Eye-Catching Microbes—Polyphasic Analysis of the Microbiota on Microscope Oculars Verifies Their Role as Fomites
Abstract
1. Introduction
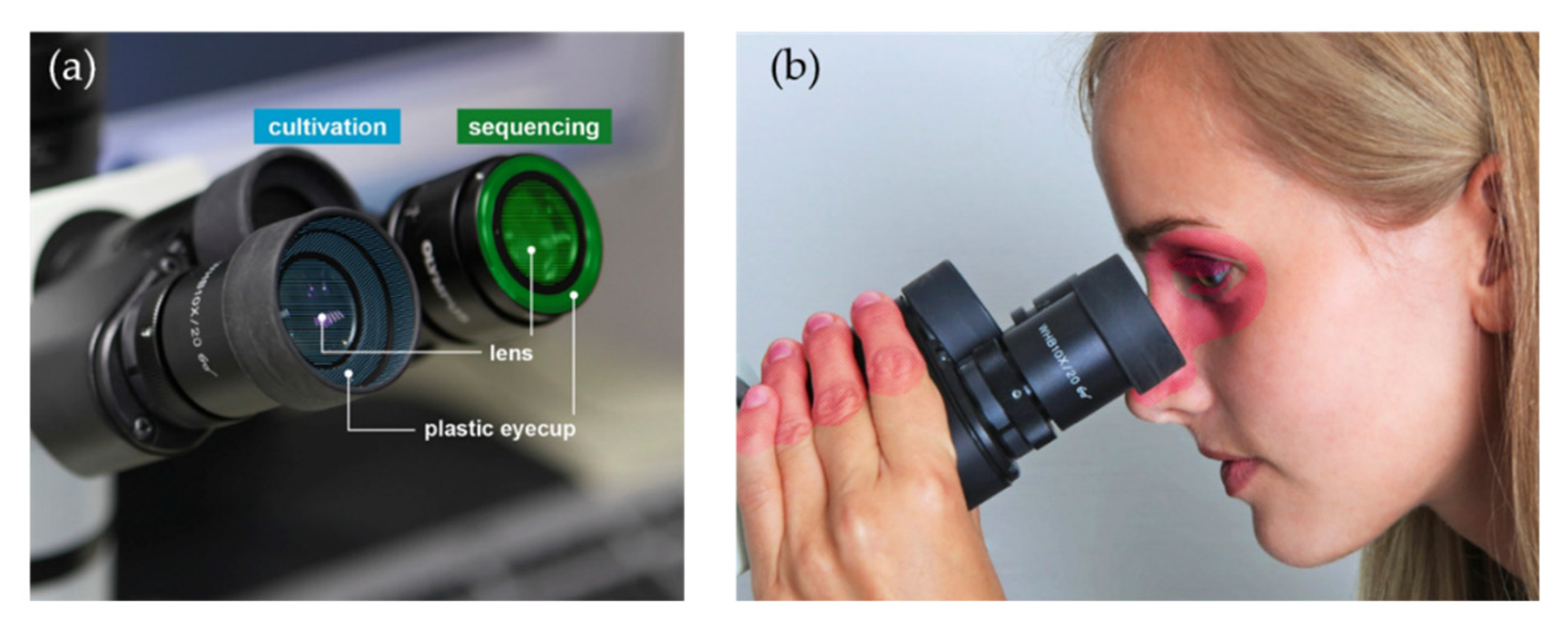
2. Materials and Methods
2.1. Cultivation-Based Analyses
2.2. Identification of Microbial Isolates by MALDI Biotyping
2.3. Sequencing-Based Analyses
2.4. DNA Extraction
2.5. Library Preparation
2.6. Sequencing
2.7. Cleaning Tests
2.8. Bioinformatics
2.9. Statistical Analyses
3. Results
3.1. Cultivation-Based Results
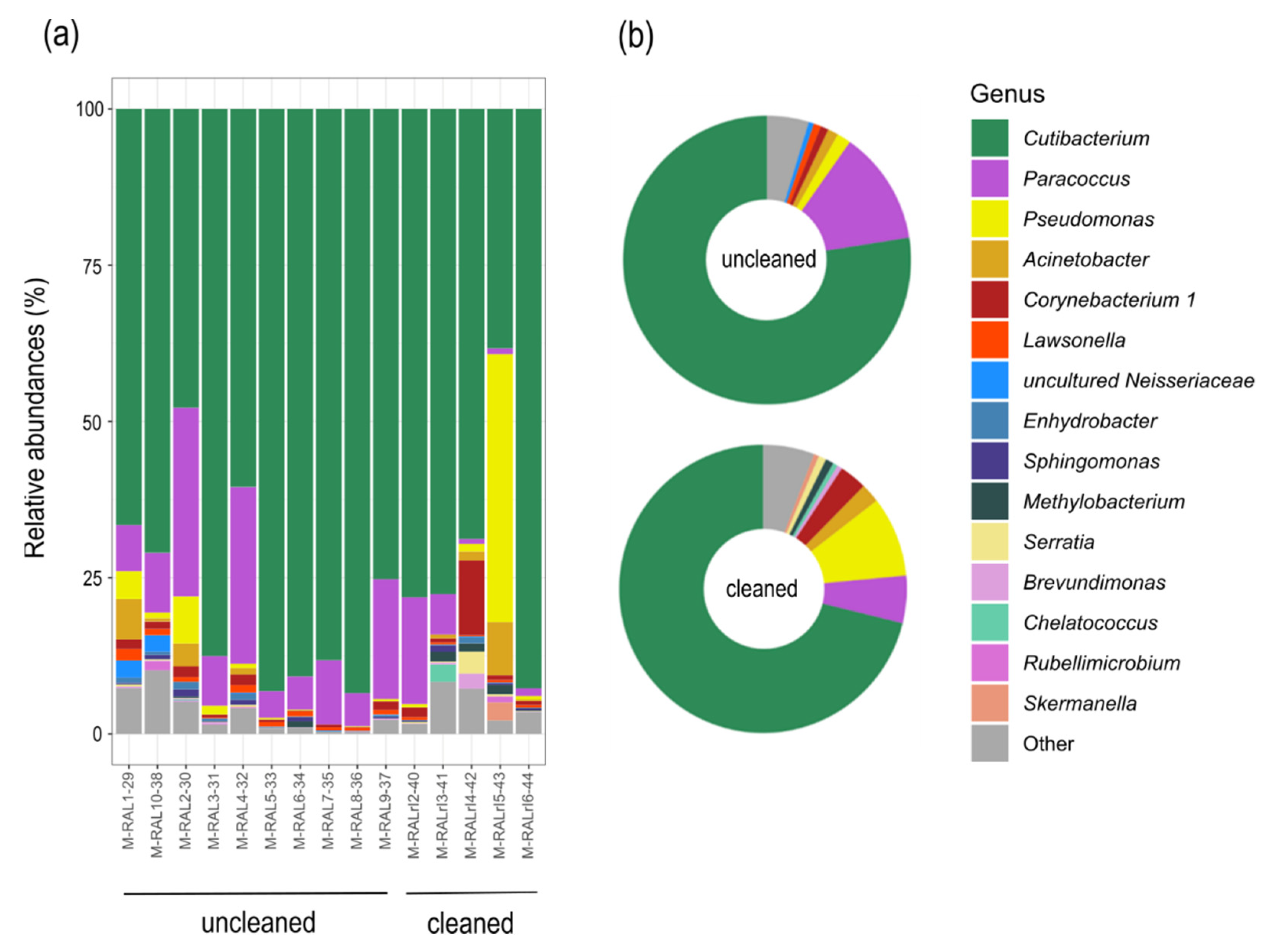
3.2. Sequencing Results
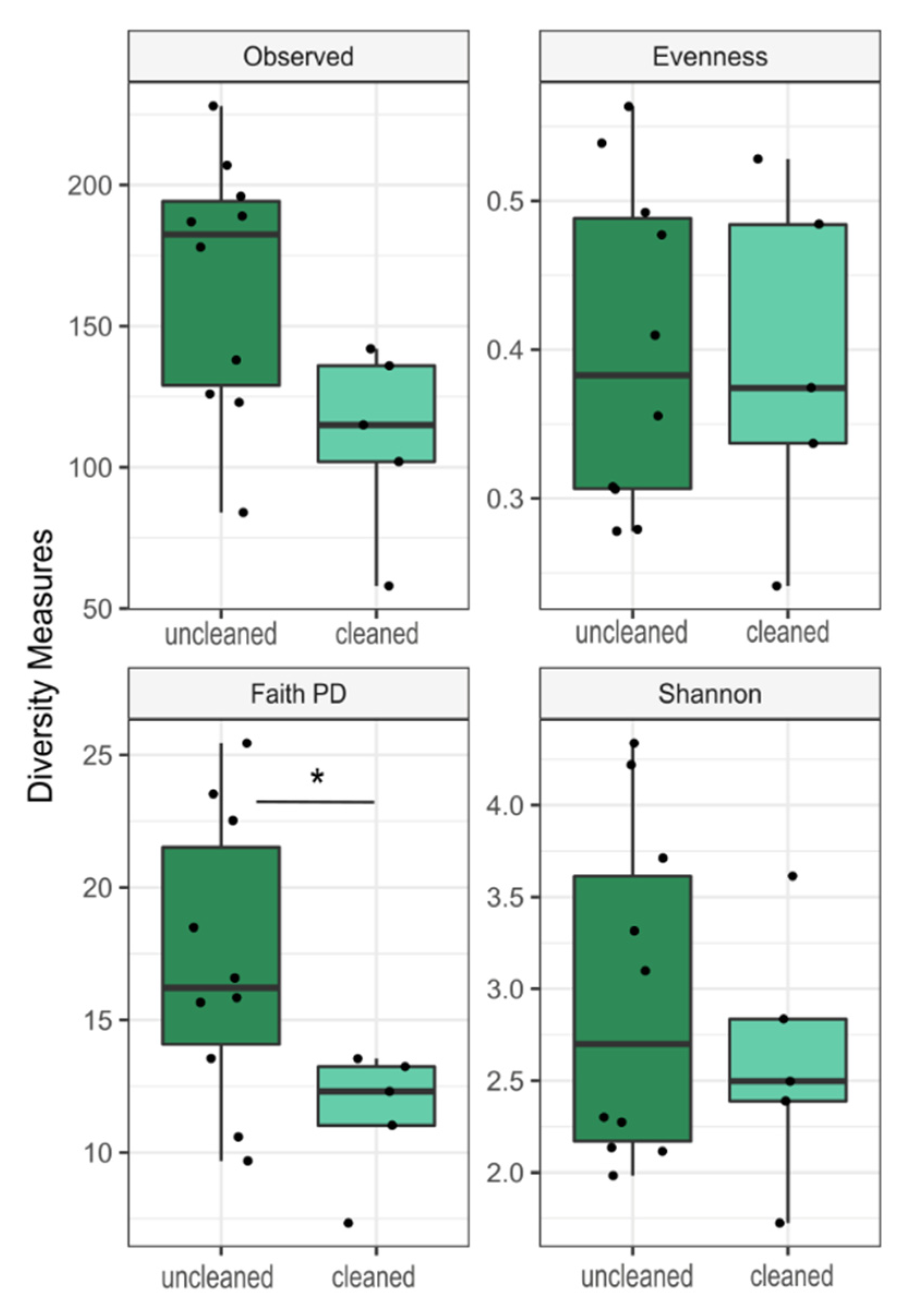
3.3. Community Composition and Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cardinale, M.; Kaiser, D.; Lueders, T.; Schnell, S.; Egert, M. Microbiome analysis and confocal microscopy of used kitchen sponges reveal massive colonization by Acinetobacter, Moraxella and Chryseobacterium species. Sci. Rep. 2017, 7, 5791. [Google Scholar] [CrossRef] [PubMed]
