Microbiome Patterns in Matched Bile, Duodenal, Pancreatic Tumor Tissue, Drainage, and Stool Samples: Association with Preoperative Stenting and Postoperative Pancreatic Fistula Development
Abstract
:1. Introduction
2. Experiment
2.1. Study Design
2.2. Sample Processing and DNA Purification
2.3. Statistical Analyses
3. Results
3.1. Patient Characteristics
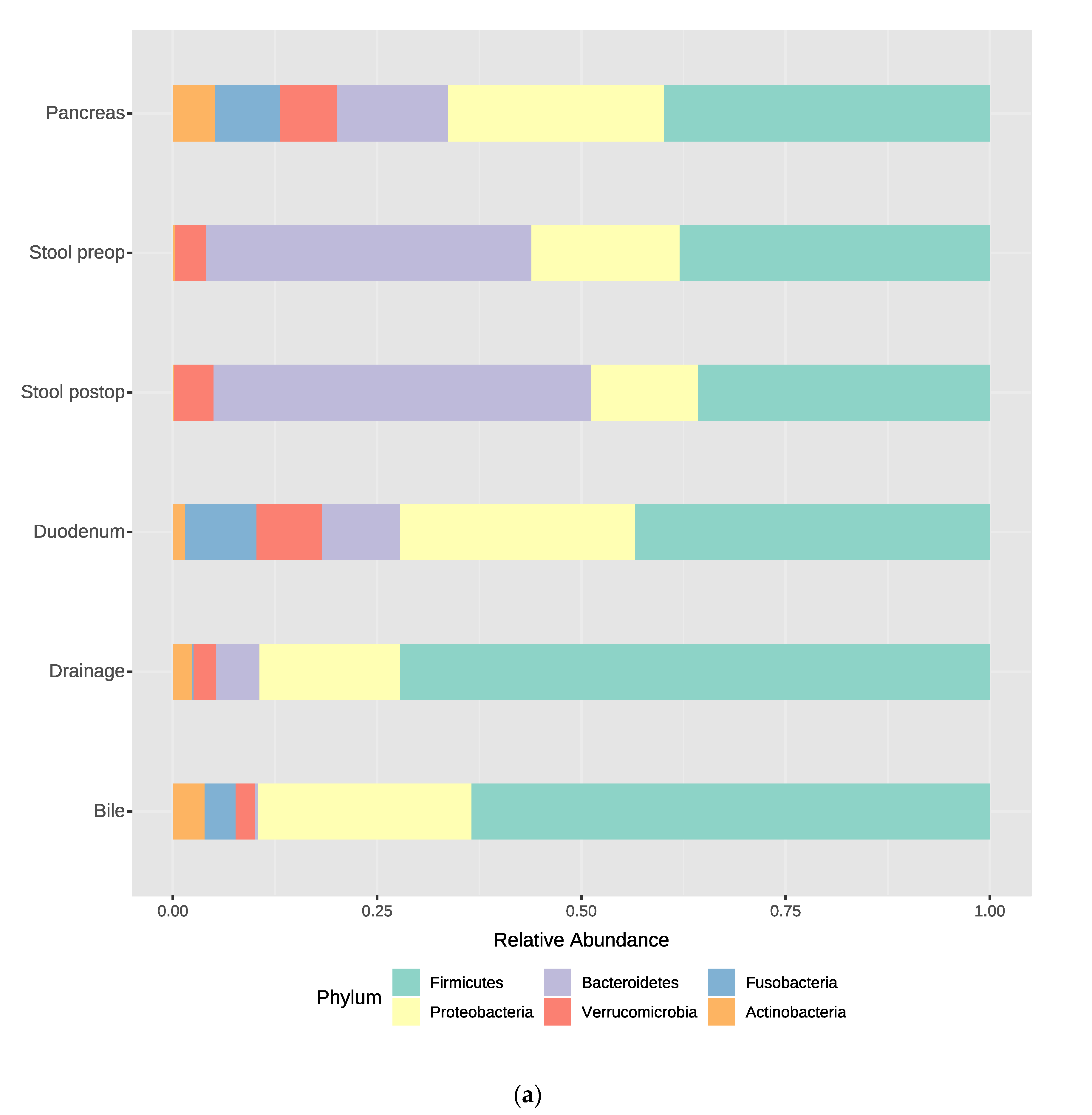
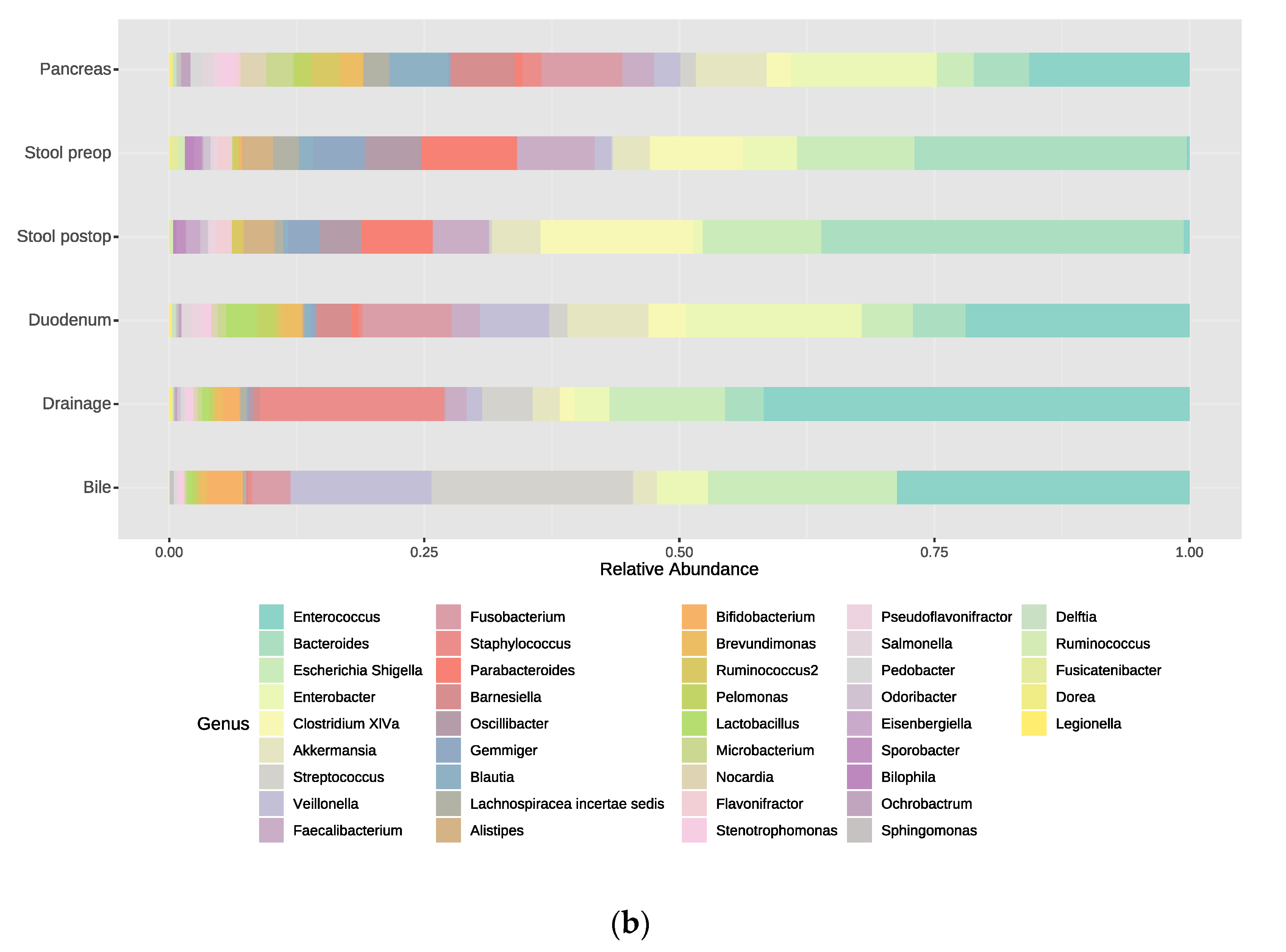
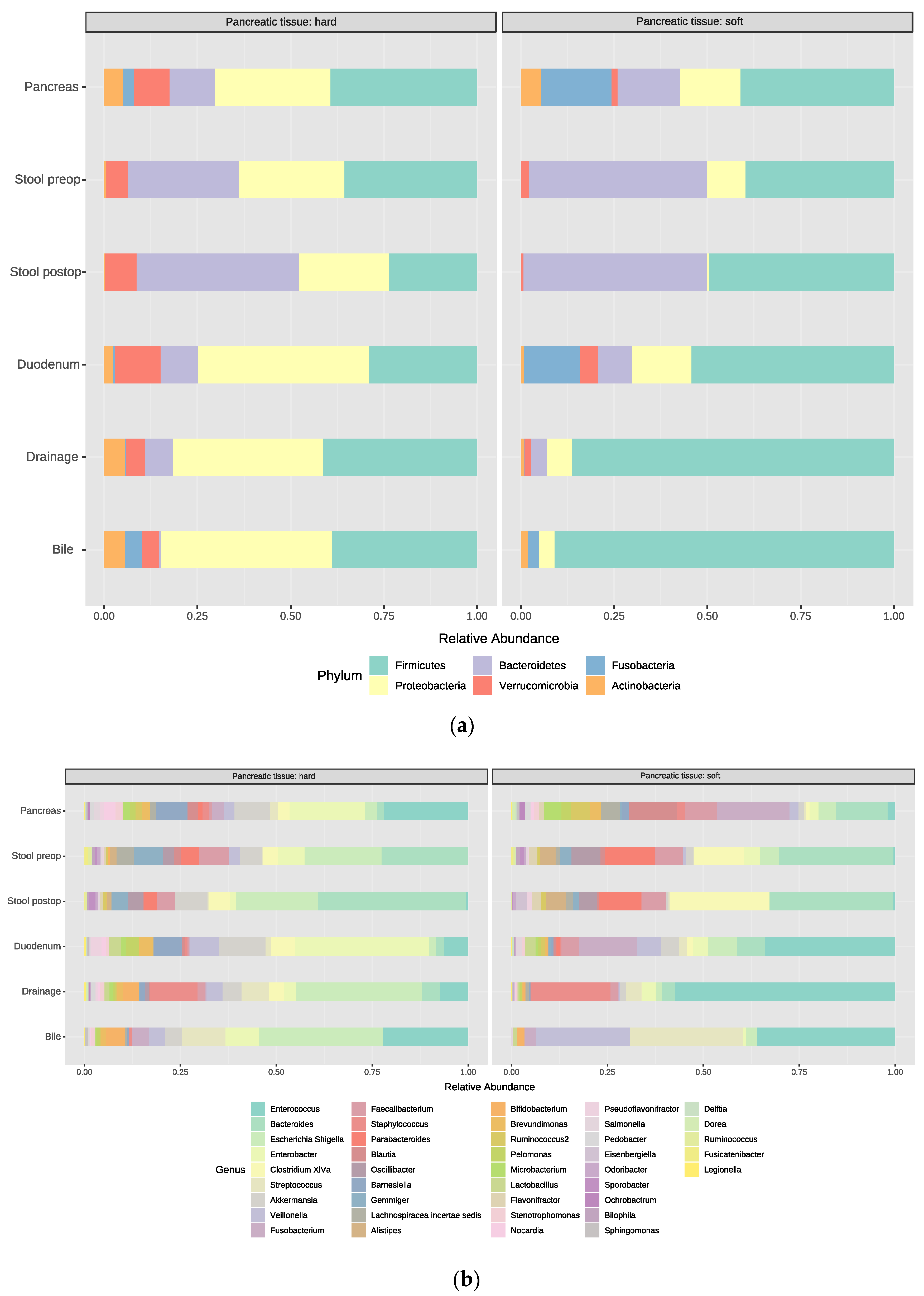
3.2. Bacterial Composition at the Phylum and Genus Levels
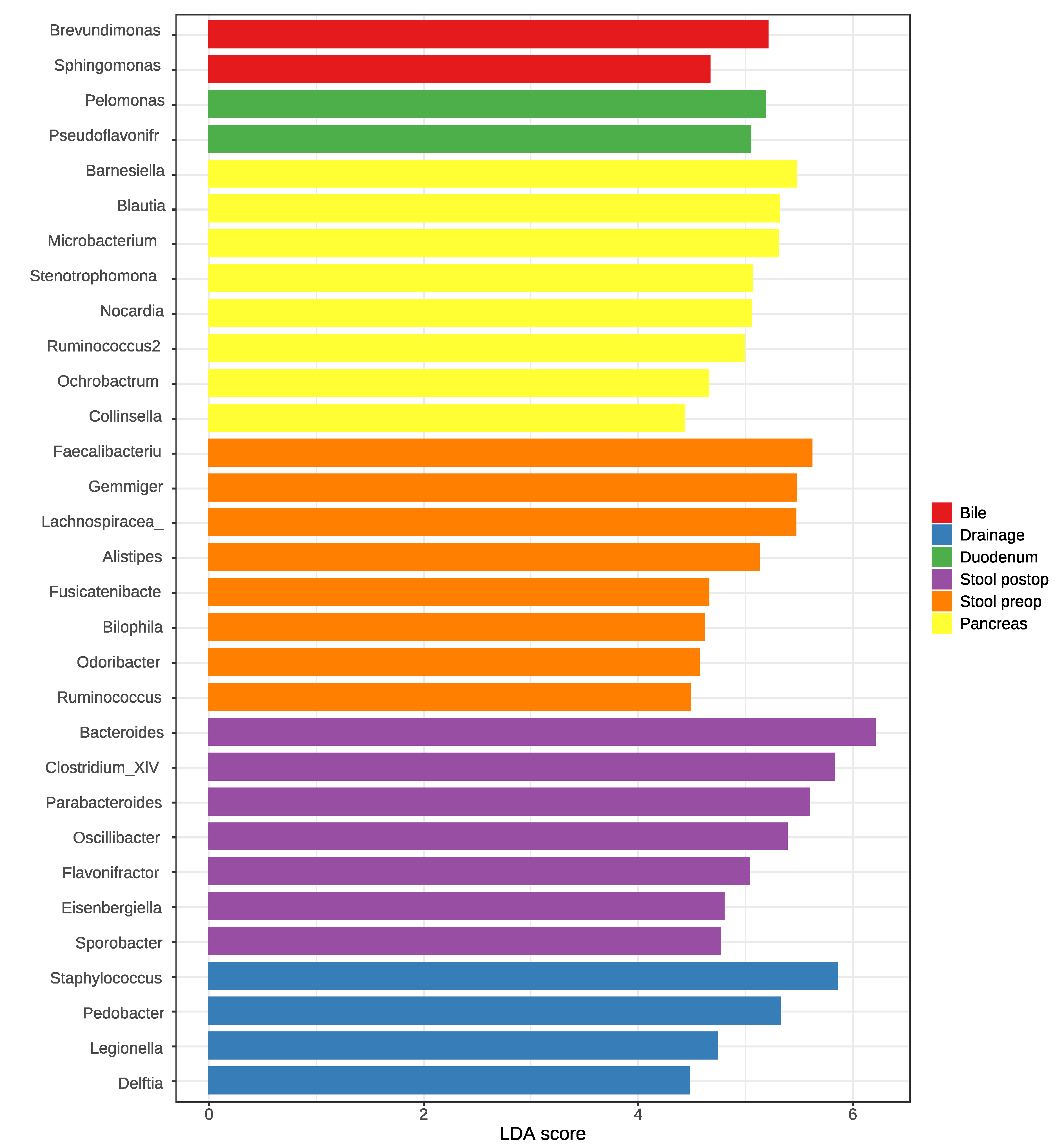
3.3. Identification of Key Taxa Associated with Different Samples
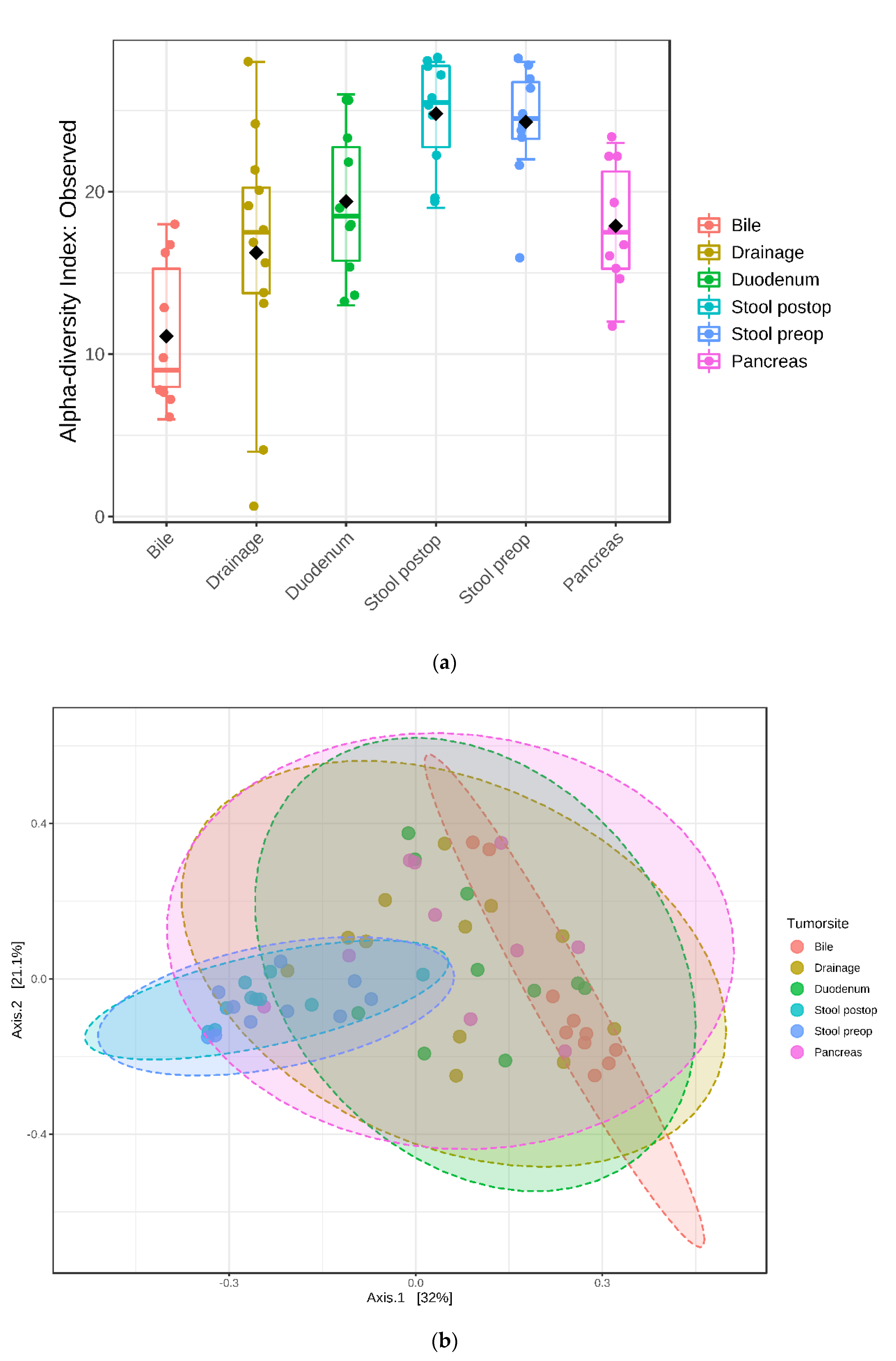
3.4. Bacterial Richness and Diversity in the Samples
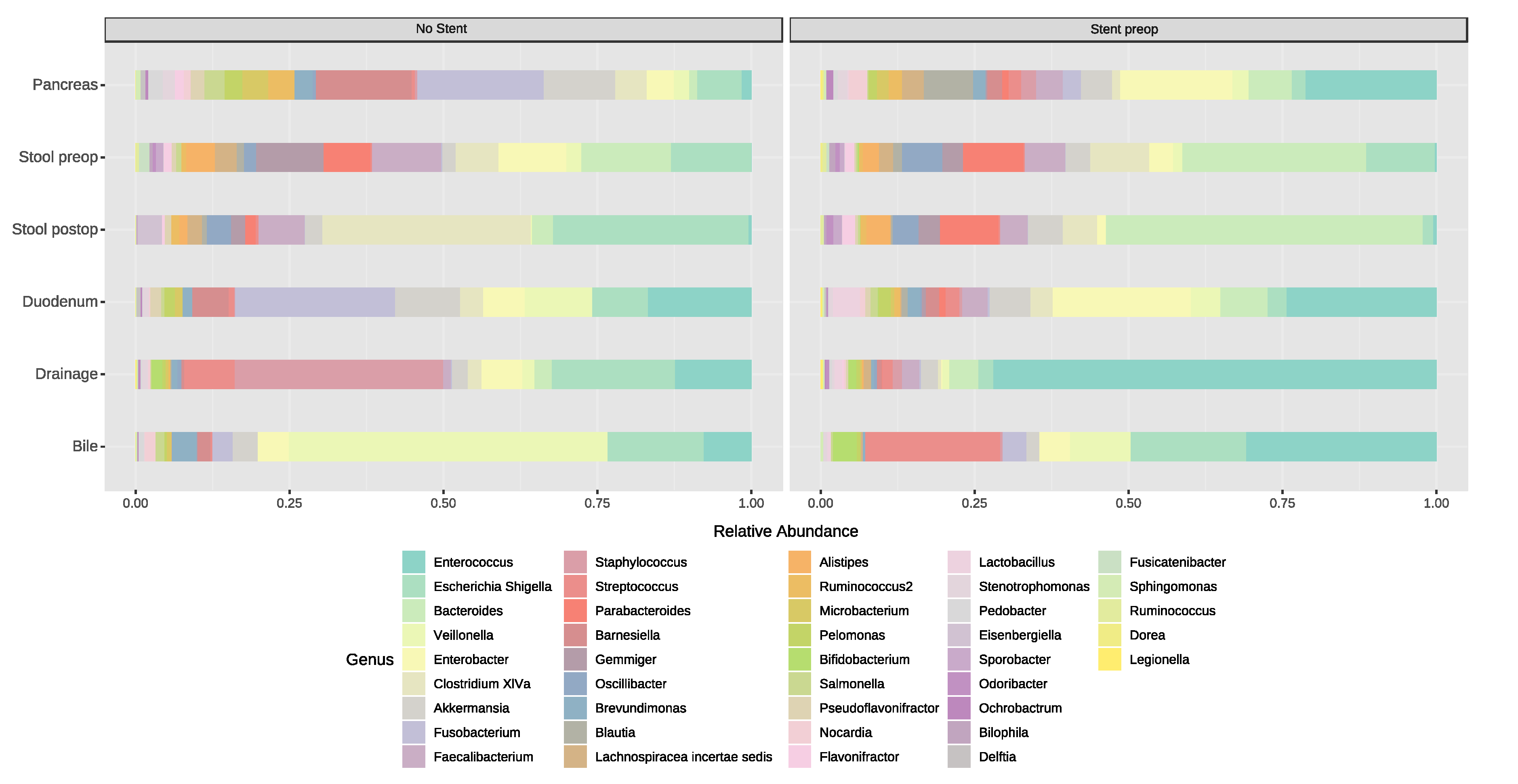
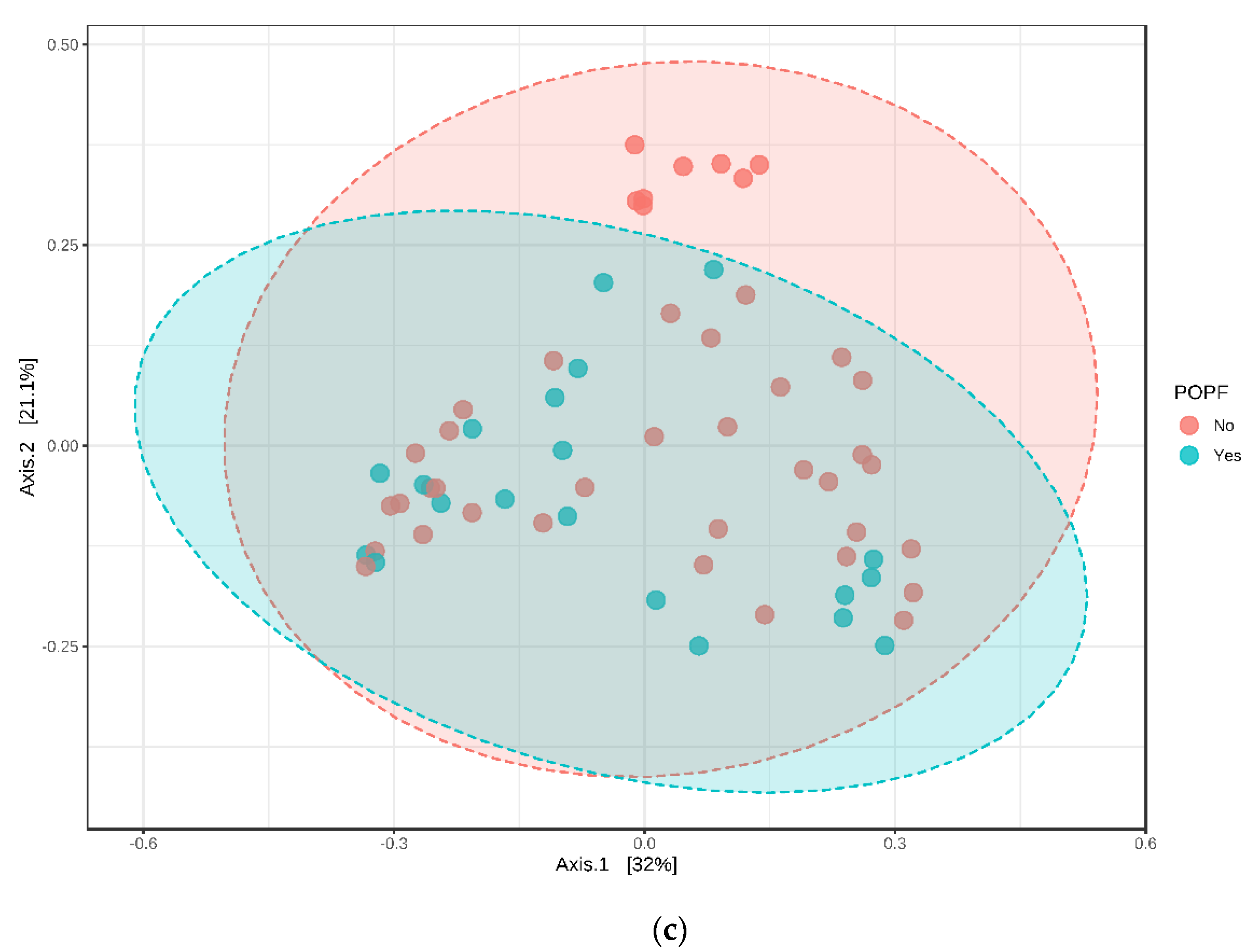
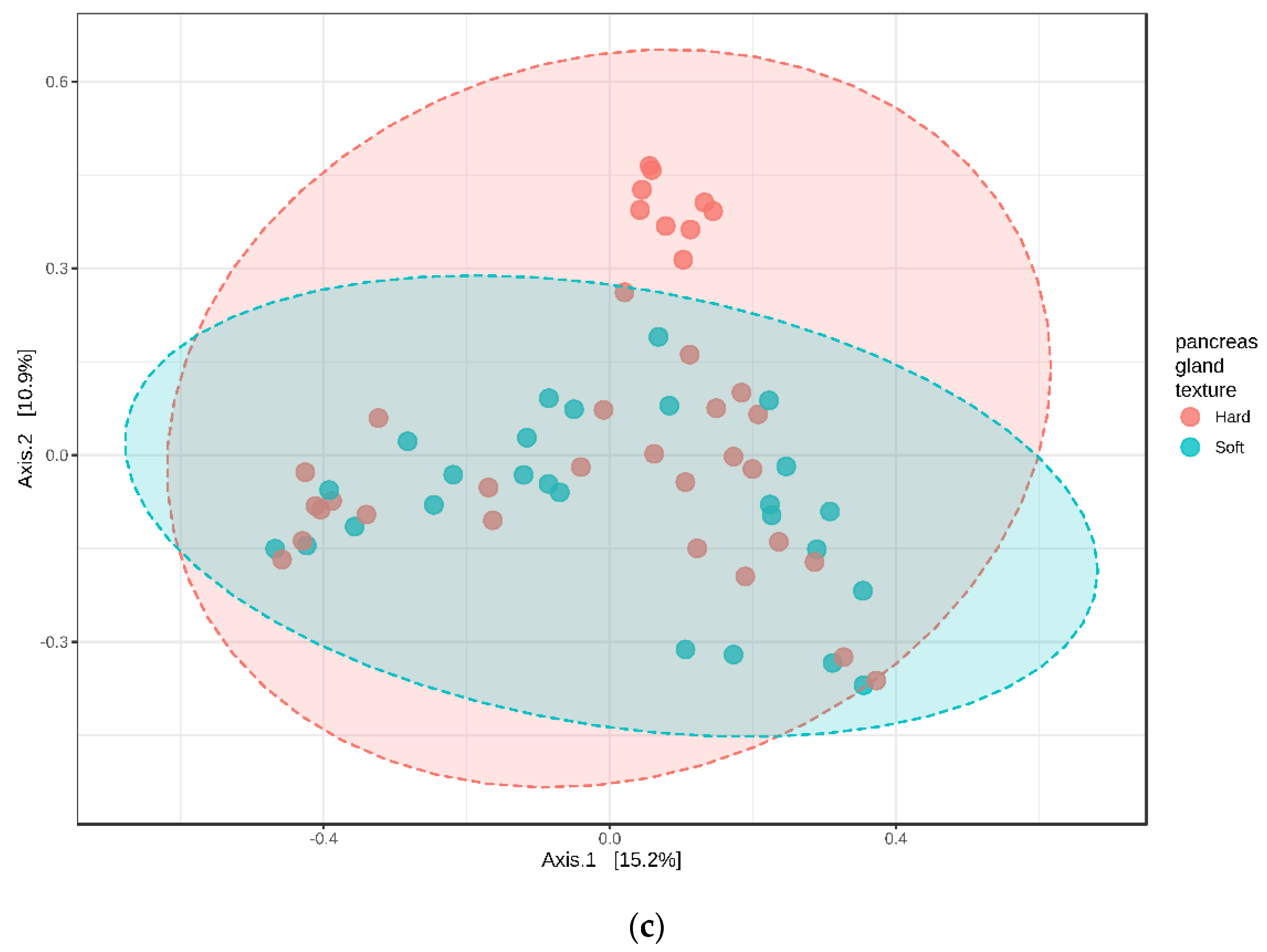
3.5. Relative Abundance and Associated Microbiome Profiles with Clinical Conditions
3.5.1. Stent Placement in the Bile Duct
3.5.2. Postoperative Complications (POPF B/C)
3.5.3. Pancreatic Tumor Tissue, Soft vs. Hard
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fitzmaurice, C. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 2006 to 2016: A systematic analysis for the Global Burden of Disease study. J. Clin. Oncol. 2018, 36, 1568. [Google Scholar] [CrossRef] [Green Version]
