Phenoconversion of Cytochrome P450 Metabolism: A Systematic Review
Abstract
1. Introduction
2. Methods
2.1. Identification of Eligible Studies
2.2. Data Extraction
3. Results
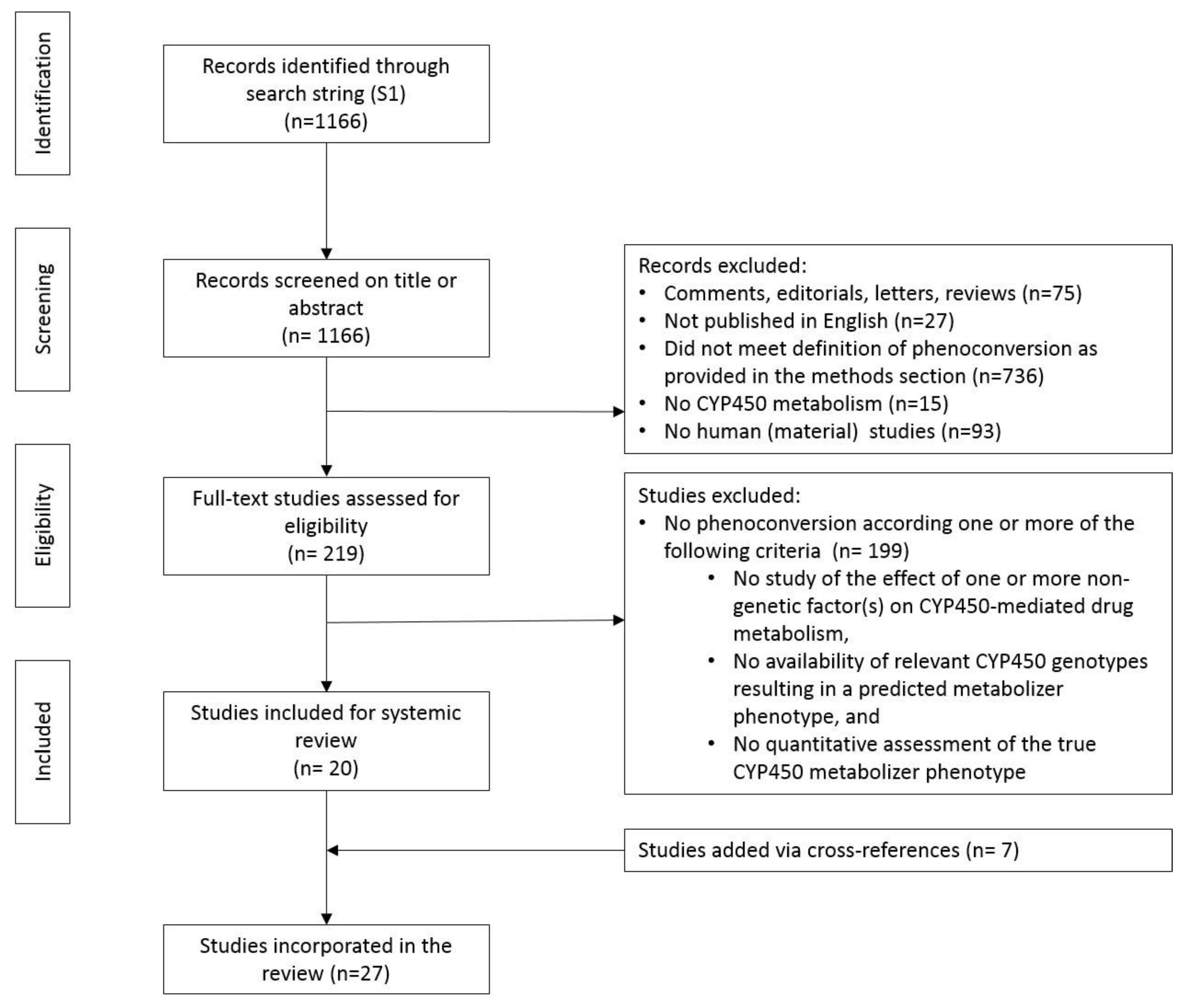
3.1. Study Selection
3.2. Impact of Studies
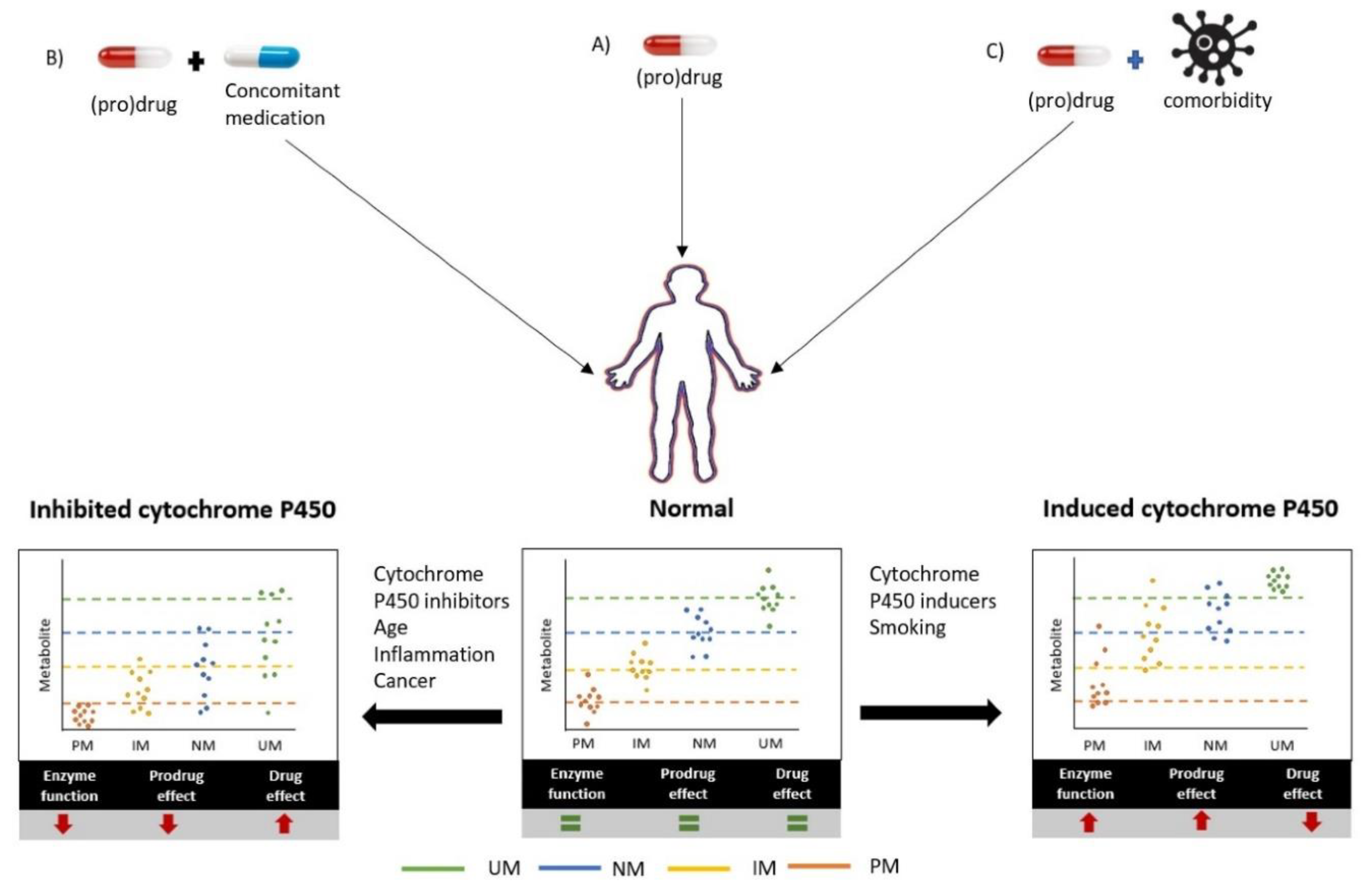
3.3. Phenoconversion of CYP450 by Concomitant Drugs and Other Extrinsic Factors
3.3.1. Anticonvulsants
3.3.2. Anticoagulants
3.3.3. Antihypertensives
3.3.4. Antimuscarinics
3.3.5. Antipsychotics
3.3.6. Antitussive and Pain Medication
3.3.7. Cardiac Drugs
3.3.8. Non-Steroidal Anti-Inflammatory Drugs (NSAIDs)
3.3.9. Proton Pump Inhibitors
3.3.10. Antiestrogenic Drugs
3.4. Phenoconversion of CYP450 by Patient- and Disease-Related Factors
3.4.1. Age
3.4.2. Cancer
3.4.3. Inflammation
3.4.4. Pregnancy
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Search Strategy
References
- Abbasi, J. Getting Pharmacogenomics into the Clinic. JAMA 2016, 316, 1533–1535. [Google Scholar] [CrossRef] [PubMed]
- Roden, D.M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; Mensah, G.A.; Peterson, J.F.; Van Driest, S.L. Pharmacogenomics. Lancet 2019, 394, 521–532. [Google Scholar] [CrossRef]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharm. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Swen, J.J.; Nijenhuis, M.; de Boer, A.; Grandia, L.; Maitland-van der Zee, A.H.; Mulder, H.; Rongen, G.A.; van Schaik, R.H.; Schalekamp, T.; Touw, D.J.; et al. Pharmacogenetics: From bench to byte—An update of guidelines. Clin. Pharm. Ther. 2011, 89, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharm. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Ieiri, I.; Yamada, S.; Seto, K.; Morita, T.; Kaneda, T.; Mamiya, K.; Tashiro, N.; Higuchi, S.; Otsubo, K. A CYP2D6 phenotype-genotype mismatch in Japanese psychiatric patients. Pharmacopsychiatry 2003, 36, 192–196. [Google Scholar] [CrossRef]
- Rost, K.L.; Brockmoller, J.; Esdorn, F.; Roots, I. Phenocopies of poor metabolizers of omeprazole caused by liver disease and drug treatment. J. Hepatol. 1995, 23, 268–277. [Google Scholar] [CrossRef]
- Kiss, A.F.; Vasko, D.; Deri, M.T.; Toth, K.; Monostory, K. Combination of CYP2C19 genotype with non-genetic factors evoking phenoconversion improves phenotype prediction. Pharm. Rep. 2018, 70, 525–532. [Google Scholar] [CrossRef]
- Shah, R.R.; Gaedigk, A.; LLerina, A.; Eichelbaum, M.; Stingl, J.; Smith, R.L. CYP450 genotype and pharmacogenetic association studies: A critical appraisal. Pharmacogenomics 2016, 17, 259–275. [Google Scholar] [CrossRef]
- Karle, J.; Bolbrinker, J.; Vogl, S.; Kreutz, R.; Denkert, C.; Eucker, J.; Wischnewsky, M.; Possinger, K.; Regierer, A.C. Influence of CYP2D6-genotype on tamoxifen efficacy in advanced breast cancer. Breast Cancer Res. Treat. 2013, 139, 553–560. [Google Scholar] [CrossRef][Green Version]
- D’Empaire, I.; Guico-Pabia, C.J.; Preskorn, S.H. Antidepressant treatment and altered CYP2D6 activity: Are pharmacokinetic variations clinically relevant? J. Psychiatr. Pract. 2011, 17, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Klein, K.; Zanger, U. Pharmacogenomics of Cytochrome P450 3A4: Recent Progress Toward the “Missing Heritability” Problem. Front. Genet. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.R.; Smith, R.L. Addressing phenoconversion: The Achilles’ heel of personalized medicine. Br. J. Clin. Pharmacol. 2015, 79, 222–240. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.R.; Smith, R.L. Inflammation-induced phenoconversion of polymorphic drug metabolizing enzymes: Hypothesis with implications for personalized medicine. Drug Metab. Dispos. 2015, 43, 400–410. [Google Scholar] [CrossRef]
- Caudle, K.E.; Sangkuhl, K.; Whirl-Carrillo, M.; Swen, J.J.; Haidar, C.E.; Klein, T.E.; Gammal, R.S.; Relling, M.V.; Scott, S.A.; Hertz, D.L.; et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 2020, 13, 116–124. [Google Scholar] [CrossRef]
- Flockhart, D. Drug Interactions: Cytochrome P450 Drug Interaction Table; Indiana University School of Medicine: Indianapolis, IN, USA, 2007. [Google Scholar]
- Birdwell, K.A.; Decker, B.; Barbarino, J.M.; Peterson, J.F.; Stein, C.M.; Sadee, W.; Wang, D.; Vinks, A.A.; He, Y.; Swen, J.J.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin. Pharm. Ther. 2015, 98, 19–24. [Google Scholar] [CrossRef]