- Meadow, J.F.; Altrichter, A.E.; Green, J.L.; Souza, V. Mobile phones carry the personal microbiome of their owners. PeerJ 2014, 2, e447. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, E.; Azhar, E.I.; Bibi, F.; Yasir, M.; Al-Ghamdi, A.K.; Ashshi, A.M.; Elshemi, A.G.; Raoult, D. Paper money and coins as potential vectors of transmissible disease. Future Microbiol. 2014, 9, 249–261. [Google Scholar] [CrossRef]
- Kang, K.; Ni, Y.; Li, J.; Imamovic, L.; Sarkar, C.; Kobler, M.D.; Heshiki, Y.; Zheng, T.; Kumari, S.; Wong, J.C.Y.; et al. The environmental exposures and inner- and intercity traffic flows of the metro system may contribute to the skin microbiome and resistome. Cell Rep. 2018, 24, 1190–1202.e5. [Google Scholar] [CrossRef] [PubMed]
- Flores, G.E.; Bates, S.T.; Knights, D.; Lauber, C.L.; Stombaugh, J.; Knight, R.; Fierer, N.; Liles, M.R. Microbial biogeography of public restroom surfaces. PLoS ONE 2011, 6, e28132. [Google Scholar] [CrossRef]
- Christoff, A.P.; Sereia, A.F.; Hernandes, C.; de Oliveira, L.F. Uncovering the hidden microbiota in hospital and built environments: New approaches and solutions. Exp. Biol. Med. 2019, 244, 534–542. [Google Scholar] [CrossRef]
- Del Campo, R.; Martínez-García, L.; Sánchez-Díaz, A.M.; Baquero, F. Biology of hand-to-hand bacterial transmission. Microbiol. Spectr. 2019, 7, 205–213. [Google Scholar] [CrossRef]
- Fritz, B.; Jenner, A.; Wahl, S.; Lappe, C.; Zehender, A.; Horn, C.; Blessing, F.; Kohl, M.; Ziemssen, F.; Egert, M. A view to a kill?—Ambient bacterial load of frames and lenses of spectacles and evaluation of different cleaning methods. PLoS ONE 2018, 13, e0207238. [Google Scholar] [CrossRef]
- Fritz, B.; März, M.; Weis, S.; Wahl, S.; Ziemssen, F.; Egert, M. Site-specific molecular analysis of the bacteriota on worn spectacles. Sci. Rep. 2020, 10, 5577. [Google Scholar] [CrossRef]
- Olcerst, R.B. Microscopes and ocular infections. Am. Ind. Hyg. Assoc. J. 1987, 48, 425–431. [Google Scholar] [CrossRef]