- Wellner, U.F.; Keck, T. Quality Indicators in Pancreatic Surgery: Lessons Learned from the German DGAV StuDoQ|Pancreas Registry. Visc. Med. 2017, 33, 126–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krautz, C.; Haase, E.; Elshafei, M.; Saeger, H.-D.; Distler, M.; Grützmann, R.; Weber, G.F. The impact of surgical experience and frequency of practice on perioperative outcomes in pancreatic surgery. BMC Surg. 2019, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xavier, J.B.; Young, V.B.; Skufca, J.; Ginty, F.; Testerman, T.; Pearson, A.T.; Macklin, P.; Mitchell, A.; Shmulevich, I.; Xie, L.; et al. The Cancer Microbiome: Distinguishing Direct and Indirect Effects Requires a Systemic View. Trends Cancer 2020, 6, 192–204. [Google Scholar] [CrossRef] [Green Version]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Shogan, B.D.; Belogortseva, N.; Luong, P.M.; Zaborin, A.; Lax, S.; Bethel, C.; Ward, M.; Muldoon, J.P.; Singer, M.; An, G.; et al. Collagen degradation and MMP9 activation by Enterococcus faecalis contribute to intestinal anastomotic leak. Sci. Transl. Med. 2015, 7, 286ra268. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, F.C.F.; Brenner, T.; Uhle, F.; Loesch, S.; Hackert, T.; Ulrich, A.; Hofer, S.; Dalpke, A.H.; Weigand, M.A.; Boutin, S. Gut microbiome patterns correlate with higher postoperative complication rates after pancreatic surgery. BMC Microbiol. 2019, 19. [Google Scholar] [CrossRef] [Green Version]