- Counsell, C. Formulating Questions and Locating Primary Studies for Inclusion in Systematic Reviews. Ann. Intern. Med. 1997, 127, 380–387. [Google Scholar] [CrossRef]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; Group, P.-P. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4. [Google Scholar] [CrossRef]
- Bank, P.C.D.; Caudle, K.E.; Swen, J.J.; Gammal, R.S.; Whirl-Carrillo, M.; Klein, T.E.; Relling, M.V.; Guchelaar, H.J. Comparison of the Guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin. Pharmacol. Ther. 2018, 103, 599–618. [Google Scholar] [CrossRef]
- Jogamoto, T.; Yamamoto, Y.; Fukuda, M.; Suzuki, Y.; Imai, K.; Takahashi, Y.; Inoue, Y.; Ohtsuka, Y. Add-on stiripentol elevates serum valproate levels in patients with or without concomitant topiramate therapy. Epilepsy Res. 2017, 130, 7–12. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Takahashi, Y.; Imai, K.; Miyakawa, K.; Nishimura, S.; Kasai, R.; Ikeda, H.; Takayama, R.; Mogami, Y.; Yamaguchi, T.; et al. Influence of CYP2C19 polymorphism and concomitant antiepileptic drugs on serum clobazam and N-desmethyl clobazam concentrations in patients with epilepsy. Ther. Drug Monit. 2013, 35, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Hata, M.; Shiono, M.; Akiyama, K.; Sezai, A.; Wakui, S.; Kimura, H.; Sekino, H. Incidence of drug interaction when using proton pump inhibitor and warfarin according to cytochrome P450 2C19 (CYP2C19) genotype in Japanese. Thorac. Cardiovasc. Surg. 2015, 63, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Llerena, A.; Berecz, R.; de la Rubia, A.; Fernández-Salguero, P.; Dorado, P. Effect of Thioridazine Dosage on the Debrisoquine Hydroxylation Phenotype in Psychiatric Patients with Different CYP2D6 Genotypes. Ther. Drug Monit. 2001, 23. [Google Scholar] [CrossRef]
- Brynne, N.; Svanstrom, C.; Aberg-Wistedt, A.; Hallen, B.; Bertilsson, L. Fluoxetine inhibits the metabolism of tolterodine-pharmacokinetic implications and proposed clinical relevance. Br. J. Clin. Pharmacol. 1999, 48, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Lisbeth, P.; Vincent, H.; Kristof, M.; Bernard, S.; Manuel, M.; Hugo, N. Genotype and co-medication dependent CYP2D6 metabolic activity: Effects on serum concentrations of aripiprazole, haloperidol, risperidone, paliperidone and zuclopenthixol. Eur. J. Clin. Pharmacol. 2016, 72, 175–184. [Google Scholar] [CrossRef]
- Kiss, A.; Menus, A.; Toth, K.; Deri, M.; Sirok, D.; Gabri, E.; Belic, A.; Csukly, G.; Bitter, I.; Monostory, K. Phenoconversion of CYP2D6 by inhibitors modifies aripiprazole exposure. Eur. Arch. Psychiatry Clin. Neurosci. 2019. [Google Scholar] [CrossRef]
- Skogh, E.; Sjodin, I.; Josefsson, M.; Dahl, M.L. High correlation between serum and cerebrospinal fluid olanzapine concentrations in patients with schizophrenia or schizoaffective disorder medicating with oral olanzapine as the only antipsychotic drug. J. Clin. Psychopharmacol. 2011, 31, 4–9. [Google Scholar] [CrossRef]
- Storelli, F.; Matthey, A.; Lenglet, S.; Thomas, A.; Desmeules, J.; Daali, Y. Impact of CYP2D6 Functional Allelic Variations on Phenoconversion and Drug-Drug Interactions. Clin. Pharmacol. Ther. 2018, 104, 148–157. [Google Scholar] [CrossRef]
- Kumar, V.; Brundage, R.C.; Oetting, W.S.; Leppik, I.E.; Tracy, T.S. Differential Genotype Dependent Inhibition of CYP2C9 in Humans. Drug Metab. Dispos. 2008, 36, 1242–1248. [Google Scholar] [CrossRef]
- Miura, M.; Tada, H.; Yasui-Furukori, N.; Uno, T.; Sugawara, K.; Tateishi, T.; Suzuki, T. Enantioselective disposition of lansoprazole in relation to CYP2C19 genotypes in the presence of fluvoxamine. Br. J. Clin. Pharmacol. 2005, 60, 61–68. [Google Scholar] [CrossRef]
- Borges, S.; Desta, Z.; Li, L.; Skaar, T.C.; Ward, B.A.; Nguyen, A.; Jin, Y.; Storniolo, A.M.; Nikoloff, D.M.; Wu, L.; et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clin Pharm. Ther. 2006, 80, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Teft, W.A.; Gong, I.Y.; Dingle, B.; Potvin, K.; Younus, J.; Vandenberg, T.A.; Brackstone, M.; Perera, F.E.; Choi, Y.H.; Zou, G.; et al. CYP3A4 and seasonal variation in vitamin D status in addition to CYP2D6 contribute to therapeutic endoxifen level during tamoxifen therapy. Breast Cancer Res. Treat. 2013, 139, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Ishizawa, Y.; Yasui-Furukori, N.; Takahata, T.; Sasaki, M.; Tateishi, T. The effect of aging on the relationship between the cytochrome P450 2C19 genotype and omeprazole pharmacokinetics. Clin. Pharmacokinet. 2005, 44, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.L.; Bhargava, P.; Cherrouk, I.; Marshall, J.L.; Flockhart, D.A.; Wainer, I.W. A discordance of the cytochrome P450 2C19 genotype and phenotype in patients with advanced cancer. Br. J. Clin. Pharmacol. 2000, 49, 485–488. [Google Scholar] [CrossRef]
