Seven Years Leptospirosis Follow-Up in a Critical Care Unit of a French Metropolitan Hospital; Role of Real Time PCR for a Quick and Acute Diagnosis
Abstract
1. Introduction
2. Experimental Section
2.1. Study Design
2.2. Real-Time PCR Method
2.3. Clinical and Biological Data
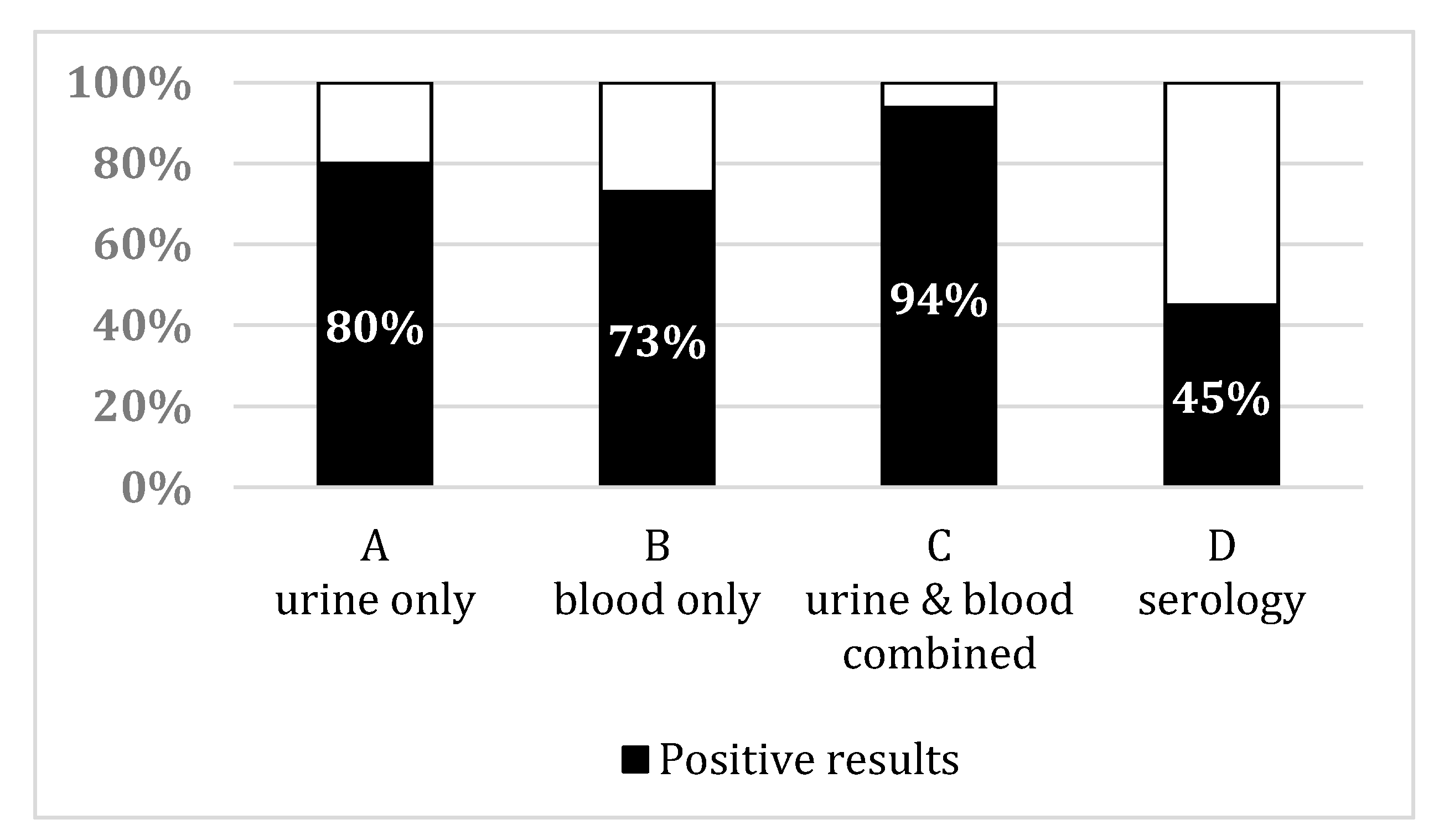
3. Results
3.1. Epidemiological Data
3.2. Clinical and Biological Characteristics
3.3. Leptospirosis Sampling Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef]
- Baranton, G.; Postic, D. Trends in leptospirosis epidemiology in France. Sixty-six years of passive serological surveillance from 1920 to 2003. Int. J. Infect. Dis. 2006, 10, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef] [PubMed]
- Haake, D.A.; Levett, P.N. Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97. [Google Scholar] [PubMed]
- Martins, M.G.; Matos, K.T.; da Silva, M.V.; de Abreu, M.T. Ocular manifestations in the acute phase of leptospirosis. Ocul. Immunol. Inflamm. 1998, 6, 75–79. [Google Scholar] [CrossRef]
- Haute Autorité de Santé-Diagnostic Biologique de la Leptospirose. Available online: https://www.has-sante.fr/portail/jcms/c_1084168/fr/diagnostic-biologique-de-la-leptospirose (accessed on 31 March 2019).