- Half, E.; Keren, N.; Reshef, L.; Dorfman, T.; Lachter, I.; Kluger, Y.; Reshef, N.; Knobler, H.; Maor, Y.; Stein, A.; et al. Fecal microbiome signatures of pancreatic cancer patients. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Li, Y.-D.; He, K.-X.; Zhu, W.-F. Correlation between invasive microbiota in margin-surrounding mucosa and anastomotic healing in patients with colorectal cancer. World J. Gastrointest. Oncol. 2019, 11, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Wiegerinck, M.; Hyoju, S.K.; Mao, J.; Zaborin, A.; Adriaansens, C.; Salzman, E.; Hyman, N.H.; Zaborina, O.; van Goor, H.; Alverdy, J.C. Novel de novo synthesized phosphate carrier compound ABA-PEG20k-Pi20 suppresses collagenase production in Enterococcus faecalis and prevents colonic anastomotic leak in an experimental model. Br. J. Surg. 2018, 105, 1368–1376. [Google Scholar] [CrossRef] [PubMed]
- Warburton, R.C.; Gelson, W.; Harper, S.; Rushbrook, S.; Narbad, A. Human bile retrieved from the normal biliary system is not sterile. J. Hepatol. 2017, 66, S65. [Google Scholar] [CrossRef]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef]