- DIN 10113-1:1997-07: Determination of Surface Colony Count on Fitment Utensils in Foodareas—Part 1: Quantitative Swab Method 1997-07. Available online: https://www.din.de/en/getting-involved/standards-committees/nal/wdc-beuth:din21:2981234 (accessed on 8 April 2020).
- Frölander, F.; Carlsson, J. Bactericidal effect of anaerobic broth exposed to atmospheric oxygen tested on Peptostreptococcus anaerobius. J. Clin. Microbiol. 1977, 6, 117–123. [Google Scholar] [PubMed]
- Bossard, D.A.; Ledergerber, B.; Zingg, P.O.; Gerber, C.; Zinkernagel, A.S.; Zbinden, R.; Achermann, Y. Optimal length of cultivation time for isolation of Propionibacterium acnes in suspected bone and joint infections is more than 7 days. J. Clin. Microbiol. 2016, 54, 3043–3049. [Google Scholar] [CrossRef]
- Bruker Daltonik GmbH. Instructions for Use: MALDI Biotarget 48: Disposable MALDI Targets for Microorganism Identification. Available online: https://www.bruker.com/fileadmin/user_upload/8-PDF-Docs/Separations_MassSpectrometry/InstructionForUse/IFU_268711_267615_226413_MALDI_Biotarget_48_Rev1.pdf (accessed on 7 February 2020).
