IMPA2 Downregulation Enhances mTORC1 Activity and Restrains Autophagy Initiation in Metastatic Clear Cell Renal Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical and Molecular Data for RCC Patients
2.2. Cell Culture
2.3. Cellular Migration Assays
2.4. Lentivirus Production and Transduction
2.5. Western Blotting Assay
2.6. Animal Experiments
2.7. Statistical Analyses
3. Results
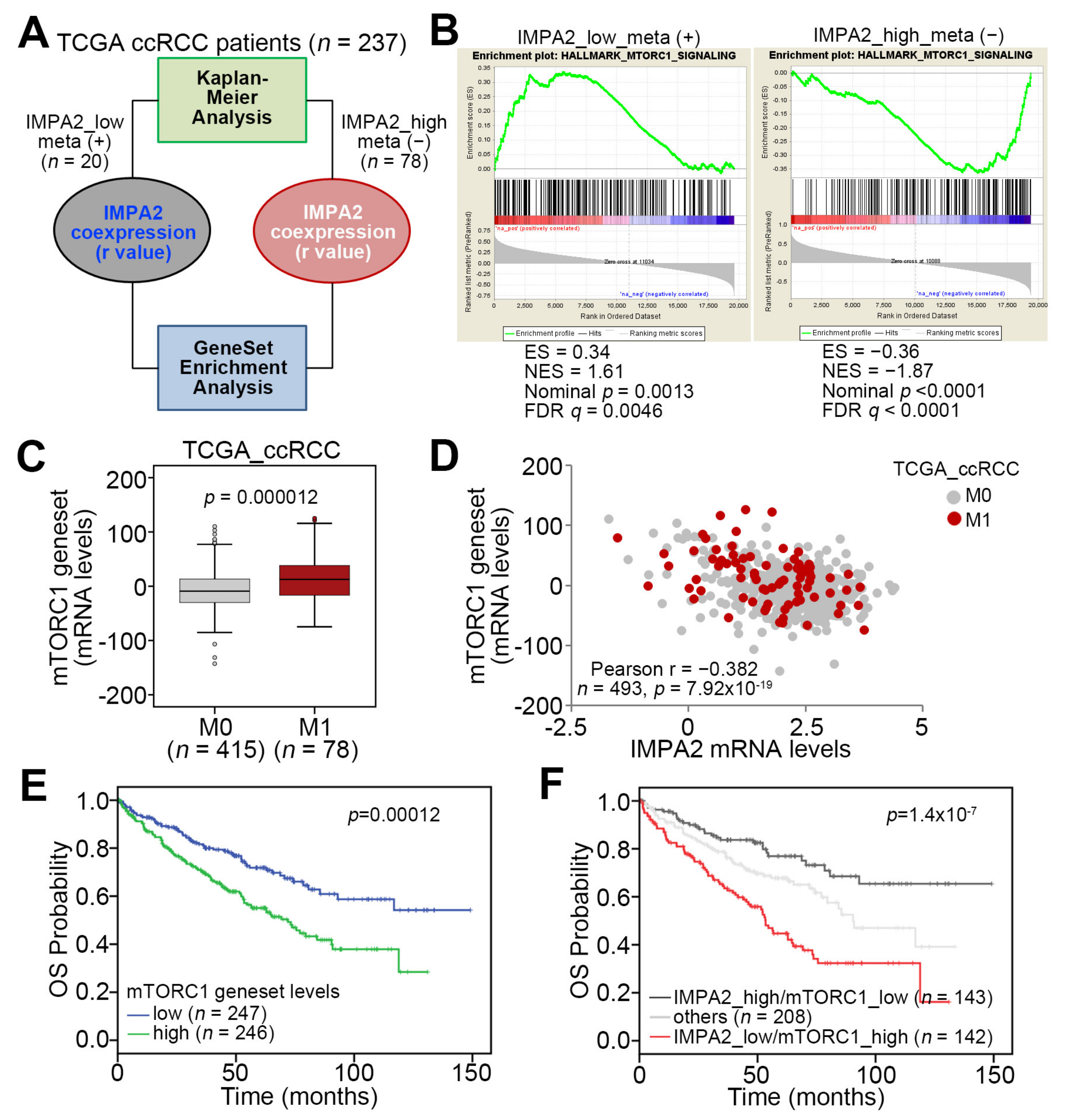
3.1. IMPA2 Downregulation Accompanied by Enhanced mTORC1 Activity Correlates with Metastatic Progression and Poor Prognosis in ccRCC Patients
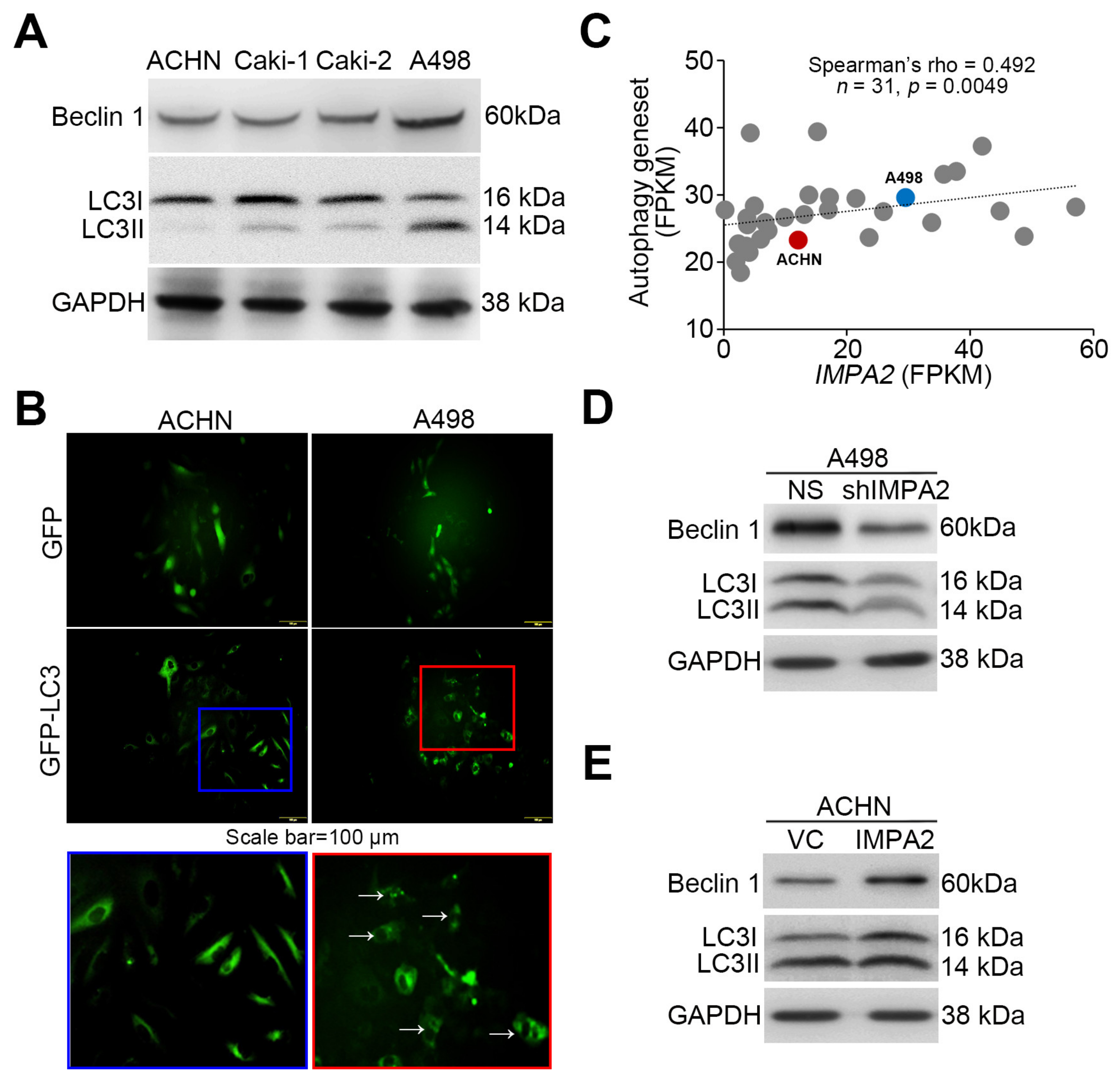
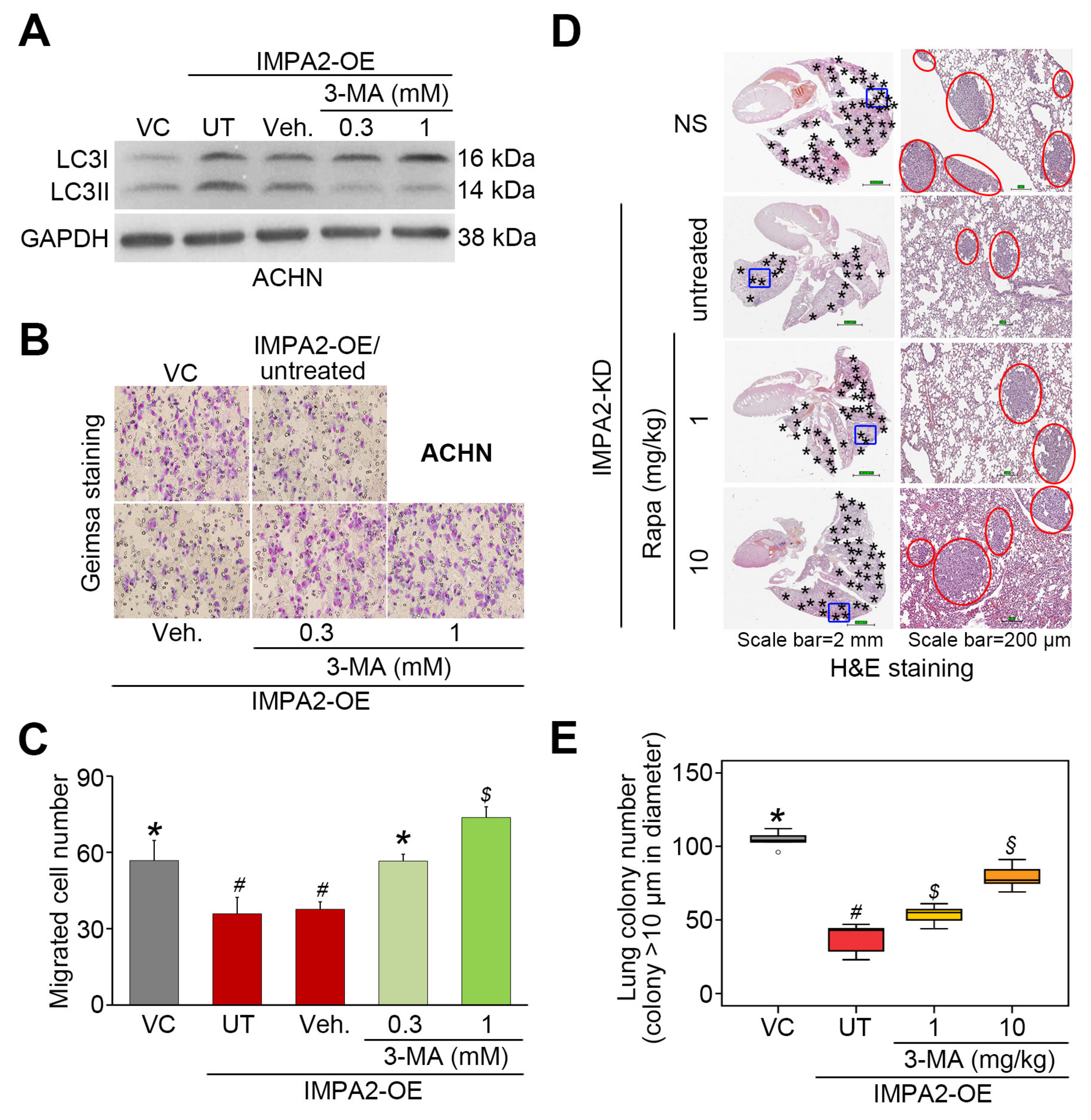
3.2. IMPA2 Expression Negatively Regulates the Activation of the Akt/mTORC1 Pathway and Associates with Autophagy Formation in ccRCC Cells
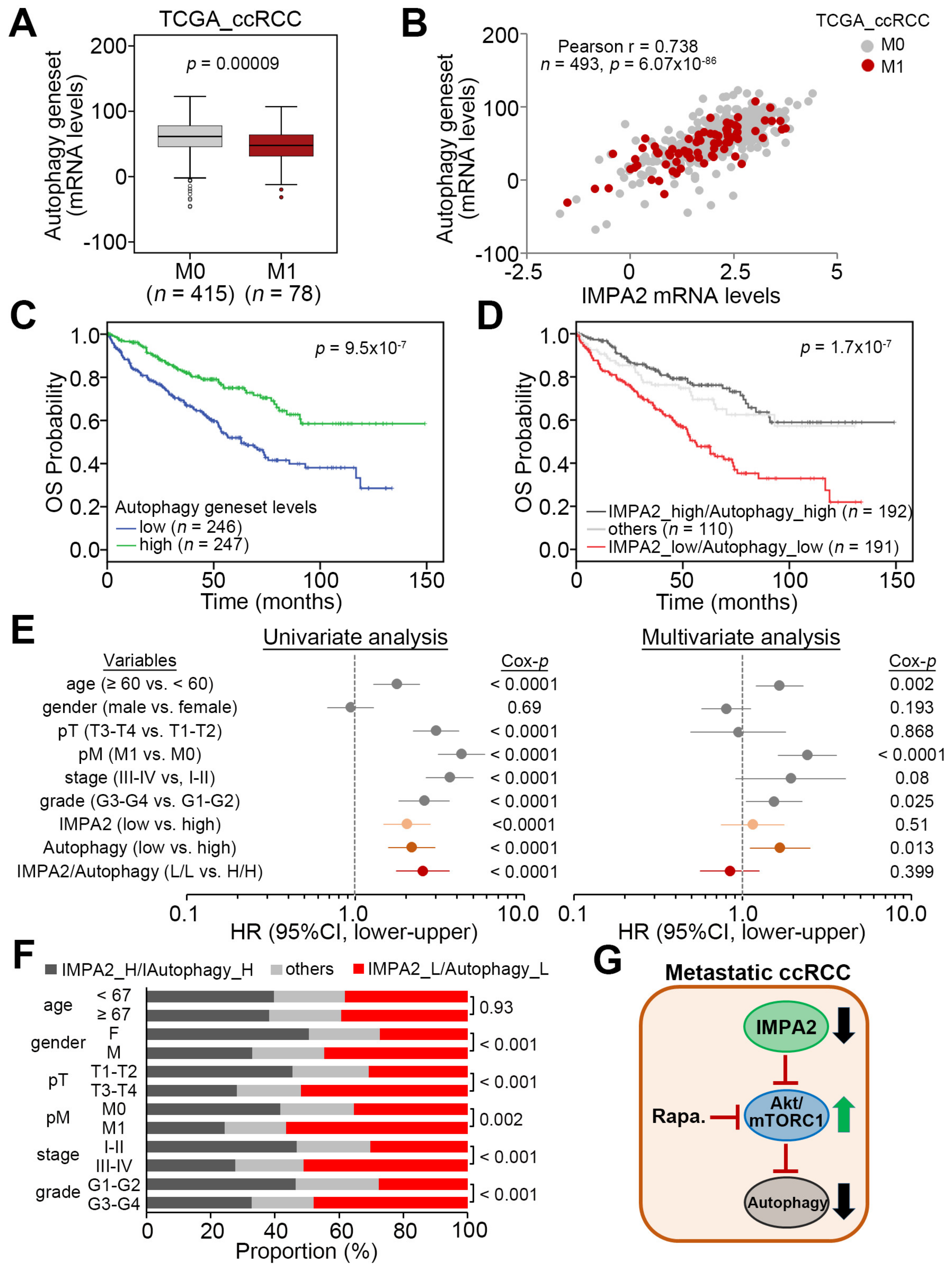
3.3. IMPA2 Downregulation Combined with Decreased Autophagy is Associated with a Poor Overall Survival Rate in ccRCC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kovacs, G.; Akhtar, M.; Beckwith, B.J.; Bugert, P.; Cooper, C.S.; Delahunt, B.; Eble, J.N.; Fleming, S.; Ljungberg, B.; Medeiros, L.J.; et al. The Heidelberg classification of renal cell tumours. J. Pathol. 1997, 183, 131–133. [Google Scholar] [CrossRef]
- Rini, B.I.; Campbell, S.C.; Escudier, B. Renal cell carcinoma. Lancet 2009, 373, 1119–1132. [Google Scholar] [CrossRef]
- Frank, I.; Blute, M.L.; Cheville, J.C.; Lohse, C.M.; Weaver, A.L.; Zincke, H. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: The SSIGN score. J. Urol. 2002, 168, 2395–2400. [Google Scholar] [CrossRef]
- Motzer, R.J.; Hutson, T.E.; McCann, L.; Deen, K.; Choueiri, T.K. Overall survival in renal-cell carcinoma with pazopanib versus sunitinib. N. Engl. J. Med. 2014, 370, 1769–1770. [Google Scholar] [CrossRef]
