RNAs and Gene Expression Predicting Postoperative Atrial Fibrillation in Cardiac Surgery Patients Undergoing Coronary Artery Bypass Grafting
Abstract
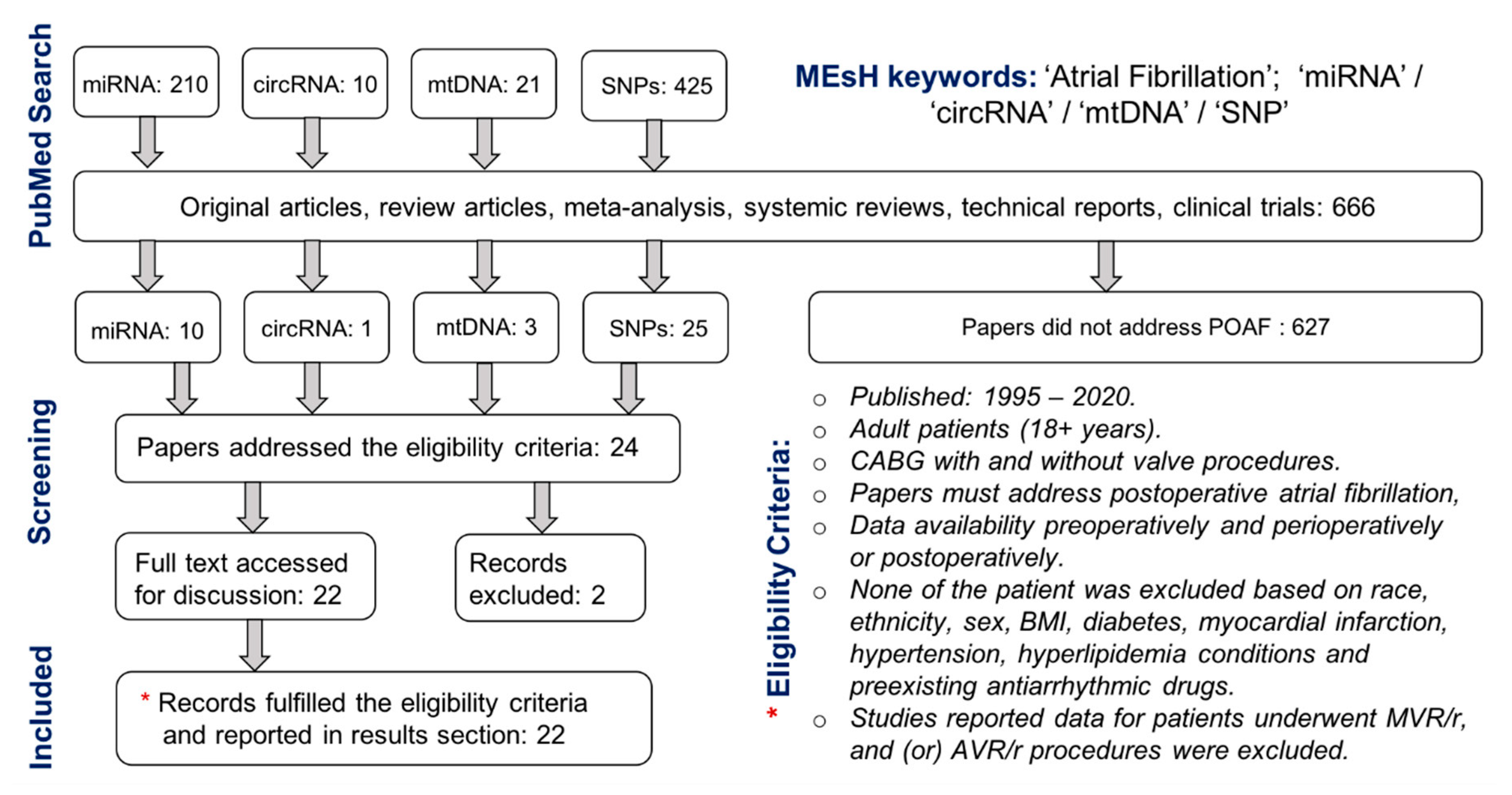
1. Introduction
2. Materials and Methods
2.1. Design of Study
2.2. Data Extraction
3. Results and Discussion
3.1. Micro-RNAs Predicting POAF
3.1.1. miRNA−483−5p
3.1.2. miRNA−29a
3.1.3. miRNA−23a and miRNA−26a
3.1.4. miRNA−199a
3.1.5. miRNA−1 and miRNA−133a
3.2. circRNA Predicting POAF
3.3. Gene Expressions Predicting POAF
3.3.1. Mitochondrial DNA (mtDNA)
3.3.2. Single Nucleotide Polymorphisms (SNPs)
4. Limitations
5. Future Perspective
- A large cohort study is warranted to investigate multivariate aforementioned parameters (miRNA, circRNA and gene expressions).