- Helsby, N.A.; Lo, W.Y.; Sharples, K.; Riley, G.; Murray, M.; Spells, K.; Dzhelai, M.; Simpson, A.; Findlay, M. CYP2C19 pharmacogenetics in advanced cancer: Compromised function independent of genotype. Br. J. Cancer 2008, 99, 1251–1255. [Google Scholar] [CrossRef]
- Burns, K.E.; Goldthorpe, M.A.; Porteus, F.; Browett, P.; Helsby, N.A. CYP2C19 genotype-phenotype discordance in patients with multiple myeloma leads to an acquired loss of drug-metabolising activity. Cancer Chemother. Pharmacol. 2014, 73, 651–655. [Google Scholar] [CrossRef]
- Goktaş, M.T.; Hatta, F.; Karaca, O.; Kalkisim, S.; Kilic, L.; Akdogan, A.; Babaoglu, M.O.; Bozkurt, A.; Helldén, A.; Bertilsson, L.; et al. Lower CYP2C9 activity in Turkish patients with Behçet’s disease compared to healthy subjects: A down-regulation due to inflammation? Eur. J. Clin. Pharm. 2015, 71, 1223–1228. [Google Scholar] [CrossRef]
- Ohnishi, A.; Murakami, S.; Akizuki, S.; Mochizuki, J.; Echizen, H.; Takagi, I. In vivo metabolic activity of CYP2C19 and CYP3A in relation to CYP2C19 genetic polymorphism in chronic liver disease. J. Clin. Pharmacol. 2005, 45, 1221–1229. [Google Scholar] [CrossRef]
- Veringa, A.; Ter Avest, M.; Span, L.F.; van den Heuvel, E.R.; Touw, D.J.; Zijlstra, J.G.; Kosterink, J.G.; van der Werf, T.S.; Alffenaar, J.C. Voriconazole metabolism is influenced by severe inflammation: A prospective study. J. Antimicrob. Chemother. 2017, 72, 261–267. [Google Scholar] [CrossRef]
- Girardin, F.; Daali, Y.; Gex-Fabry, M.; Rebsamen, M.; Roux-Lombard, P.; Cerny, A.; Bihl, F.; Binek, J.; Moradpour, D.; Negro, F.; et al. Liver kidney microsomal type 1 antibodies reduce the CYP2D6 activity in patients with chronic hepatitis C virus infection. J. Viral Hepat. 2012, 19, 568–573. [Google Scholar] [CrossRef]
- Suzuki, Y.; Muraya, N.; Fujioka, T.; Sato, F.; Tanaka, R.; Matsumoto, K.; Sato, Y.; Ohno, K.; Mimata, H.; Kishino, S.; et al. Factors involved in phenoconversion of CYP3A using 4beta-hydroxycholesterol in stable kidney transplant recipients. Pharmacol. Rep. Pract. 2019, 71, 276–281. [Google Scholar] [CrossRef]
- Wadelius, M.; Darj, E.; Frenne, G.; Rane, A. Induction of CYP2D6 in pregnancy. Clin. Pharmacol. Ther. 1997, 62, 400–407. [Google Scholar] [CrossRef]
- Ververs, F.F.; Voorbij, H.A.; Zwarts, P.; Belitser, S.V.; Egberts, T.C.; Visser, G.H.; Schobben, A.F. Effect of cytochrome P450 2D6 genotype on maternal paroxetine plasma concentrations during pregnancy. Clin. Pharmacokinet. 2009, 48, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.; Rey, E.; Pons, G.; Rousseau, M.; d’Athis, P.; Olive, G.; Mather, G.G.; Bishop, F.E.; Wurden, C.J.; Labroo, R.; et al. Influence of stiripentol on cytochrome P450-mediated metabolic pathways in humans: In vitro and in vivo comparison and calculation of in vivo inhibition constants. Clin. Pharmacol. Ther. 1997, 62, 490–504. [Google Scholar] [CrossRef]
- Giraud, C.; Tran, A.; Rey, E.; Vincent, J.; Tréluyer, J.M.; Pons, G. In vitro characterization of clobazam metabolism by recombinant cytochrome P450 enzymes: Importance of CYP2C19. Drug Metab. Dispos. 2004, 32, 1279–1286. [Google Scholar] [CrossRef]
- Kroon, L. Drug interactions with smoking. Am. J. Health Syst. Pharm. 2007, 64, 1917–1921. [Google Scholar] [CrossRef]
- Meek, M.D.; Finch, G.L. Diluted Mainstream Cigarette Smoke Condensates Activate Estrogen Receptor and Aryl Hydrocarbon Receptor-Mediated Gene Transcription. Environ. Res. 1999, 80, 9–17. [Google Scholar] [CrossRef]
- Storelli, F.; Desmeules, J.; Daali, Y. Genotype-sensitive reversible and time-dependent CYP2D6 inhibition in human liver microsomes. Basic Clin. Pharmacol. Toxicol. 2019, 124, 170–180. [Google Scholar] [CrossRef]
- Doki, K.; Sekiguchi, Y.; Kuga, K.; Aonuma, K.; Homma, M. Serum flecainide S/R ratio reflects the CYP2D6 genotype and changes in CYP2D6 activity. Drug Metab. Pharmacokinet. 2015, 30, 257–262. [Google Scholar] [CrossRef]
- Schmiedlin-Ren, P.; Thummel, K.E.; Fisher, J.M.; Paine, M.F.; Lown, K.S.; Watkins, P.B. Expression of enzymatically active CYP3A4 by Caco-2 cells grown on extracellular matrix-coated permeable supports in the presence of 1alpha,25-dihydroxyvitamin D3. Mol. Pharm. 1997, 51, 741–754. [Google Scholar] [CrossRef]