- Shamir, A.; Sjoholt, G.; Ebstein, R.P.; Agam, G.; Steen, V.M. Characterization of two genes, Impa1 and Impa2 encoding mouse myo-inositol monophosphatases. Gene 2001, 271, 285–291. [Google Scholar] [CrossRef]
- Canales, J.; Buitrago, F.; Faraldo, A.; Avalos, M.; Cameselle, J.C. Identification of rat liver glucose-3-phosphatase as an inositol monophosphatase inhibited by lithium. Arch. Biochem. Biophys. 1997, 343, 27–34. [Google Scholar] [CrossRef]
- Vadnal, R.; Parthasarathy, R. Myo-inositol monophosphatase: Diverse effects of lithium, carbamazepine, and valproate. Neuropsychopharmacology 1995, 12, 277–285. [Google Scholar] [CrossRef]
- Seelan, R.S.; Parthasarathy, L.K.; Parthasarathy, R.N. Lithium modulation of the human inositol monophosphatase 2 (IMPA2) promoter. Biochem. Biophys. Res.Commun. 2004, 324, 1370–1378. [Google Scholar] [CrossRef]
- Ohnishi, T.; Ohba, H.; Seo, K.C.; Im, J.; Sato, Y.; Iwayama, Y.; Furuichi, T.; Chung, S.K.; Yoshikawa, T. Spatial expression patterns and biochemical properties distinguish a second myo-inositol monophosphatase IMPA2 from IMPA1. J. Biol. Chem. 2007, 282, 637–646. [Google Scholar] [CrossRef]
- Damri, O.; Sade, Y.; Toker, L.; Bersudsky, Y.; Belmaker, R.H.; Agam, G.; Azab, A.N. Molecular effects of lithium are partially mimicked by inositol-monophosphatase (IMPA)1 knockout mice in a brain region-dependent manner. Eur. Neuropsychopharmacol. 2015, 25, 425–434. [Google Scholar] [CrossRef]
- Cryns, K.; Shamir, A.; Shapiro, J.; Daneels, G.; Goris, I.; Van, C.H.; Straetemans, R.; Belmaker, R.H.; Agam, G.; Moechars, D.; et al. Lack of lithium-like behavioral and molecular effects in IMPA2 knockout mice. Neuropsychopharmacology 2007, 32, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, T.; Watanabe, A.; Ohba, H.; Iwayama, Y.; Maekawa, M.; Yoshikawa, T. Behavioral analyses of transgenic mice harboring bipolar disorder candidate genes, IMPA1 and IMPA2. Neurosci. Res. 