- Prospective study should be established with cardiac surgery patients with no preoperative AF history.
- Multicenter studies should enroll cardiac surgery patients regardless of their race, ethnicity, sex, BMI, diabetes, COPD, hypertension, hyperlipidemia, PVD, PAD, myocardial infarction, PCI, TIA and CAD.
- For each patient, pre-, intra- and post-operative antiarrhythmic drug record must be reported.
- Each patient blood sample must be collected preoperatively at two time points (24 h and 6 h) before surgery. RAA tissue sample can also collected intraoperatively.
- Same technique (qPCR, eQTL or OMNI1-Quad BeadChip) must be implemented to test assay.
- Electrophysiology findings such as conduction velocity and refractive period should be co-related with the levels for miRNA, circRNA and SNPs.
- Patients must be categorized as ‘no-POAF’ for those who do not develop post-surgery atrial fibrillation and ‘POAF’ for those who develop post-surgery AF within 1–4 days cardiac surgery.
- Results must be reported for both discovery and validated groups.
Author Contributions
Funding
Conflicts of Interest
References
- Gillinov, A.M.; Bagiella, E.; Moskowitz, A.J.; Raiten, J.M.; Groh, M.A.; Bowdish, M.E.; Ailawadi, G.; Kirkwood, K.A.; Perrault, L.P.; Parides, M.K.; et al. Rate Control versus Rhythm Control for Atrial Fibrillation after Cardiac Surgery. N. Engl. J. Med. 2016, 374, 1911–1921. [Google Scholar] [CrossRef]
- Bidar, E.; Bramer, S.; Maesen, B.; Maessen, J.G.; Schotten, U. Post-operative atrial fibrillation-Pathophysiology, treatment and prevention. J. Atr. Fibrillation 2013, 5, 781. [Google Scholar] [PubMed]
- Bessissow, A.; Khan, J.; Devereaux, P.J.; Alvarez-Garcia, J.; Alonso-Coello, P. Postoperative atrial fibrillation in non-cardiac and cardiac surgery: An overview. J. Thromb. Haemost. 2015, 13, S304–S312. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Ling, X.; Zhang, Y.; Shen, H.; Min, J.; Xi, W.; Wang, J.; Wang, Z. CHADS2 and CHA2DS2-VASc Scoring Systems for Predicting Atrial Fibrillation following Cardiac Valve Surgery. PLoS ONE 2015, 10, e0123858. [Google Scholar] [CrossRef] [PubMed]
- Aranki, S.F.; Shaw, D.P.; Adams, D.H.; Rizzo, R.J.; Couper, G.S.; VanderVliet, M.; Collins, J.J.; Cohn, L.H.; Burstin, H.R. Predictors of Atrial Fibrillation After Coronary Artery Surgery: Current Trends and Impact on Hospital Resources. Circulation 1996, 94, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, J.S. Postoperative atrial fibrillation: A billion-dollar problem. J. Am. Coll. Cardiol. 2004, 43, 1001–1003. [Google Scholar] [CrossRef]
- Badhwar, V.; Rankin, J.S.; Damiano, R.J.; Gillinov, A.M.; Bakaeen, F.G.; Edgerton, J.R.; Philpott, J.M.; McCarthy, P.M.; Bolling, S.F.; Roberts, H.G.; et al. The Society of Thoracic Surgeons 2017 Clinical Practice Guidelines for the Surgical Treatment of Atrial Fibrillation. Ann. Thorac. Surg. 2017, 103, 329–341. [Google Scholar] [CrossRef]
- Mavroudis, C.; Deal, B.J. Prophylactic arrhythmia surgery in association with congenital heart disease. Transl. Pediatrics 2016, 5, 148–159. [Google Scholar] [CrossRef]
- Stulak, J.M.; Suri, R.M.; Dearani, J.A.; Sundt, T.M.; Schaff, H.V. When Should Prophylactic Maze Procedure Be Considered in Patients Undergoing Mitral Valve Surgery? Ann. Thorac. Surg. 2010, 89, 1395–1401. [Google Scholar] [CrossRef]
- Yamashita, K.; Selzman, C.; Ranjan, R.; Hu, N.; Dosdall, D. Clinical Risk Factors for Post-operative Atrial Fibrillation among Patients after Cardiac Surgery. Thorac. Cardiovasc. Surg. 2019, 67, 107–116. [Google Scholar]