- McAllister, F.; Khan, M.A.W.; Helmink, B.; Wargo, J.A. The Tumor Microbiome in Pancreatic Cancer: Bacteria and Beyond. Cancer Cell 2019, 36, 577–579. [Google Scholar] [CrossRef]
- Wei, M.-Y.; Shi, S.; Liang, C.; Meng, Q.-C.; Hua, J.; Zhang, Y.-Y.; Liu, J.; Zhang, B.; Xu, J.; Yu, X.-J. The microbiota and microbiome in pancreatic cancer: More influential than expected. Mol. Cancer 2019, 18. [Google Scholar] [CrossRef]
- Schierwagen, R.; Alvarez-Silva, C.; Madsen, M.S.A.; Kolbe, C.C.; Meyer, C.; Thomas, D.; Uschner, F.E.; Magdaleno, F.; Jansen, C.; Pohlmann, A.; et al. Circulating microbiome in blood of different circulatory compartments. Gut 2019, 68, 578–580. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R.M.; Jobin, C. Microbiota in pancreatic health and disease: The next frontier in microbiome research. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 53–64. [Google Scholar] [CrossRef]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. SINTAX: A Simple non-Bayesian Taxonomy Classifier for 16S and ITS Sequences; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2016. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Shu, Y.; Gu, Y. The potential role of bacteria in pancreatic cancer: A systematic review. Carcinogenesis 2020, 41, 397–404. [Google Scholar] [CrossRef]