- Thummel, K.E.; Brimer, C.; Yasuda, K.; Thottassery, J.; Senn, T.; Lin, Y.; Ishizuka, H.; Kharasch, E.; Schuetz, J.; Schuetz, E. Transcriptional control of intestinal cytochrome P-4503A by 1alpha,25-dihydroxy vitamin D3. Mol. Pharm. 2001, 60, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Lanchote, V.L.; Almeida, R.; Barral, A.; Barral-Netto, M.; Marques, M.P.; Moraes, N.V.; da Silva, A.M.; Souza, T.M.; Suarez-Kurtz, G. Impact of visceral leishmaniasis and curative chemotherapy on cytochrome P450 activity in Brazilian patients. Br. J. Clin. Pharm. 2015, 80, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, N.E.; Shields, P.G.; Landi, M.T.; Shaw, G.L.; Tucker, M.A.; Hoover, R.; Sugimura, H.; Weston, A.; Harris, C.C. The debrisoquine metabolic phenotype and DNA-based assays: Implications of misclassification for the association of lung cancer and the debrisoquine metabolic phenotype. Environ. Health Perspect. 1992, 98, 101–105. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Velenosi, T.J.; Feere, D.A.; Sohi, G.; Hardy, D.B.; Urquhart, B.L. Decreased nuclear receptor activity and epigenetic modulation associates with down-regulation of hepatic drug-metabolizing enzymes in chronic kidney disease. FASEB J. 2014, 28, 5388–5397. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, Z.; Wang, L.; Gao, Y. Upregulation of nuclear factor-κB activity mediates CYP24 expression and reactive oxygen species production in indoxyl sulfate-induced chronic kidney disease. Nephrology 2016, 21, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Takahashi, Y.; Imai, K.; Mogami, Y.; Matsuda, K.; Nakai, M.; Kagawa, Y.; Inoue, Y. Interaction between sulthiame and clobazam: Sulthiame inhibits the metabolism of clobazam, possibly via an action on CYP2C19. Epilepsy Behav. 2014, 34, 124–126. [Google Scholar] [CrossRef]
- Frye, R.F.; Zgheib, N.K.; Matzke, G.R.; Chaves-Gnecco, D.; Rabinovitz, M.; Shaikh, O.S.; Branch, R.A. Liver disease selectively modulates cytochrome P450–mediated metabolism. Clin. Pharmacol. Ther. 2006, 80, 235–245. [Google Scholar] [CrossRef]
- Pageaux, G.P.; Micallef, J.; Nataf, M.B.; Levron, J.C.; Lacarelle, B.; Le Moing, J.P.; Bouhours, P.; Blin, O. Pharmacokinetics of sabeluzole and dextromethorphan oxidation capacity in patients with severe hepatic dysfunction and healthy volunteers. Br. J. Clin. Pharmacol. 2001, 51, 164–168. [Google Scholar] [CrossRef]
- Shao, J.G.; Jiang, W.; Li, K.Q.; Lu, J.R.; Sun, Y.Y. Blood concentration of pantoprazole sodium is significantly high in hepatogenic peptic ulcer patients, especially those with a poor CYP2C19 metabolism. J. Dig. Dis. 2009, 10, 55–60. [Google Scholar] [CrossRef]
- Okubo, M.; Murayama, N.; Miura, J.; Chiba, Y.; Yamazaki, H. Effects of cytochrome P450 2D6 and 3A5 genotypes and possible coadministered medicines on the metabolic clearance of antidepressant mirtazapine in Japanese patients. Biochem. Pharmacol. 2015, 93, 104–109. [Google Scholar] [CrossRef]
- Lesche, D.; Mostafa, S.; Everall, I.; Pantelis, C.; Bousman, C.A. Impact of CYP1A2, CYP2C19, and CYP2D6 genotype- and phenoconversion-predicted enzyme activity on clozapine exposure and symptom severity. Pharm. J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, L.B.; Sorensen, R.N.; Miners, J.O.; Somogyi, A.A.; Grgurinovich, N.; Birkett, D.J. Polymorphic hydroxylation of perhexiline in vitro. Br. J. Clin. Pharmacol. 2003, 55, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Corbett, J.L.; Duncan, S.A. iPSC-Derived Hepatocytes as a Platform for Disease Modeling and Drug Discovery. Front. Med. 2019, 6, 265. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.P.; Hyland, R.; Youdim, K. Minimizing polymorphic metabolism in drug discovery: Evaluation of the utility of in vitro methods for predicting pharmacokinetic consequences associated with CYP2D6 metabolism. Drug Metab. Dispos. Biol. Fate Chem. 2006, 34, 1516–1522. [Google Scholar] [CrossRef]
- Yoshihara, M.; Oguchi, A.; Murakawa, Y. Genomic Instability of iPSCs and Challenges in Their Clinical Applications. Adv. Exp. Med. Biol. 2019, 1201, 23–47. [Google Scholar] [CrossRef]
- Dorr, C.R.; Remmel, R.P.; Muthusamy, A.; Fisher, J.; Moriarity, B.S.; Yasuda, K.; Wu, B.; Guan, W.; Schuetz, E.G.; Oetting, W.S.; et al. CRISPR/Cas9 Genetic Modification of CYP3A5 *3 in HuH-7 Human Hepatocyte Cell Line Leads to Cell Lines with Increased Midazolam and Tacrolimus Metabolism. Drug Metab. Dispos. 2017, 45, 957–965. [Google Scholar] [CrossRef]