- Reckman, Y.J.; Creemers, E.E. Circulating circles predict postoperative atrial fibrillation. J. Am. Heart Assoc. 2018, 7, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Yamashita, K.; Sharma, V.; Ranjan, R.; Selzman, C.H.; Dosdall, D.J. Perioperative Biomarkers Predicting Postoperative Atrial Fibrillation Risk After Coronary Artery Bypass Grafting: A Narrative Review. J. Cardiothorac. Vasc. Anesth. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kondkar, A.A.; Abu-Amero, K.K. Utility of circulating MicroRNAs as clinical biomarkers for cardiovascular diseases. BioMed Res. Int. 2015, 2015, 821823. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.S.M.; Xia, K.; Salma, U.; Yang, T.; Peng, J. Diagnosis, prognosis and therapeutic role of circulating miRNAs in cardiovascular diseases. Heart Lung Circ. 2014, 23, 503–510. [Google Scholar] [CrossRef]
- Ono, K.; Kuwabara, Y.; Han, J. MicroRNAs and cardiovascular diseases. FEBS J. 2011, 278, 1619–1633. [Google Scholar] [CrossRef]
- Yamac, A.H.; Kucukbuzcu, S.; Ozansoy, M.; Gok, O.; Oz, K.; Erturk, M.; Yilmaz, E.; Ersoy, B.; Zeybek, R.; Goktekin, O.; et al. Altered expression of micro-RNA 199a and increased levels of cardiac SIRT1 protein are associated with the occurrence of atrial fibrillation after coronary artery bypass graft surgery. Cardiovasc. Pathol. 2016, 25, 232–236. [Google Scholar] [CrossRef]
- Slagsvold, K.H.; Rognmo, O.; Hoydal, M.; Wisloff, U.; Wahba, A. Remote ischemic preconditioning preserves mitochondrial function and influences myocardial MicroRNA expression in atrial myocardium during coronary bypass surgery. Circ. Res. 2014, 114, 851–859. [Google Scholar] [CrossRef]
- McManus, D.D.; Lin, H.; Tanriverdi, K.; Quercio, M.; Yin, X.; Larson, M.G.; Ellinor, P.T.; Levy, D.; Freedman, J.E.; Benjamin, E.J. Relations between circulating microRNAs and atrial fibrillation: Data from the Framingham Offspring Study. Heart Rhythm 2014, 11, 663–669. [Google Scholar] [CrossRef]
- Gurha, P. MicroRNAs in cardiovascular disease. Curr. Opin. Cardiol. 2016, 31, 249–254. [Google Scholar] [CrossRef]
- Zhou, S.-S.; Jin, J.-P.; Wang, J.-Q.; Zhang, Z.-G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084. [Google Scholar] [CrossRef]
- Claudia, B.; Fiedler, J.; Thum, T. Cardiovascular Importance of the MicroRNA−23/27/24 Family. Microcirculation 2012, 19, 208–214. [Google Scholar]
- Van Rooij, E.; Sutherland, L.B.; Liu, N.; Williams, A.H.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA 2006, 103, 18255–18260. [Google Scholar] [CrossRef] [PubMed]
- Grueter, C.E.; Van Rooij, E.; Johnson, B.A.; Deleon, S.M.; Sutherland, L.B.; Qi, X.; Gautron, L.; Elmquist, J.K.; Bassel-Duby, R.; Olson, E.N. A cardiac MicroRNA governs systemic energy homeostasis by regulation of MED13. Cell 2012, 149, 671–683. [Google Scholar] [CrossRef]
- Yang, B.; Lin, H.; Xiao, J.; Lu, Y.; Luo, X.; Li, B.; Zhang, Y.; Xu, C.; Bai, Y.; Wang, H.; et al. The muscle-specific microRNA miR−1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007, 13, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lu, Y.; Yang, B. MicroRNAs and atrial fibrillation: New fundamentals. Cardiovasc. Res. 2011, 89, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, N.W.E.; Kawasaki, M.; Berger, W.R.; Neefs, J.; Meulendijks, E.; Tijsen, A.J.; de Groot, J.R. MicroRNAs in Atrial Fibrillation: From Expression Signatures to Functional Implications. Cardiovasc. Drugs Ther. 2017, 2017, 345–365. [Google Scholar] [CrossRef] [PubMed]