- Elinav, E.; Garrett, W.S.; Trinchieri, G.; Wargo, J. The cancer microbiome. Nat. Rev. Cancer 2019, 19, 371–376. [Google Scholar] [CrossRef]
- Kiss, B.; Mikó, E.; Sebő, É.; Toth, J.; Ujlaki, G.; Szabó, J.; Uray, K.; Bai, P.; Árkosy, P. Oncobiosis and Microbial Metabolite Signaling in Pancreatic Adenocarcinoma. Cancers 2020, 12, 1068. [Google Scholar] [CrossRef]
- Liwinski, T.; Zenouzi, R.; John, C.; Ehlken, H.; Rühlemann, M.C.; Bang, C.; Groth, S.; Lieb, W.; Kantowski, M.; Andersen, N.; et al. Alterations of the bile microbiome in primary sclerosing cholangitis. Gut 2019, 69, 665–672. [Google Scholar] [CrossRef] [Green Version]
- Pereira, P.; Aho, V.; Arola, J.; Boyd, S.; Jokelainen, K.; Paulin, L.; Auvinen, P.; Färkkilä, M. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS ONE 2017, 12, e0182924. [Google Scholar] [CrossRef]
- Ducreux, M.; Cuhna, A.S.; Caramella, C.; Hollebecque, A.; Burtin, P.; Goéré, D.; Seufferlein, T.; Haustermans, K.; Van Laethem, J.L.; Conroy, T.; et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, v56–v68. [Google Scholar] [CrossRef] [PubMed]
- Arkadopoulos, N.; Kyriazi, M.A.; Papanikolaou, I.S.; Vasiliou, P.; Theodoraki, K.; Lappas, C.; Oikonomopoulos, N.; Smyrniotis, V. Preoperative biliary drainage of severely jaundiced patients increases morbidity of pancreaticoduodenectomy: Results of a case-control study. World J. Surg. 2014, 38, 2967–2972. [Google Scholar] [CrossRef] [PubMed]