2010, 67, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.F.; Chou, J.L.; Chang, J.S.; Chiu, I.J.; Chiu, H.W.; Lin, Y.F. Dysregulation of the miR-25-IMPA2 axis promotes metastatic progression in clear cell renal cell carcinoma. EBioMedicine 2019, 45, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Floto, R.A.; Berger, Z.; Imarisio, S.; Cordenier, A.; Pasco, M.; Cook, L.J.; Rubinsztein, D.C. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 2005, 170, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- O’Donovan, T.R.; Rajendran, S.; O’Reilly, S.; O’Sullivan, G.C.; McKenna, S.L. Lithium Modulates Autophagy in Esophageal and Colorectal Cancer Cells and Enhances the Efficacy of Therapeutic Agents In Vitro and In Vivo. PLoS ONE 2015, 10, e0134676. [Google Scholar] [CrossRef]
- de Araujo, W.M.; Robbs, B.K.; Bastos, L.G.; de Souza, W.F.; Vidal, F.C.; Viola, J.P.; Morgado-Diaz, J.A. PTEN Overexpression Cooperates With Lithium to Reduce the Malignancy and to Increase Cell Death by Apoptosis via PI3K/Akt Suppression in Colorectal Cancer Cells. J. Cell Biochem. 2016, 117, 458–469. [Google Scholar] [CrossRef]
- Ljungberg, B.; Campbell, S.C.; Choi, H.Y.; Jacqmin, D.; Lee, J.E.; Weikert, S.; Kiemeney, L.A. The epidemiology of renal cell carcinoma. Eur. Urol. 2011, 60, 615–621. [Google Scholar] [CrossRef]
- Motzer, R.J.; Bukowski, R.M. Targeted therapy for metastatic renal cell carcinoma. J. Clin. Oncol. 2006, 24, 5601–5608. [Google Scholar] [CrossRef]
- Powles, T.; Albiges, L.; Staehler, M.; Bensalah, K.; Dabestani, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; Lam, T.B.; et al. Updated European Association of Urology Guidelines Recommendations for the Treatment of First-line Metastatic Clear Cell Renal Cancer. Eur. Urol. 2018, 73, 311–315. [Google Scholar] [CrossRef]