- World Health Organization. Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control. 2003. Available online: https://apps.who.int/iris/handle/10665/42667 (accessed on 17 July 2020).
- Merien, F.; Baranton, G.; Perolat, P. Comparison of polymerase chain reaction with microagglutination test and culture for diagnosis of leptospirosis. J. Infect. Dis. 1995, 172, 281–285. [Google Scholar] [CrossRef]
- Bourhy, P.; Bremont, S.; Zinini, F.; Giry, C.; Picardeau, M. Comparison of real-time PCR assays for detection of pathogenic Leptospira spp. in blood and identification of variations in target sequences. J. Clin. Microbiol. 2011, 49, 2154–2160. [Google Scholar] [CrossRef]
- Agampodi, S.B.; Matthias, M.A.; Moreno, A.C.; Vinetz, J.M. Utility of Quantitative Polymerase Chain Reaction in Leptospirosis Diagnosis: Association of Level of Leptospiremia and Clinical Manifestations in Sri Lanka. Clin. Infect. Dis. 2012, 54, 1249–1255. [Google Scholar] [CrossRef]
- Goarant, C.; Laumond-Barny, S.; Perez, J.; Vernel-Pauillac, F.; Chanteau, S.; Guigon, A. Outbreak of leptospirosis in New Caledonia: Diagnosis issues and burden of disease. Trop. Med. Int. Health 2009, 14, 926–929. [Google Scholar] [CrossRef]
- Pailhoriès, H.; Buzelé, R.; Picardeau, M.; Robert, S.; Mercier, E.; Mereghetti, L.; Lanotte, P. Molecular characterization of Leptospira sp by multilocus variable number tandem repeat analysis (MLVA) from clinical samples: A case report. Int. J. Infect. Dis. 2015, 37, 119–121. [Google Scholar] [CrossRef][Green Version]
- Slack, A.; Symonds, M.; Dohnt, M.; Harris, C.; Brookes, D.; Smythe, L. Evaluation of a modified Taqman assay detecting pathogenic Leptospira spp. against culture and Leptospira-specific IgM enzyme-linked immunosorbent assay in a clinical environment. Diagn. Microbiol. Infect. Dis. 2007, 57, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Engelberts, M.F.M.; Boer, K.R.; Ahmed, N.; Hartskeerl, R.A. Development and validation of a real-time PCR for detection of pathogenic leptospira species in clinical materials. PLoS ONE 2009, 4, e7093. [Google Scholar] [CrossRef] [PubMed]
- Thaipadunpanit, J.; Chierakul, W.; Wuthiekanun, V.; Limmathurotsakul, D.; Amornchai, P.; Boonslip, S.; Smythe, L.D.; Limpaiboon, R.; Hoffmaster, A.R.; Day, N.P.J.; et al. Diagnostic Accuracy of Real-Time PCR Assays Targeting 16S rRNA and lipl32 Genes for Human Leptospirosis in Thailand: A Case-Control Study. PLoS ONE 2011, 6, e16236. [Google Scholar] [CrossRef] [PubMed]
- Villumsen, S.; Pedersen, R.; Borre, M.B.; Ahrens, P.; Jensen, J.S.; Krogfelt, K.A. Novel TaqMan® PCR for detection of Leptospira species in urine and blood: Pit-falls of in silico validation. J. Microbiol. Methods 2012, 91, 184–190. [Google Scholar] [CrossRef]
- Merien, F.; Portnoi, D.; Bourhy, P.; Charavay, F.; Berlioz-Arthaud, A.; Baranton, G. A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol. Lett. 2005, 249, 139–147. [Google Scholar] [CrossRef]
- Guernier, V.; Lagadec, E.; Cordonin, C.; Le Minter, G.; Gomard, Y.; Pagès, F.; Jaffar-Bandjee, M.-C.; Michault, A.; Tortosa, P.; Dellagi, K. Human Leptospirosis on Reunion Island, Indian Ocean: Are Rodents the (Only) Ones to Blame? PLoS Negl. Trop. Dis. 2016, 10, e0004733. [Google Scholar] [CrossRef]