- Castelino, M.; Eyre, S.; Moat, J.; Fox, G.; Martin, P.; Ho, P.; Upton, M.; Barton, A. Optimisation of methods for bacterial skin microbiome investigation: Primer selection and comparison of the 454 versus MiSeq platform. BMC Microbiol. 2017, 17, 23. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.-C.; Chou, C.-Y.; Chen, L.-L.; Kuo, C.-H. Bacterial community dynamics in a swine wastewater anaerobic reactor revealed by 16S rDNA sequence analysis. J. Biotechnol. 2015, 194, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Illumina. 16S Metagenomic Sequencing Library Preparation: Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 7 February 2019).
- Rottenfusser, R.; Wilson, E.E.; Davidsen, M.W. Education in Microscopy and Digital Imaging: Microscope Cleaning and Maintenance. Available online: http://zeiss-campus.magnet.fsu.edu/articles/basics/care.html (accessed on 4 December 2019).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McDonald, D.; Clemente, J.C.; Kuczynski, J.; Rideout, J.R.; Stombaugh, J.; Wendel, D.; Wilke, A.; Huse, S.; Hufnagle, J.; Meyer, F.; et al. The Biological Observation Matrix (BIOM) format or: How I learned to stop worrying and love the ome-ome. GigaScience 2012, 1, 7. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.-i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Classification of Prokaryotes (Bacteria and Archaea) into Risk Groups. Technical Rule for Biological Agents, TRBA 466. 2015. Available online: https://www.baua.de/EN/Service/Legislative-texts-and-technical-rules/Rules/TRBA/TRBA-466.html (accessed on 4 February 2020).
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R Package Version 2.5-4. Available online: https://CRAN.R-project.org/package=vegan (accessed on 4 February 2020).
- Zeileis, A.; Wiel, M.A.; Hornik, K.; Hothorn, T. Implementing a class of permutation tests: The coin Package. J. Stat. Soft. 2008, 28, 1–23. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Dordrecht, The Netherlands; New York, NY, USA, 2009. [Google Scholar]
- Bisanz, J.E. qiime2R-Importing QIIME2 Artifacts and Associated Data into R Sessions. Version 0.99.13. Available online: https://rdrr.io/github/jbisanz/qiime2R/ (accessed on 4 February 2020).
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots; R-package Version 0.2.3. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 4 February 2020).
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143. [Google Scholar] [CrossRef]
- Platsidaki, E.; Dessinioti, C. Recent advances in understanding Propionibacterium acnes (Cutibacterium acnes) in acne. F1000Research 2018, 7, 1953. [Google Scholar] [CrossRef]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef]
- Yang, J.; Tsukimi, T.; Yoshikawa, M.; Suzuki, K.; Takeda, T.; Tomita, M.; Fukuda, S. Cutibacterium acnes (Propionibacterium acnes) 16S rRNA Genotyping of microbial samples from possessions contributes to owner identification. mSystems 2019, 4. [Google Scholar] [CrossRef]
- Di Lodovico, S.; Del Vecchio, A.; Cataldi, V.; Di Campli, E.; Di Bartolomeo, S.; Cellini, L.; Di Giulio, M. Microbial contamination of smartphone touchscreens of italian university students. Curr. Microbiol. 2018, 75, 336–342. [Google Scholar] [CrossRef]
- Gerba, C.P.; Wuollet, A.L.; Raisanen, P.; Lopez, G.U. Bacterial contamination of computer touch screens. Am. J. Infect. Control. 2016, 44, 358–360. [Google Scholar] [CrossRef]
