassinosteroids Signaling. PLoS ONE 2008, 3, e3521. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Fan, X.; Song, W.; Zhang, Y.; Xu, G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1: Auxin efflux transporter gene OsPIN2. Plant Biotechnol. J. 2012, 10, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tang, D.; Li, M.; Wang, K.; Cheng, Z. Loose Plant Architecture1, an INDETERMINATE DOMAIN Protein Involved in Shoot Gravitropism, Regulates Plant Architecture in Rice. Plant Physiol. 2013, 161, 317–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, D.; Chen, D.; Liu, G.; Liang, Y.; Huang, L.; Meng, X.; Chu, J.; Sun, X.; Dong, G.; Yuan, Y.; et al. Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 11199–11204. [Google Scholar] [CrossRef] [Green Version]
- Okamura, M.; Hirose, T.; Hashida, Y.; Ohsugi, R.; Aoki, N. Suppression of starch synthesis in rice stems splays tiller angle due to gravitropic insensitivity but does not affect yield. Funct. Plant. Biol. 2015, 42, 31. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Piao, W.; Lim, J.-H.; Han, S.-H.; Kim, Y.-S.; An, G.; Paek, N.-C. Rice ONAC106 Inhibits Leaf Senescence and Increases Salt Tolerance and Tiller Angle. Plant. Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef]
- Zeng, D.; Tian, Z.; Rao, Y.; Dong, G.; Yang, Y.; Huang, L.; Leng, Y.; Xu, J.; Sun, C.; Zhang, G.; et al. Rational design of high-yield and superior-quality rice. Nat. Plants 2017, 3, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.-J.; Won, Y.J.; Cha, K.W.; Lee, J.C.; Park, S.Z.; Heu, M.H. Inheritance of Tillering Angle in Rice. Korean J. Breed. 1997, 29, 431–435. [Google Scholar]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 Family Gene, OsPIN1, involved in Auxin-dependent Adventitious Root Emergence and Tillering in Rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yao, W.; Ouyang, Y.; Yang, W.; Wang, G.; Lian, X.; Xing, Y.; Chen, L.; Xie, W. RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 2015, 43, D1018–D1022. [Google Scholar] [CrossRef] [PubMed]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.-C.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008, 9, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef]
- Groemping, U. Relative Importance for Linear Regression in R: The Package relaimpo. J. Stat. Softw. 2006, 17, 1–27. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Wu, D.; Guo, Z.; Ye, J.; Feng, H.; Liu, J.; Chen, G.; Zheng, J.; Yan, D.; Yang, X.; Xiong, X.; et al. Combining high-throughput micro-CT-RGB phenotyping and genome-wide association study to dissect the genetic architecture of tiller growth in rice. J. Exp. Bot. 2018, 70, 545–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Yang, S.; Gong, J.; Zhao, Q.; Feng, Q.; Zhan, Q.; Zhao, Y.; Li, W.; Cheng, B.; Xia, J.; et al. Genomic architecture of heterosis for yield traits in rice. Nature 2016, 537, 629–633. [Google Scholar] [CrossRef]

| QTL/Gene | SNP Marker | Binary Code of Allele | Estimate | Standard Deviation | t-Value | |
|---|---|---|---|---|---|---|
| 1 | 0 | |||||
| OsPIN2 | PIN2-1 | G (N) | A (W) | −8.4 | 1.06 | −7.92 *** |
| OsLIC1 | LIC1-1 | G (W) | A (N) | 2.93 | 0.5 | 5.82 *** |
| LIC1-2 | A (N) | G (W) | −1.96 | 0.66 | −2.97 *** | |
| D3 | D3-2 | T (N) | C (W) | −1.5 | 0.37 | −4.09 *** |
| D17 | D17-2 | T (W) | C (N) | 6.6 | 1.13 | 5.83 *** |
| qTA8a | qTA8a-1 | G (W) | T (N) | 4.64 | 1.08 | 4.31 *** |
| TAC3 | TAC3 | A (W) | T (N) | 1.68 | 0.81 | 2.06 * |
| TAC1 | TAC1 | C (N) | T (W) | −3.24 | 0.91 | −3.58 *** |
| Intercept | 5.29 | 0.97 | 5.48 *** | |||
| Regression equation | Tiller angle = 5.29 − 8.4 (PIN2-1) + 2.93 (LIC1-1) − 1.96 (LIC1-2) − 1.5 (D3-2) + 6.6 (D17-2) + 4.64 (qTA8a-1) + 1.68 (TAC3) − 3.24 (TAC1) | |||||
| R2 | 0.53 | |||||
| Adjusted R2 | 0.51 | |||||
| F-statistic | 36.92 | |||||
| p-value | <0.001 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, S.; Kang, Y.S.; Lee, Y.K.; Koh, H.-J. Evaluating Multiple Allelic Combination to Determine Tiller Angle Variation in Rice. Agriculture 2020, 10, 428. https://doi.org/10.3390/agriculture10100428
Jang S, Kang YS, Lee YK, Koh H-J. Evaluating Multiple Allelic Combination to Determine Tiller Angle Variation in Rice. Agriculture. 2020; 10(10):428. https://doi.org/10.3390/agriculture10100428
Chicago/Turabian StyleJang, Su, Yoo Seok Kang, Yoon Kyung Lee, and Hee-Jong Koh. 2020. "Evaluating Multiple Allelic Combination to Determine Tiller Angle Variation in Rice" Agriculture 10, no. 10: 428. https://doi.org/10.3390/agriculture10100428
APA StyleJang, S., Kang, Y. S., Lee, Y. K., & Koh, H.-J. (2020). Evaluating Multiple Allelic Combination to Determine Tiller Angle Variation in Rice. Agriculture, 10(10), 428. https://doi.org/10.3390/agriculture10100428