- Fornazari, F.; Langoni, H.; Marson, P.M.; Nóbrega, D.B.; Teixeira, C.R. Leptospira reservoirs among wildlife in Brazil: Beyond rodents. Acta Trop. 2018, 178, 205–212. [Google Scholar] [CrossRef]
- Thibeaux, R.; Iraola, G.; Ferrés, I.; Bierque, E.; Girault, D.; Soupé-Gilbert, M.-E.; Picardeau, M.; Goarant, C. Deciphering the unexplored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb. Genom. 2018, 4, e000144. [Google Scholar] [CrossRef]
- Hochedez, P.; Escher, M.; Decoussy, H.; Pasgrimaud, L.; Martinez, R.; Rosine, J.; Théodose, R.; Bourhy, P.; Picardeau, M.; Olive, C.; et al. Outbreak of leptospirosis among canyoning participants, Martinique, 2011. Euro. Surveill. 2013, 18, 20472. [Google Scholar]
- Esteves, L.M.; Bulhões, S.M.; Branco, C.C.; Carreira, T.; Vieira, M.L.; Gomes-Solecki, M.; Mota-Vieira, L. Diagnosis of Human Leptospirosis in a Clinical Setting: Real-Time PCR High Resolution Melting Analysis for Detection of Leptospira at the Onset of Disease. Sci. Rep. 2018, 8, 9213. [Google Scholar] [CrossRef]
- Bal, A.E.; Gravekamp, C.; Hartskeerl, R.A.; De Meza-Brewster, J.; Korver, H.; Terpstra, W.J. Detection of leptospires in urine by PCR for early diagnosis of leptospirosis. J. Clin. Microbiol. 1994, 32, 1894–1898. [Google Scholar] [CrossRef] [PubMed]
- Dadon, Y.; Haas, E.J.; Kaliner, E.; Anis, E.; Singer, S.R.; Atiya-Nasagi, Y.; Cohen-Dar, M.; Avramovich, E.; King, R.; Sued, O.; et al. Outbreak of human leptospirosis linked to contaminated water bodies in Northern Israel, June to August 2018. Euro Surveill. 2018, 23, 180486. [Google Scholar] [CrossRef] [PubMed]
- Riediger, I.N.; Stoddard, R.A.; Ribeiro, G.S.; Nakatani, S.M.; Moreira, S.D.R.; Skraba, I.; Biondo, A.W.; Reis, M.G.; Hoffmaster, A.R.; Vinetz, J.M.; et al. Rapid, actionable diagnosis of urban epidemic leptospirosis using a pathogenic Leptospira lipL32-based real-time PCR assay. PLoS Negl. Trop. Dis. 2017, 11, e0005940. [Google Scholar] [CrossRef] [PubMed]
- Miailhe, A.-F.; Mercier, E.; Maamar, A.; Lacherade, J.-C.; Le Thuaut, A.; Gaultier, A.; Asfar, P.; Argaud, L.; Ausseur, A.; Ben Salah, A.; et al. Severe leptospirosis in non-tropical areas: A nationwide, multicentre, retrospective study in French ICUs. Intensive Care Med. 2019, 45, 1763–1773. [Google Scholar] [CrossRef] [PubMed]
| Age | Gender | Date (M/Y) | Main Symptoms | Biological Data | Samples (Results) | Delay between First Symptom and Sampling (Days) | Duration of ICU Hospitalization (Days) | Treatment Received | Outcome | Epidemiological Data |
|---|---|---|---|---|---|---|---|---|---|---|
| 60 | M | September 2011 | Fever, myalgia; acute respiratory distress and acute kidney failure | Creatininemia: 516 µmol/L | PCR on blood and urine (positive)/no serology performed | 3 | 1 | Ceftriaxone | Death | Contact with stagnant water |
| Elevated liver enzymes | ||||||||||
| Thrombocytopenia: 81 G/L | ||||||||||
| 60 | F | April 2012 | Fever; vomiting; acute respiratory distress syndrome due to intra-alveolar hemorrhage, acute kidney failure and jaundice | Creatininemia: 405µmol/L | PCR on blood, urine, lung and liver post mortem biopsies (positive)/serology on day 9 (positive) | 7 | 2 | Ceftriaxone | Death | Unknown |
| Elevated liver enzymes | ||||||||||