- Krüger, C.M.; Adam, U.; Adam, T.; Kramer, A.; Heidecke, C.D.; Makowiec, F.; Riediger, H. Bacterobilia in pancreatic surgery-conclusions for perioperative antibiotic prophylaxis. World J. Gastroenterol. 2019, 25, 6238–6247. [Google Scholar] [CrossRef] [PubMed]
- Matthews, J.B. Infection After Pancreatic Surgery. JAMA Surg. 2017, 152, 1030. [Google Scholar] [CrossRef]
- Miller, B.C.; Christein, J.D.; Behrman, S.W.; Drebin, J.A.; Pratt, W.B.; Callery, M.P.; Vollmer, C.M. A Multi-Institutional External Validation of the Fistula Risk Score for Pancreatoduodenectomy. J. Gastrointest. Surg. 2013, 18, 172–180. [Google Scholar] [CrossRef]
- Callery, M.P.; Pratt, W.B.; Kent, T.S.; Chaikof, E.L.; Vollmer, C.M. A Prospectively Validated Clinical Risk Score Accurately Predicts Pancreatic Fistula after Pancreatoduodenectomy. J. Am. Coll. Surg. 2013, 216, 1–14. [Google Scholar] [CrossRef]
- Pratt, W.B.; Callery, M.P.; Vollmer, C.M. Risk Prediction for Development of Pancreatic Fistula Using the ISGPF Classification Scheme. World J. Surg. 2008, 32, 419–428. [Google Scholar] [CrossRef]


| Demographics | Overall (n = 10) | POPF | |
|---|---|---|---|
| Yes (n = 3) | No (n = 7) | ||
| Age | 44–89 | 66–79 | 44–89 |
| Sex | |||
| Male | 8 | 3 | 5 |