- Hendriks, D.F.G.; Vorrink, S.U.; Smutny, T.; Sim, S.C.; Nordling, Å.; Ullah, S.; Kumondai, M.; Jones, B.C.; Johansson, I.; Andersson, T.B.; et al. Clinically Relevant Cytochrome P450 3A4 Induction Mechanisms and Drug Screening in Three-Dimensional Spheroid Cultures of Primary Human Hepatocytes. Clin. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef]
- Vorrink, S.U.; Ullah, S.; Schmidt, S.; Nandania, J.; Velagapudi, V.; Beck, O.; Ingelman-Sundberg, M.; Lauschke, V.M. Endogenous and xenobiotic metabolic stability of primary human hepatocytes in long-term 3D spheroid cultures revealed by a combination of targeted and untargeted metabolomics. FASEB J. 2017, 31, 2696–2708. [Google Scholar] [CrossRef]
- Dickschen, K.; Willmann, S.; Thelen, K.; Lippert, J.; Hempel, G.; Eissing, T. Physiologically Based Pharmacokinetic Modeling of Tamoxifen and its Metabolites in Women of Different CYP2D6 Phenotypes Provides New Insight into the Tamoxifen Mass Balance. Front. Pharmacol. 2012, 3, 92. [Google Scholar] [CrossRef]
- Andreu, F.; Colom, H.; Elens, L.; van Gelder, T.; van Schaik, R.H.N.; Hesselink, D.A.; Bestard, O.; Torras, J.; Cruzado, J.M.; Grinyo, J.M.; et al. A New CYP3A5*3 and CYP3A4*22 Cluster Influencing Tacrolimus Target Concentrations: A Population Approach. Clin. Pharmacokinet. 2017, 56, 963–975. [Google Scholar] [CrossRef]
- Woillard, J.B.; Mourad, M.; Neely, M.; Capron, A.; van Schaik, R.H.; van Gelder, T.; Lloberas, N.; Hesselink, D.A.; Marquet, P.; Haufroid, V.; et al. Tacrolimus Updated Guidelines through popPK Modeling: How to Benefit More from CYP3A Pre-emptive Genotyping Prior to Kidney Transplantation. Front. Pharmacol. 2017, 8, 358. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, D.; Tang, W.; Zhou, W.; Al-Huniti, N.; Masson, E. Physiologically Based Pharmacokinetic Modeling to Evaluate the Systemic Exposure of Gefitinib in CYP2D6 Ultrarapid Metabolizers and Extensive Metabolizers. J. Clin. Pharmacol. 2018, 58, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Moj, D.; Britz, H.; Burhenne, J.; Stewart, C.F.; Egerer, G.; Haefeli, W.E.; Lehr, T. A physiologically based pharmacokinetic and pharmacodynamic (PBPK/PD) model of the histone deacetylase (HDAC) inhibitor vorinostat for pediatric and adult patients and its application for dose specification. Cancer Chemother. Pharmacol. 2017, 80, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Titze, M.I.; Schaaf, O.; Hofmann, M.H.; Sanderson, M.P.; Zahn, S.K.; Quant, J.; Lehr, T. A comprehensive pharmacokinetic/pharmacodynamics analysis of the novel IGF1R/INSR inhibitor BI 893923 applying in vitro, in vivo and in silico modeling techniques. Cancer Chemother. Pharmacol. 2016, 77, 1303–1314. [Google Scholar] [CrossRef] [PubMed]
| Pharmacological Class | Drug | Enzyme | Cause of Phenoconversion | Type of Interaction | Drug/Factor Responsible for Phenoconversion | Type of Study | Genotype Assessment | Phenotype Assessment | Effect of Phenoconversion | Subjects | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Anticonvulsants | Valproate | CYP2C19 | Comedication | Inhibition | Stiripentol | 2 | CYP2C9*1/*2/*3 | Concentration-to-dose ratio of valproate | NM: Valproate serum concentration ↑↑ | 28 | [21] |
| CYP2C19*1/*2/*3 | PM: Valproate serum concentration ↑ | (Dravet syndrome patients) | |||||||||
| Clobazam | CYP2C19 | Comedication | Inhibition | Stiripentol/zonisamide | 2 | CYP2C19*1/*2/*3 | Clobazam and N-desmethyl-clobazam via HPLC | 22 NM zonisamide and stiripentol users (total 97 NM patients): N-desmethyl-clobazam concentration dose ratio ↑ | 238 | [22] | |
| NM: *1/*1 | 25 IM zonisamide and stiripentol users (total 133 IM patients): N-desmethyl-clobazam concentration dose ratio ↑ | (epilepsy patients) | |||||||||
| IM: *1/*2 or *1/*3 | 17 PM zonisamide and stiripentol users (total 72 PM patients): N-desmethyl-clobazam concentration dose ratio = | ||||||||||
| PM: *2/*2, *2/*3 or *3/*3 | |||||||||||
| Clobazam | CYP2C19 | Comedication | Induction | Phenytoin/carbamazepine | 2 | CYP2C19*1/*2/*3 | Clobazam and N-desmethyl-clobazam via HPLC | 51 NM phenytoin/carbamazepine users (total 97 NM patients): clobazam concentration dose ratio ↓ | 238 | [22] | |
| NM: *1/*1 | N-desmethyl-clobazam concentration dose ratio ↑ | (epilepsy patients) | |||||||||
| IM: *1/*2 or *1/*3 | 75 IM phenytoin/carbamazepine users (total 133 IM patients): clobazam concentration dose ratio ↓ | ||||||||||