| Bilirubinemia: 341µmol/L | ||||||||||
| Thrombocytopenia: 8 G/L | ||||||||||
| 55 | M | July 2013 | Fever; myalgia, acute respiratory distress; acute kidney failure and jaundice | Creatininemia: 168 µmol/L | PCR on blood and urine (positive)/serology on day 6 (negative) | 6 | 3 | Ceftriaxone | Complete recovery | Swimming in a river |
| Bilirubinemia: 706 µmol/L Thrombocytopenia: 12 G/L | ||||||||||
| 20 | M | August 2013 | Fever; myalgia, acute respiratory distress with intra alveolar hemorrhage; acute kidney failure and jaundice | Creatininemia: 124 µmol/L | PCR on blood and urine (positive)/serology on day 2 (negative) | 2 | 8 | Ceftriaxone | Complete recovery | Swimming in a river/skin lesions |
| Bilirubinemia: 217 µmol/L Thrombocytopenia: 25 G/L | ||||||||||
| 43 | M | June 2014 | Fever, myalgia, arthralgia, diarrhea, cutaneous rash; Acute Kidney Failure | Creatininemia: 140 µmol/L | PCR on blood (positive) and urine (negative)/serology on day 7 (negative) | 4 | 8 | Ceftriaxone followed by Amoxicillin | Complete recovery | Consumption of stagnant water |
| Elevated liver enzymes | ||||||||||
| Rhabdomyolysis | ||||||||||
| 39 | M | July 2014 | Fever and jaundice | Bilirubinemia: 53 µmol/L | PCR on blood (positive) no urine sampling/serology on day 8 (negative) | 7 | 8 | Ceftriaxone followed by Doxycycline | Complete recovery | Swimming in a river |
| Elevated liver enzymes Thrombocytopenia: 87 G/L | ||||||||||
| 49 | M | August 2014 | Fever, myalgia, diarrhea, acute respiratory distress with intra-alveolar hemorrhage requiring oro-tracheal intubation; acute kidney failure and jaundice | Creatininemia: 223 µmol/L | PCR on urine (positive) no plasma sampling/serology on day 7 (negative) and day 22 (positive) | 7 | 14 | Ceftriaxone | Complete recovery | Fishing |
| Elevated liver enzyme | ||||||||||
| Bilirubinemia: 405 µmol/L | ||||||||||
| Anemia | ||||||||||
| Thrombocytopenia: 10 G/L | ||||||||||
| 71 | F | August 2014 | Fever, diarrhea, acute kidney failure and jaundice | Creatininemia: 164 µmol/L | PCR on blood and urine (positive)/no serology performed | 4 | 6 | Ceftriaxone followed by Doxycycline | Complete recovery | Contact with stagnant water and consumption of water from a well |
| Elevated liver enzymes | ||||||||||
| Bilirubinemia: 278 µmol/L | ||||||||||
| Thrombocytopenia: 57 G/L | ||||||||||
| 65 | M | November 2014 | Fever, acute kidney failure and jaundice | Creatininemia: 411 µmol/L | PCR on blood and urine (positive)/serology on day 30 (positive) | 6 | 3 | Ceftriaxone followed by Amoxicillin | Complete recovery | Hunting |
| Elevated liver enzymes | ||||||||||
| Bilirubinemia: 46 µmol/L | ||||||||||
| Thrombocytopenia: 10 G/L | ||||||||||
| 58 | M | December 2014 | Fever, myalgia, arthralgia, acute kidney failure and jaundice | Creatininemia: 327 µmol/L | PCR on blood, urine and CSF (negative)/serology on day 17 (positive) | 10 | 4 | Amoxicillin and Ofloxacin | Complete recovery | Swimming in a river |
| Elevated liver enzymes | ||||||||||
| Bilirubinemia: 139 µmol/L | ||||||||||
| Thrombocytopenia: 83 G/L | ||||||||||
| 37 | M | May 2015 | Fever, myalgia, rhabdomyolysis, acute kidney failure | Creatininemia: 210 µmol/L | PCR on blood (negative) and urine (positive)/serology on day 6 (positive) | 6 | 4 | Ceftriaxone | Complete recovery | Swimming in a river |