| Female | 2 | 0 | 2 |
| Body mass index (BMI) | |||
| 18.8–24.9 normal | 1 | 0 | 1 |
| 25.0–29.9 pre obesity | 6 | 3 | 3 |
| 30.0–34.9 obesity class 1 | 2 | 0 | 2 |
| 35.0–39.9 obesity class II | 1 | 0 | 1 |
| Nicotine | |||
| Yes | 1 | 0 | 1 |
| No | 9 | 3 | 6 |
| Proton pump inhibitors | |||
| Yes | 4 | 3 | 1 |
| No | 6 | 0 | 6 |
| Histology | |||
| Benign | 3 | 1 | 2 |
| Malignant | 7 | 2 | 5 |
| Physical ASA status (American Society of Anästhesiologists) | |||
| 1 | 0 | 0 | 0 |
| 2 | 4 | 0 | 4 |
| 3 | 6 | 3 | 3 |
| Stent | |||
| Yes | 7 | 3 | 4 |
| No | 3 | 0 | 3 |
| Gland texture | |||
| Soft | 4 | 2 | 2 |
| Hard | 7 | 1 | 6 |
| Diameter of the pancreatic duct | |||
| 1 mm | 1 (soft) | 1 (hard) | |
| 2 mm | 0 | 2 | |
| 3 mm | 0 | 1 | |
| 4 mm | 1 (hard) | 1 (hard) | |
| 5 mm | 1 (soft) | 1 (hard) | |
| 6 mm | 0 | 0 | |
| 7 mm | 0 | 1 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Langheinrich, M.; Wirtz, S.; Kneis, B.; Gittler, M.M.; Tyc, O.; Schierwagen, R.; Brunner, M.; Krautz, C.; Weber, G.F.; Pilarsky, C.; et al. Microbiome Patterns in Matched Bile, Duodenal, Pancreatic Tumor Tissue, Drainage, and Stool Samples: Association with Preoperative Stenting and Postoperative Pancreatic Fistula Development. J. Clin. Med. 2020, 9, 2785. https://doi.org/10.3390/jcm9092785