- Heng, Y.; Xie, W.; Regan, M.M.; Harshman, L.C.; Bjarnason, G.A.; Vaishampayan, U.N.; Mackenzie, M.; Wood, L.; Donskov, F.; Tan, M.H.; et al. External validation and comparison with other models of the International Metastatic Renal-Cell Carcinoma Database Consortium prognostic model: A population-based study. Lancet Oncol. 2013, 14, 141–148. [Google Scholar]
- Hudes, G.R. mTOR as a target for therapy of renal cancer. Clin. Adv. Hematol. Oncol. 2007, 5, 772–774. [Google Scholar] [PubMed]
- Hudes, G.; Carducci, M.; Tomczak, P.; Dutcher, J.; Figlin, R.; Kapoor, A.; Staroslawska, E.; Sosman, J.; McDermott, D.; Bodrogi, I.; et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N. Engl. J. Med. 2007, 356, 2271–2281. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Zhang, B.; Mao, Y.; Zhang, H.; Hong, W. Long non-coding RNA OECC promotes cell proliferation and metastasis through the PI3K/Akt/mTOR signaling pathway in human lung cancer. Oncol. Lett. 2019, 18, 3017–3024. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhao, Y.; Hu, Z.; Li, J.; Chu, D.; Zhang, J.; Li, Z.; Chen, B.; Zhang, X.; Pan, H.; et al. MetaLnc9 Facilitates Lung Cancer Metastasis via a PGK1-Activated AKT/mTOR Pathway. Cancer Res. 2017, 77, 5782–5794. [Google Scholar] [CrossRef]
- Yang, X.; Song, X.; Wang, X.; Liu, X.; Peng, Z. Downregulation of TM7SF4 inhibits cell proliferation and metastasis of A549 cells through regulating the PI3K/AKT/mTOR signaling pathway. Mol. Med. Rep. 2017, 16, 6122–6127. [Google Scholar] [CrossRef]
- Yue, S.J.; Zhang, P.X.; Zhu, Y.; Li, N.G.; Chen, Y.Y.; Li, J.J.; Zhang, S.; Jin, R.Y.; Yan, H.; Shi, X.Q.; et al. A Ferulic Acid Derivative FXS-3 Inhibits Proliferation and Metastasis of Human Lung Cancer A549 Cells via Positive JNK Signaling Pathway and Negative ERK/p38, AKT/mTOR and MEK/ERK Signaling Pathways. Molecules 2019, 24, 2165. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, R.; Li, Q.; Jie, X.; Hong, J.; Zong, Y.; Dong, X.; Zhang, S.; Li, Z.; Wu, G. Cardamonin inhibits the proliferation and metastasis of non-small-cell lung cancer cells by suppressing the PI3K/Akt/mTOR pathway. Anticancer Drugs 2019, 30, 241–250. [Google Scholar] [CrossRef]