| PM: *2/*2, *2/*3 or *3/*3 | N-desmethyl-clobazam concentration dose ratio ↑ | ||||||||||
| 36 PM phenytoin/carbamazepine users (total 72 PM patients): clobazam concentration dose ratio ↓ | |||||||||||
| N-desmethyl-clobazam concentration dose ratio = | |||||||||||
| Mephenytoin | CYP2C19 | Dietary | Inhibition | Alcohol consumption | 2 | CYP2C19*1/*2/*3/*4/*17 | Mephenytoin 4′-hydroxylation and mephenytoin via HPLC | The genotype-predicted phenotype overestimated (47%) the measured phenotype. For the overestimated group, 7 were alcohol users, resulting in lower CYP2C19 activity. | 114 | [8] | |
| PM: two loss of function alleles (*2, *3 or *4) | CYP2C19 activity: | (liver microsomes) | |||||||||
| IM: one functional and one loss of function allele | PM: <8 pmol/(mg protein*min) | ||||||||||
| NM: *1/*1 | UM: >75 pmol/(mg protein*min) | ||||||||||
| UM: one or two *17 alleles | IM: between 8 and 23 pmol/(mg protein*min) | ||||||||||
| (except *2/*17: IM or NM are accepted) | NM: between 23 and 75 pmol/(mg protein*min) | ||||||||||
| Anticoagulants | Warfarin | CYP2C19 | Comedication | Inhibition | Lansoprazole | 2 | CYP2C19 NM, IM and PM | * | There were no bleeding events in the rabeprazole control group (n = 41). Percentage of bleeding events in lansoprazole group (n = 41): | 82 | [23] |
| NM: 12% ↑ | (open heart surgery patients) | ||||||||||
| IM: 40% ↑↑ | |||||||||||
| PM: 22% ↑ | |||||||||||
| Showing de interaction between warfarin, lansoprazole and CYP2C19, (DDGI) | |||||||||||
| Antihypertensive | Debrisoquine | CYP2D6 | Comedication | Inhibition | Thioridazine | 1 | CYP2D6*1/*3/*4/*5/*6 | Phenotyping by debrisoquine, debrisoquine and 4-hydroxydebrisoquine ratios via GC | 8 NM→PM | 16 | [24] |
| 2 PM = PM | (psychiatric patients) | ||||||||||
| Antimuscarinics | Tolterodine | CYP2D6 | Comedication | Inhibition | Fluoxetine | 2 | CYP2D6*1/*3/*4 | Dealkyl and carboxyl tolterodine LC-ESI-MS/MS | NM: oral clearance of tolterodine ↓ (80%) | 9 | [25] |
| Fluoxetine and norfluoxetine via HPLC-UV | IM: oral clearance of tolterodine ↓↓ (93%) | (depressed patients) | |||||||||
| Possible: NM→PM | |||||||||||
| Antipsychotics | Aripiprazole, haloperidol, risperidone, paliperidone zuclopenthixol | CYP2D6 | Comedication | Inhibition | Strong inhibitors (paroxetine/bupropion) | 1 | CYP2D6*1/*2/*3/*4/*5/*6/*7/*8/*9/*10/*11/*15/*17/*29/*41, deletions and duplications | Concentration of parent compounds antipsychotics via HPLC-MS/MS | 8 patients taking CYP2D6 inhibitors | 82 | [26] |
| Moderate inhibitors (sertraline/duloxetine) | Strong inhibitors: | (psychiatric patients) | |||||||||
| 4 NM→PM | |||||||||||
| 1 IM→PM | |||||||||||
| Moderate inhibitors: | |||||||||||
| 3 NM→IM | |||||||||||
| Aripiprazole | CYP2D6 | Comedication | Inhibition | CYP2D6 inhibitors | 2 | CYP2D6*1/*3/*4/*5/*6/*10/*41 and duplication, CYP3A4*1/*1B/*22, and CYP3A5*1/*3 | Aripiprazole, dehydroaripiprazole, N-dealkyl-aripiprazole, monohydroxy-aripiprazole and dehydroaripiprazole via LC-MS/MS | NM: aripiprazole concentration ↑↑ (50%) | 93 | [27] | |
| UM: aripiprazole concentration ↑ (20%) | (psychiatric patients) | ||||||||||
| Olanzapine | CYP1A2 | Smoking | Induction | Polycyclic aromatic hydrocarbons | 2 | CYP2D6*1/*3/*4/*5/*6/*41 and duplication | Olanzapine and desmethylolanzapine via LD-MS | CYP1A2*1F/*1F smokers: 4′-N-desmethylolanzapine/olanzapine ratio ↑: | 37 | [28] | |
| PM: two defective alleles (*3, *4, *5 or *6 ) | 132% compared to smokers with other CYP1A2 polymorphisms and, 107% compared to nonsmokers that are homozygous carriers of CYP1A2*1F | (schizophrenia or schizoaffective disorder patients) | |||||||||
| IM: one defective allele | |||||||||||
| NM: *1/*1 | |||||||||||
| UM: gene duplication together with *1 | |||||||||||
| ABCB1 | |||||||||||
| CYP1A2*1/*1C/*1D/*1K/*1F | |||||||||||
| Antitussive and pain medication | Dextromethorphan, tramadol | CYP2D6 | Comedication | Inhibition | Duloxetine/paroxetine | 1 | CYP2D6*1/*2/*3/*4/*6/*7/*8/*9/*10/*17/*29/*35/*41/*5 (deletion) and duplication | Phenotyping of dextromethorphan and tramadol by LC–MS/MS followed by LLE | Duloxetine: | 17 | [29] |
| PM: UMRDEM/DOR > 0.3 | 71% NM→IM | (healthy volunteers) | |||||||||
| IM: 0.03 < UMR < 0.3 | 25% NM→PM | ||||||||||
| NM: 0.003 < UMR < 0.03 | Paroxetine: | ||||||||||
| UM: UMR < 0.003 | 94% heterozygous NM→PM | ||||||||||
| 56% homozygous NM→PM | |||||||||||