- Bayston, R.; Ashraf, W.; Barker-Davies, R.; Tucker, E.; Clement, R.; Clayton, J.; Freeman, B.J.C.; Nuradeen, B. Biofilm formation by Propionibacterium acnes on biomaterials in vitro and in vivo: Impact on diagnosis and treatment. J. Biomed. Mater. Res. 2007, 81, 705–709. [Google Scholar] [CrossRef]
- Courjaret, J.-C.; Drancourt, M.; Hoffart, L. Paracoccus yeei keratitis in a contact lens wearer. Eye Contact Lens 2014, 40, e21–e22. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-D.; Lee, H.B.; Yi, H.; Kim, Y.; Bae, K.S.; Choi, J.-E.; Jung, H.S.; Chun, J. Pseudomonas panacis sp. nov. isolated from the surface of rusty roots of Korean ginseng. Int. J. Syst. Evol. Microbiol. 2005, 55, 1721–1724. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baur, C.; Krewinkel, M.; Kutzli, I.; Kranz, B.; von Neubeck, M.; Huptas, C.; Wenning, M.; Scherer, S.; Stoeckel, M.; Hinrichs, J.; et al. Isolation and characterisation of a heat-resistant peptidase from Pseudomonas panacis withstanding general UHT processes. Int. Dairy J. 2015, 49, 46–55. [Google Scholar] [CrossRef]
- Tauch, A.; Fernández-Natal, I.; Soriano, F. A microbiological and clinical review on Corynebacterium kroppenstedtii. Int. J. Infect. Dis. 2016, 48, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, S.; Zbinden, R.; Pagano, M.; Fischler, M.; Speich, R. Central venous catheter infection with Brevibacterium sp. in an immunocompetent woman: Case report and review of the literature. Infection 2006, 34, 103–106. [Google Scholar] [CrossRef]
- Kandi, V.; Palange, P.; Vaish, R.; Bhatti, A.B.; Kale, V.; Kandi, M.R.; Bhoomagiri, M.R. Emerging bacterial infection: Identification and clinical significance of Kocuria species. Cureus 2016, 8, e731. [Google Scholar] [CrossRef]
- Lasek, R.; Szuplewska, M.; Mitura, M.; Decewicz, P.; Chmielowska, C.; Pawłot, A.; Sentkowska, D.; Czarnecki, J.; Bartosik, D. Genome structure of the opportunistic pathogen Paracoccus yeei (Alphaproteobacteria) and identification of putative virulence factors. Front. Microbiol. 2018, 9, 2553. [Google Scholar] [CrossRef]
- Tak, E.J.; Kim, P.S.; Hyun, D.-W.; Kim, H.S.; Lee, J.-Y.; Kang, W.; Sung, H.; Shin, N.-R.; Kim, M.-S.; Whon, T.W.; et al. Phenotypic and genomic properties of Brachybacterium vulturis sp. nov. and Brachybacterium avium sp. nov. Front. Microbiol. 2018, 9, 1809. [Google Scholar] [CrossRef]
- Merino, N.; Zhang, S.; Tomita, M.; Suzuki, H. Comparative genomics of bacteria commonly identified in the built environment. BMC Genom. 2019, 20, 92. [Google Scholar] [CrossRef]
- Wu, D.C.; Chan, W.W.; Metelitsa, A.I.; Fiorillo, L.; Lin, A.N. Pseudomonas skin infection. Am. J. Clin. Dermatol. 2011, 12, 157–169. [Google Scholar] [CrossRef]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and pathophysiological overview of acinetobacter infections: A century of challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Schwebke, I.; Kampf, G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 2006, 6, 130. [Google Scholar] [CrossRef] [PubMed]
- Missiakas, D.M.; Schneewind, O. Growth and laboratory maintenance of Staphylococcus aureus. Curr. Protoc. Microbiol. 2013, 28, 9C-1. [Google Scholar] [CrossRef]
- Forney, L.J.; Zhou, X.; Brown, C.J. Molecular microbial ecology: Land of the one-eyed king. Curr. Opin. Microbiol. 2004, 7, 210–220. [Google Scholar] [CrossRef]
- Willcox, M.D.P. Characterization of the normal microbiota of the ocular surface. Exp. Eye Res. 2013, 117, 99–105. [Google Scholar] [CrossRef]