- Lv, X.; Li, C.Y.; Han, P.; Xu, X.Y. MicroRNA-520a-3p inhibits cell growth and metastasis of non-small cell lung cancer through PI3K/AKT/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2321–2327. [Google Scholar]
- Mao, H.Y.; Liu, S.P.; Kong, G.M.; Xu, Y.; Xu, Y.C.; Sun, G.Z.; Wang, Z.; Han, F.; Tong, J.D.; Bo, P. FBLN3 inhibited the invasion and metastasis of colorectal cancer through the AKT/mTOR pathway. Neoplasma 2019, 66, 336–342. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, C.; Zhao, Q.; Wei, W.; Dong, Z.; Shao, L.; Li, J.; Wu, W.; Zhang, H.; Huang, H.; et al. The miR-873/NDFIP1 axis promotes hepatocellular carcinoma growth and metastasis through the AKT/mTOR-mediated Warburg effect. Am. J. Cancer Res. 2019, 9, 927–944. [Google Scholar]
- Xu, X.; Kong, X.; Liu, T.; Zhou, L.; Wu, J.; Fu, J.; Wang, Y.; Zhu, M.; Yao, S.; Ding, Y.; et al. Metastasis-associated protein 1, modulated by miR-30c, promotes endometrial cancer progression through AKT/mTOR/4E-BP1 pathway. Gynecol. Oncol. 2019, 154, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Wang, W.J.; Zhang, Y. Aclidinium inhibits proliferation and metastasis of ovarian cancer SKOV3 cells via downregulating PI3K/AKT/mTOR signaling pathway. Oncol. Lett. 2018, 16, 6417–6422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.R.; Wang, S.Y.; Sun, W.; Wei, C. Isoliquiritigenin inhibits proliferation and metastasis of MKN28 gastric cancer cells by suppressing the PI3K/AKT/mTOR signaling pathway. Mol. Med. Rep. 2018, 18, 3429–3436. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.Y.; Wu, M.L.; Chang, C.I.; Liu, C.I.; Cheng, T.C.; Wu, Y.J. Bornyl cis-4-Hydroxycinnamate Suppresses Cell Metastasis of Melanoma through FAK/PI3K/Akt/mTOR and MAPK Signaling Pathways and Inhibition of the Epithelial-to-Mesenchymal Transition. Int. J. Mol. Sci. 2018, 19, 2152. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.H.; Zhang, C.; Bai, Y.; Li, Y.; Kang, X.; Chen, J.X.; Yao, K.; Jiang, T.; Zhong, X.S.; Li, W.B. Antiglioma effects of cytarabine on leptomeningeal metastasis of high-grade glioma by targeting the PI3K/Akt/mTOR pathway. Drug Des Devel. Ther. 2017, 11, 1905–1915. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tan, X.; Liu, P.; Wu, Y.; Qian, S.; Zhang, X. Phosphoglycerate Mutase 1 (PGAM1) Promotes Pancreatic Ductal Adenocarcinoma (PDAC) Metastasis by Acting as a Novel Downstream Target of the PI3K/Akt/mTOR Pathway. Oncol. Res. 2018, 26, 1123–1131. [Google Scholar] [CrossRef]
- Wang, M.H.; Sun, R.; Zhou, X.M.; Zhang, M.Y.; Lu, J.B.; Yang, Y.; Zeng, L.S.; Yang, X.Z.; Shi, L.; Xiao, R.W.; et al. Epithelial cell adhesion molecule overexpression regulates epithelial-mesenchymal transition, stemness and metastasis of nasopharyngeal carcinoma cells via the PTEN/AKT/mTOR pathway. Cell Death. Dis. 2018, 9, 2. [Google Scholar] [CrossRef]
- Ma, H.; Su, R.; Feng, H.; Guo, Y.; Su, G. Long noncoding RNA UCA1 promotes osteosarcoma metastasis through CREB1-mediated epithelial-mesenchymal transition and activating PI3K/AKT/mTOR pathway. J. Bone Oncol. 2019, 16, 100228. [Google Scholar] [CrossRef]
- Xia, E.; Zhou, X.; Bhandari, A.; Zhang, X.; Wang, O. Synaptopodin-2 plays an important role in the metastasis of breast cancer via PI3K/Akt/mTOR pathway. Cancer Manag. Res. 2018, 10, 1575–1583. [Google Scholar] [CrossRef]