| Dextromethorphan | CYP2D6 | Comedication | Inhibition | Levomeprazine/biperiden | 1 | CYP2D6*1/*2/*3/*4/*5/*8/*10/*18/*21 | Dextromethorphan and dextrorphan via HPLC | 14 patients: | 104 | [6] | |
| Levomepromazine and biperiden via GCMS | 3 NM→PM | (90 healthy volunteers) | |||||||||
| 2 NM→IM | (14 schizophrenia patients) | ||||||||||
| 9 stayed IM | |||||||||||
| Cardiac drugs | Flecainide | CYP2D6 | Comedication | Inhibition | Bepridil | 1 | CYP2D6*1/*2/*4/*5/*10/*14/*21/*36 | Flecainide enantiomers via HPLC | All 17 concomitant treated NM patients showed a serum flecainide S/R ratio of PM subjects: NM→PM | 143 | [22] |
| NM: *1 and *2 | (supraventricular tachyarrhythmia patients) | ||||||||||
| IM: *10 | |||||||||||
| PM: *4/*5/*14/*21/*36 | |||||||||||
| NSAID | Flurbiprofen | CYP2C9 | Comedication | Inhibition | Fluconazole | 1 | CYP2C9*1/*2/*3 | Flurbiprofen, 4′-hydroxyflurbiprofen and fluconazole via HPLC | 200 mg fluconazole: | 189 | [30] |
| 11 NM→IM | (healthy subjects) | ||||||||||
| 8 IM→PM | |||||||||||
| 2 PM = PM | |||||||||||
| 400 mg fluconazole: | |||||||||||
| 11 NM→PM | |||||||||||
| 8 IM→PM | |||||||||||
| 2 PM = PM | |||||||||||
| Proton pump inhibitors | Lansoprazole | CYP2C19 | Comedication | Inhibition | Fluvoxamine | 2 | CYP2C19*1/*2/*3 | Lansoprazole enantiomers and lansoprazole sulphone via HPLC | NM: (R)-lansoprazole AUC ↑↑ (903%) | 18 | [31] |
| NM: *1/*1 | (S)-lansoprazole AUC ↑↑ (1664%) | (healthy volunteers) | |||||||||
| IM: *1/*2 or *1/*3 | IM: (R)-lansoprazole AUC ↑ (462%) | ||||||||||
| PM: *2/*2 or *2/*3 | (S)-lansoprazole AUC ↑ (781%) | ||||||||||
| Antiestrogens | Tamoxifen | CYP2D6 | Comedication | Inhibition | Potent inhibitors: paroxetine and fluoxetine | 1 | CYP2D6*1/*3/*4/*6/*7/*8/*10/*11/*14/*15/*17/*19/*20/*25/*26/*29/*31/*35/*36/*40/*41 and duplications | Tamoxifen and metabolites via HPLC | 42 CYP2D6 subjects with CYP2D6 inhibitor compared to 85 subjects without concomitant medication. | 158 | [32] |
| Weak inhibitors: sertraline, citalopram, celecoxib, diphenhydramine, and chlorpheniramine | Mean group endoxifen plasma concentrations by potent inhibitor usage: | (breast cancer patients) | |||||||||
| 1 UM→IM | |||||||||||
| 5 NM→PM | |||||||||||
| 11 IM→PM | |||||||||||
| Mean group endoxifen plasma concentrations by weak inhibitor usage: | |||||||||||
| 3 UM→NM or IM | |||||||||||
| 10 NM→IM | |||||||||||
| 12 IM→PM | |||||||||||
| Tamoxifen | CYP3A4 | Other | Induction | Vitamin D | 2 | CYP3A4*1/*22, POR*28, CYP2C9*1/*2/*3, CYP2B6*1/*4/*5/*6, MDR1 c.3435C > T, BCRP c.421C > A/c.34G > A, CYP3A5*1/*3, CYP2D6*1/*3/*4/*5/*9/*10/*41 | Tamoxifen, NDM-tamoxifen, 4-OH-tamoxifen, Z-endoxifen, Z-3-OH tamoxifen and Z-α-OH-tamoxifen via LC-MS/MS normalized to 4-β-OH-cholesterol via UHPLC-MS/MS and 25-OH-vitamin D via ELISA | During winter months endoxifen levels ↓ (20%) | 196 | [33] | |
| (breast cancer patients) |
| Cause of Phenoconversion | Drug | Enzyme | Result | Type of Study | Genotype Assessment | Phenotype Assessment | Effect of Phenoconversion | Subjects | Ref |
|---|---|---|---|---|---|---|---|---|---|
| Age | Omeprazole | CYP2C19 | Reduced activity | 1 | CYP2C19*1/*2/*3 | Omeprazole (10 mg for elderly and 20 mg for young), omeprazole, 5-hydroxyomeprazole and omeprazole sulfone via LC | 38% NM→PM | 51 | [34] |
| 42% IM→PM | (healthy volunteers: older age | ||||||||
| (elderly 66–85 years, younger 21–36)) | |||||||||
| Cancer | Omeprazole | CYP2C19 | Reduced activity | 1 | CYP2C19*1/*2 | Phenotyping via the prodrug omeprazole (20 mg) measured in plasma. | 25% NM→PM | 16 | [35] |
| NM: log10 [OM]/[OH-OM] <1 | (advanced cancer patients) | ||||||||
| PM: log10 [OM]/[OH-OM] ≥1 | |||||||||
| Omeprazole | CYP2C19 | Reduced activity | 1 | CYP2C19*1/*2/*3 | Phenotyping via the prodrug omeprazole (20 mg) measured in plasma. | 37% NM→PM | 33 | [36] | |
| NM: log10 [OM]/[OH-OM] <1 | (30 NM patients) | (advanced cancer patients) | |||||||
| PM: log10 [OM]/[OH-OM] ≥1 | |||||||||
| Proguanil | CYP2C19 | Reduced activity | 1 | CYP2C19*1/*2/*3/*17 | Proguanil (PG) (200 mg), PG and cycloguanil (CG) via HPLC | 27% NM→PM | 25 | [37] | |
| NM: PG/CG ≥ 1 | 53% IM→PM | (multiple myeloma patients) | |||||||
| PM: PG/CG < 1 | |||||||||
| Inflammation | Losartan | CYP2C9 | Reduced activity | 1 | CYP2C9*1/*2/*3 | Losartan and metabolite (E-3174) via HPLC | Mean group metabolic ratio in patients: | 51 | [38] |
| 31 NM→IM | (patients with Behçet’s disease) | ||||||||
| 20 IM→PM | 96 | ||||||||