Langheinrich M, Wirtz S, Kneis B, Gittler MM, Tyc O, Schierwagen R, Brunner M, Krautz C, Weber GF, Pilarsky C, et al. Microbiome Patterns in Matched Bile, Duodenal, Pancreatic Tumor Tissue, Drainage, and Stool Samples: Association with Preoperative Stenting and Postoperative Pancreatic Fistula Development. Journal of Clinical Medicine. 2020; 9(9):2785. https://doi.org/10.3390/jcm9092785
Chicago/Turabian StyleLangheinrich, Melanie, Stefan Wirtz, Barbara Kneis, Matthias M. Gittler, Olaf Tyc, Robert Schierwagen, Maximilian Brunner, Christian Krautz, Georg F. Weber, Christian Pilarsky, and et al. 2020. "Microbiome Patterns in Matched Bile, Duodenal, Pancreatic Tumor Tissue, Drainage, and Stool Samples: Association with Preoperative Stenting and Postoperative Pancreatic Fistula Development" Journal of Clinical Medicine 9, no. 9: 2785. https://doi.org/10.3390/jcm9092785
APA StyleLangheinrich, M., Wirtz, S., Kneis, B., Gittler, M. M., Tyc, O., Schierwagen, R., Brunner, M., Krautz, C., Weber, G. F., Pilarsky, C., Trebicka, J., Agaimy, A., Grützmann, R., & Kersting, S. (2020). Microbiome Patterns in Matched Bile, Duodenal, Pancreatic Tumor Tissue, Drainage, and Stool Samples: Association with Preoperative Stenting and Postoperative Pancreatic Fistula Development. Journal of Clinical Medicine, 9(9), 2785. https://doi.org/10.3390/jcm9092785