- Ni, J.; Cozzi, P.; Hao, J.; Beretov, J.; Chang, L.; Duan, W.; Shigdar, S.; Delprado, W.; Graham, P.; Bucci, J.; et al. Epithelial cell adhesion molecule (EpCAM) is associated with prostate cancer metastasis and chemo/radioresistance via the PI3K/Akt/mTOR signaling pathway. Int. J. Biochem. Cell Biol. 2013, 45, 2736–2748. [Google Scholar] [CrossRef]
- Fang, Z.; Tang, Y.; Fang, J.; Zhou, Z.; Xing, Z.; Guo, Z.; Guo, X.; Wang, W.; Jiao, W.; Xu, Z.; et al. Simvastatin inhibits renal cancer cell growth and metastasis via AKT/mTOR, ERK and JAK2/STAT3 pathway. PLoS ONE 2013, 8, e62823. [Google Scholar] [CrossRef]
- Xie, J.; Lin, W.; Huang, L.; Xu, N.; Xu, A.; Chen, B.; Watanabe, M.; Liu, C.; Huang, P. Bufalin suppresses the proliferation and metastasis of renal cell carcinoma by inhibiting the PI3K/Akt/mTOR signaling pathway. Oncol. Lett. 2018, 16, 3867–3873. [Google Scholar] [CrossRef] [PubMed]
- Mowers, E.E.; Sharifi, M.N.; Macleod, K.F. Autophagy in cancer metastasis. Oncogene 2017, 36, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, S.; Chua, M.S.; Li, H.; Luo, D.; Wang, S.; Zhang, S.; Han, B.; Sun, C. SOCS5 inhibition induces autophagy to impair metastasis in hepatocellular carcinoma cells via the PI3K/Akt/mTOR pathway. Cell Death. Dis. 2019, 10, 612. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.S.; Gao, Z.R.; Zhang, Q.; Tang, X.F.; Lv, Y.F.; Zhang, Z.S.; Zhang, Y.; Tan, Q.L.; Peng, D.B.; Jiang, D.M.; et al. TSSC3 promotes autophagy via inactivating the Src-mediated PI3K/Akt/mTOR pathway to suppress tumorigenesis and metastasis in osteosarcoma, and predicts a favorable prognosis. J. Exp. Clin. Cancer Res. 2018, 37, 188. [Google Scholar] [CrossRef]
- Singla, M.; Bhattacharyya, S. Autophagy as a potential therapeutic target during epithelial to mesenchymal transition in renal cell carcinoma: An in vitro study. Biomed. Pharmacother 2017, 94, 332–340. [Google Scholar] [CrossRef]