| (healthy volunteers) | |||||||||
| Omeprazole | CYP2C19 | Reduced activity | 1 | CYP2C19*1/*2/*3 | Omeprazole and 5--hydroxy omeprazole via LC | Mean group metabolic ratio of omeprazole/5-hydroxy omeprazole in patients: | 31 | [39] | |
| NM: *1/*1 | NM→PM | (patients with chronic hepatitis or cirrhosis that tested positive for hepatitis C virus (HCV)) | |||||||
| IM: *1/*2 or *1/*3 | IM→PM | 30 | |||||||
| PM: *2/*2, *2/*3 or *3/*3 | PM = PM | (healthy volunteers) | |||||||
| Voriconazole | CYP2C19 | Reduced activity | 2 | CYP2C19*1/*2/*3/*17 | Voriconazole and voriconazole-N-oxide concentrations were measured via LC-MS/MS | 20 patients applicable for genotyping: | 36 | [40] | |
| UM: *1/*17 | Metabolic ratio: | (patients treated with intravenous or oral voriconazole) | |||||||
| NM: *1/*1 | UM = −0.994147N | ||||||||
| IM: *1/*2 or *1/*3 | NM = −0.991972N | ||||||||
| PM: *2/*2, *2/*3 or *3/*3 | IM = −0.986512N | ||||||||
| Voriconazole trough concentration: | |||||||||
| UM = +1.003685N | |||||||||
| NM = +1.004965N | |||||||||
| IM = +1.009365N | |||||||||
| N = CRP concentration | |||||||||
| Mephenytoin | CYP2C19 | Reduced activity | 2 | CYP2C19*1/*2/*3/*4/*17 | Mephenytoin 4′-hydroxylation and mephenytoin via HPLC | The genotype-predicted phenotype overestimated (47%) the measured phenotype. For the overestimated group, 9 had inflammatory diseases (rheumatoid arthritis and gastrointestinal perforation), resulting in lower CYP2C19 activity. | 114 | [8] | |
| PM: two loss of function alleles (*2, *3 or *4) | CYP2C19 activity: | (liver microsomes) | |||||||
| IM: one functional and one loss of function allele | PM: <8 pmol/(mg protein*min) | ||||||||
| NM: *1/*1 | UM: >75 pmol/(mg protein*min) | ||||||||
| UM: one or two *17 alleles | IM: between 8 and 23 pmol/(mg protein*min) | ||||||||
| (except *2/*17: IM or NM is accepted) | NM: between 23 and 75 pmol/(mg protein*min) | ||||||||
| Dextromethorphan | CYP2D6 | Reduced activity- | 1 | CYP2D6 | Dextromethorphan (DEM) (2.5 mg), DEM and dextrorphan (DOR) via HPLC | 10 LKM-1 positive subjects: | 1723 | [41] | |
| 33 allelic variants (AmpliChip®) | UM: DEM/DOR < 0.003 | 6 NM→IM | (chronic hepatitis C patients) | ||||||
| NM: 0.003 < DEM/DOR < 0.03 | 1 NM→PM | ||||||||
| IM: 0.03 < DEM/DOR < 0.3 | CYP2D6 metabolic function is reduced by LK-1 antibody presence | ||||||||
| PM: DEM/DOR > 0.3 | |||||||||
| Immunosuppressants (tacrolimus or cyclosporine A) | CYP3A5 | Reduced activity | 1 | CYP3A5*1/*3 | 4β-hydroxycholesterol via GCMS | 10 CYP3A5*1 recipients had low CYP3A5 activity (phenoconversion), might be due to increased indoxyl sulfate concentrations | 63 | [42] | |
| (kidney transplant recipients) | |||||||||
| Pregnancy | Dextromethorphan | CYP2D6 | Reduced activity | 2 | CYP2D6*1/*3/*4 | Dextromethorphan (30 mg) | NM: metabolic ratio dextromethorphan/dextrorphan ↓ (29%) | 140 | [43] |
| CYP2D6: O-demethylated → dextrorphan (dextromethorphan/dextrorphan) | IM: metabolic ratio dextromethorphan/dextrorphan ↓ (63%) | (pregnant women) | |||||||
| CYP3A: N-demethylated to 3-methoxymorphinan (dextromethorphan/3-demethoxymorphinon) | |||||||||
| Paroxetine | CYP2D6 | Increased activity | 2 | CYP2D6*1/*3/*4/*5/*6/*9/*10/*41 and duplication | Paroxetine via HPLC-UV or LC-MS/MS | NM/UM: plasma paroxetine concentration ↓ | 74 | [44] | |
| NM: *1/*1 | PM plasma paroxetine concentration ↑ | (pregnant women) | |||||||
| UM: duplication | |||||||||
| IM: heterozygous for nonfunctional allele | |||||||||
| PM: homozygous for nonfunctional allele |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klomp, S.D.; Manson, M.L.; Guchelaar, H.-J.; Swen, J.J. Phenoconversion of Cytochrome P450 Metabolism: A Systematic Review. J. Clin. Med. 2020, 9, 2890. https://doi.org/10.3390/jcm9092890
Klomp SD, Manson ML, Guchelaar H-J, Swen JJ. Phenoconversion of Cytochrome P450 Metabolism: A Systematic Review. Journal of Clinical Medicine. 2020; 9(9):2890. https://doi.org/10.3390/jcm9092890
Chicago/Turabian StyleKlomp, Sylvia D., Martijn L. Manson, Henk-Jan Guchelaar, and Jesse J. Swen. 2020. "Phenoconversion of Cytochrome P450 Metabolism: A Systematic Review" Journal of Clinical Medicine 9, no. 9: 2890. https://doi.org/10.3390/jcm9092890
APA StyleKlomp, S. D., Manson, M. L., Guchelaar, H.-J., & Swen, J. J. (2020). Phenoconversion of Cytochrome P450 Metabolism: A Systematic Review. Journal of Clinical Medicine, 9(9), 2890. https://doi.org/10.3390/jcm9092890