| Elevated liver enzymes | ||||||||||
| Thrombocytopenia: 139 G/L | ||||||||||
| 72 | M | June 2015 | Fever, myalgia, acute kidney failure requiring extra-renal purification, and jaundice | Creatininemia: 404 µmol/L Elevated liver enzymes Bilirubinemia: 177µmol/L Thrombocytopenia: 12 G/L Rhabdomyolysis | PCR on blood and urine (positive)/serology on day 3 (negative) | 3 | 8 | Ceftriaxone | Complete recovery | Hunting |
| 61 | M | July 2015 | Fever, myalgia, acute respiratory distress requiring oro-tracheal intubation, acute kidney failure with hematuria, and jaundice | Creatininemia: 656 µmol/L | PCR on blood and urine (positive)/no serology performed | 5 | 9 | Ceftriaxone followed by Amoxicillin | Complete recovery | Hunting and fishing |
| Elevated liver enzymes | ||||||||||
| Bilirubinemia: 406 µmol/L | ||||||||||
| Thrombocytopenia: 15 G/L | ||||||||||
| Anemia | ||||||||||
| 51 | M | July 2015 | Fever, myalgia, arthralgia, acute kidney failure and jaundice | Creatininemia: 257 µmol/L | PCR on blood (negative) and urine (positive)/serology on day 6 (negative) | 6 | 3 | Ceftriaxone | Complete recovery | Fishing |
| Elevated liver enzymes | ||||||||||
| Bilirubinemia: 31 µmol/L Thrombocytopenia: 102 G/L | ||||||||||
| 15 | M | September 2016 | Fever, myalgia, arthralgia, acute kidney failure and jaundice | Creatininemia: 137 µmol/L | PCR on blood (positive) and urine (negative)/no serology performed | 5 | 4 | Ceftriaxone | Complete recovery | Swimming in a river |
| Elevated liver enzymes | ||||||||||
| Bilirubinemia: 52 µmol/L Thrombocytopenia: 22 G/L | ||||||||||
| 62 | M | January 2017 | Fever, myalgia, arthralgia, rhabdomyolysis, acute kidney failure and jaundice | Creatininemia: 331 µmol/L | PCR on blood (negative) and urine (positive)/no serology performed | 8 | 2 | Ceftriaxone | Complete recovery | Gardening |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahuaud, O.; Pastuszka, A.; Le Brun, C.; Ehrmann, S.; Lanotte, P. Seven Years Leptospirosis Follow-Up in a Critical Care Unit of a French Metropolitan Hospital; Role of Real Time PCR for a Quick and Acute Diagnosis. J. Clin. Med. 2020, 9, 3011. https://doi.org/10.3390/jcm9093011
Bahuaud O, Pastuszka A, Le Brun C, Ehrmann S, Lanotte P. Seven Years Leptospirosis Follow-Up in a Critical Care Unit of a French Metropolitan Hospital; Role of Real Time PCR for a Quick and Acute Diagnosis. Journal of Clinical Medicine. 2020; 9(9):3011. https://doi.org/10.3390/jcm9093011
Chicago/Turabian StyleBahuaud, Olivier, Adeline Pastuszka, Cécile Le Brun, Stephan Ehrmann, and Philippe Lanotte. 2020. "Seven Years Leptospirosis Follow-Up in a Critical Care Unit of a French Metropolitan Hospital; Role of Real Time PCR for a Quick and Acute Diagnosis" Journal of Clinical Medicine 9, no. 9: 3011. https://doi.org/10.3390/jcm9093011
APA StyleBahuaud, O., Pastuszka, A., Le Brun, C., Ehrmann, S., & Lanotte, P. (2020). Seven Years Leptospirosis Follow-Up in a Critical Care Unit of a French Metropolitan Hospital; Role of Real Time PCR for a Quick and Acute Diagnosis. Journal of Clinical Medicine, 9(9), 3011. https://doi.org/10.3390/jcm9093011