- Li, F.; Ma, Z.; Guan, Z.; Chen, Y.; Wu, K.; Guo, P.; Wang, X.; He, D.; Zeng, J. Autophagy induction by silibinin positively contributes to its anti-metastatic capacity via AMPK/mTOR pathway in renal cell carcinoma. Int. J. Mol. Sci. 2015, 16, 8415–8429. [Google Scholar] [CrossRef]
- Liu, S.; Xie, F.; Wang, H.; Liu, Z.; Liu, X.; Sun, L.; Niu, Z. Ubenimex inhibits cell proliferation, migration and invasion in renal cell carcinoma: The effect is autophagy-associated. Oncol. Rep. 2015, 33, 1372–1380. [Google Scholar] [CrossRef]
- Byun, S.; Lee, E.; Lee, K.W. Therapeutic Implications of Autophagy Inducers in Immunological Disorders, Infection, and Cancer. Int. J. Mol. Sci. 2017, 18, 1959. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuei, C.-H.; Lin, H.-Y.; Lee, H.-H.; Lin, C.-H.; Zheng, J.-Q.; Chen, K.-C.; Lin, Y.-F. IMPA2 Downregulation Enhances mTORC1 Activity and Restrains Autophagy Initiation in Metastatic Clear Cell Renal Cell Carcinoma. J. Clin. Med. 2020, 9, 956. https://doi.org/10.3390/jcm9040956
Kuei C-H, Lin H-Y, Lee H-H, Lin C-H, Zheng J-Q, Chen K-C, Lin Y-F. IMPA2 Downregulation Enhances mTORC1 Activity and Restrains Autophagy Initiation in Metastatic Clear Cell Renal Cell Carcinoma. Journal of Clinical Medicine. 2020; 9(4):956. https://doi.org/10.3390/jcm9040956
Chicago/Turabian StyleKuei, Chia-Hao, Hui-Yu Lin, Hsun-Hua Lee, Che-Hsuan Lin, Jing-Quan Zheng, Kuan-Chou Chen, and Yuan-Feng Lin. 2020. "IMPA2 Downregulation Enhances mTORC1 Activity and Restrains Autophagy Initiation in Metastatic Clear Cell Renal Cell Carcinoma" Journal of Clinical Medicine 9, no. 4: 956. https://doi.org/10.3390/jcm9040956
APA StyleKuei, C.-H., Lin, H.-Y., Lee, H.-H., Lin, C.-H., Zheng, J.-Q., Chen, K.-C., & Lin, Y.-F. (2020). IMPA2 Downregulation Enhances mTORC1 Activity and Restrains Autophagy Initiation in Metastatic Clear Cell Renal Cell Carcinoma. Journal of Clinical Medicine, 9(4), 956. https://doi.org/10.3390/jcm9040956