- Lee, S.H.; Oh, D.H.; Jung, J.Y.; Kim, J.C.; Jeon, C.O. Comparative ocular microbial communities in humans with and without blepharitis. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5585–5593. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Cheng, C.; Yi, H.; Lin, L.; Wu, K. Quantitative analysis of the bacteria in blepharitis with Demodex infestation. Front. Microbiol. 2018, 9, 1719. [Google Scholar] [CrossRef]
- Ovodenko, B.; Seedor, J.A.; Ritterband, D.C.; Shah, M.; Yang, R.; Koplin, R.S. The prevalence and pathogenicity of Propionibacterium acnes keratitis. Cornea 2009, 28, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. J. Clin. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Bodena, D.; Teklemariam, Z.; Balakrishnan, S.; Tesfa, T. Bacterial contamination of mobile phones of health professionals in Eastern Ethiopia: Antimicrobial susceptibility and associated factors. Trop. Med. Health. 2019, 47, 15. [Google Scholar] [CrossRef]
- Cave, R.; Misra, R.; Chen, J.; Wang, S.; Mkrtchyan, H.V. Whole genome sequencing revealed new molecular characteristics in multidrug resistant staphylococci recovered from high frequency touched surfaces in London. Sci. Rep. 2019, 9, 9637. [Google Scholar] [CrossRef] [PubMed]
- Esteves, D.C.; Pereira, V.C.; Souza, J.M.; Keller, R.; Simões, R.D.; Winkelstroter Eller, L.K.; Rodrigues, M.V.P. Influence of biological fluids in bacterial viability on different hospital surfaces and fomites. Am. J. Infect. Control. 2016, 44, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Szentmáry, N.; Daas, L.; Shi, L.; Laurik, K.L.; Lepper, S.; Milioti, G.; Seitz, B. Acanthamoeba keratitis Clinical signs, differential diagnosis and treatment. J. Curr. Ophthalmol. 2019, 31, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Azher, T.N.; Yin, X.-T.; Tajfirouz, D.; Huang, A.J.; Stuart, P.M. Herpes simplex keratitis: Challenges in diagnosis and clinical management. Clin. Ophthalmol. 2017, 11, 185–191. [Google Scholar] [CrossRef]
- Watson, S.; Cabrera-Aguas, M.; Khoo, P. Common eye infections. Aust. Prescr. 2018, 41, 67–72. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Ihira, M.; Suzuki, K.; Suga, S.; Tomitaka, A.; Ueda, H.; Asano, Y. Rapid contamination of the environments with varicella-zoster virus DNA from a patient with herpes zoster. J. Med. Virol. 2001, 63, 64–66. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fritz, B.; Schäfer, K.; März, M.; Wahl, S.; Ziemssen, F.; Egert, M. Eye-Catching Microbes—Polyphasic Analysis of the Microbiota on Microscope Oculars Verifies Their Role as Fomites. J. Clin. Med. 2020, 9, 1572. https://doi.org/10.3390/jcm9051572
Fritz B, Schäfer K, März M, Wahl S, Ziemssen F, Egert M. Eye-Catching Microbes—Polyphasic Analysis of the Microbiota on Microscope Oculars Verifies Their Role as Fomites. Journal of Clinical Medicine. 2020; 9(5):1572. https://doi.org/10.3390/jcm9051572
Chicago/Turabian StyleFritz, Birgit, Karin Schäfer, Melanie März, Siegfried Wahl, Focke Ziemssen, and Markus Egert. 2020. "Eye-Catching Microbes—Polyphasic Analysis of the Microbiota on Microscope Oculars Verifies Their Role as Fomites" Journal of Clinical Medicine 9, no. 5: 1572. https://doi.org/10.3390/jcm9051572
APA StyleFritz, B., Schäfer, K., März, M., Wahl, S., Ziemssen, F., & Egert, M. (2020). Eye-Catching Microbes—Polyphasic Analysis of the Microbiota on Microscope Oculars Verifies Their Role as Fomites. Journal of Clinical Medicine, 9(5), 1572. https://doi.org/10.3390/jcm9051572