- Gomes Da Silva, A.M.; Silbiger, V.N. MiRNAs as biomarkers of atrial fibrillation. Biomarkers 2014, 19, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Vester, B.; Wengel, J. MicroRNA assay methods: A review of current technologies. Biochemistry 2004, 43, 13233–13241. [Google Scholar] [CrossRef]
- Moody, L.; He, H.; Pan, Y.-X.; Chen, H. Methods and novel technology for microRNA quantification in colorectal cancer screening. Clin. Epigenetics 2017, 9, 119. [Google Scholar] [CrossRef]
- Garcia-Elias, A.; Alloza, L.; Puigdecanet, E.; Nonell, L.; Tajes, M.; Curado, J.; Enjuanes, C.; Díaz, O.; Bruguera, J.; Martí-Almor, J.; et al. Defining quantification methods and optimizing protocols for microarray hybridization of circulating microRNAs. Sci. Rep. 2017, 7, 7725. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Tan, C.; Liu, X. Circular RNAs: A new frontier in the study of human diseases. J. Med. Genet. 2016, 53, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Luo, F.; Liu, Q.; Xu, D. Regulatory non-coding RNAs in acute myocardial infarction. J. Cell. Mol. Med. 2017, 21, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Carnes, C.A.; Janssen, P.M.L.; Ruehr, M.L.; Nakayama, H.; Nakayama, T.; Haase, H.; Bauer, J.A.; Chung, M.K.; Fearon, I.M.; Gillinov, A.M.; et al. Atrial Glutathione Content, Calcium Current, and Contractility. J. Biol. Chem. 2007, 282, 28063–28073. [Google Scholar] [CrossRef] [PubMed]
- Harling, L.; Lambert, J.; Ashrafian, H.; Darzi, A.; Gooderham, N.J.; Athanasiou, T. Elevated serum microRNA 483−5p levels may predict patients at risk of post-operative atrial fibrillation. Eur. J. Cardiothorac Surg. 2017, 51, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, F.; Mirza, M.; Olet, S.; Albrecht, M.; Edwards, S.; Emelyanova, L.; Kress, D.; Ross, G.R.; Holmuhamedov, E.; Tajik, A.J.; et al. Noninvasive biomarker-based risk stratification for development of new onset atrial fibrillation after coronary artery bypass surgery. Int. J. Cardiol. 2020. [Google Scholar] [CrossRef]
- Feldman, A.; Moreira, D.A.R.; Gun, C.; Wang, H.-T.L.; Hirata, M.H.; de Freitas Germano, J.; Leite, G.G.S.; Farsky, P. Analysis of Circulating miR−1, miR−23a, and miR−26a in Atrial Fibrillation Patients Undergoing Coronary Bypass Artery Grafting Surgery. Ann. Hum. Genet. 2017, 81, 99–105. [Google Scholar] [CrossRef]
- Tsoporis, J.N.; Fazio, A.; Rizos, I.K.; Izhar, S.; Proteau, G.; Salpeas, V.; Rigopoulos, A.; Sakadakis, E.; Toumpoulis, I.K.; Parker, T.G. Increased right atrial appendage apoptosis is associated with differential regulation of candidate MicroRNAs 1 and 133A in patients who developed atrial fibrillation after cardiac surgery. J. Mol. Cell. Cardiol. 2018, 121, 25–32. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Y.; Xu, S.; Liu, Y.; Yu, L.; Li, Z.; Xue, X.; Wang, H. Plasma Circular RNAs, Hsa_circRNA_025016, Predict Postoperative Atrial Fibrillation After Isolated Off-Pump Coronary Artery Bypass Grafting. J. Am. Heart Assoc. 2018, 7, e006642. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, S.; Xu, Y.; Liu, Y.; Li, Z.; Zhang, Y.; Jin, Y.; Xue, X.; Wang, H. Relation of Mitochondrial DNA Copy Number in Peripheral Blood to Postoperative Atrial Fibrillation After Isolated Off-Pump Coronary Artery Bypass Grafting. Am. J. Cardiol. 2017, 119, 473–477. [Google Scholar] [CrossRef]
- Sandler, N.; Kaczmarek, E.; Itagaki, K.; Zheng, Y.; Otterbein, L.; Khabbaz, K.; Liu, D.; Senthilnathan, V.; Gruen, R.L.; Hauser, C.J. Mitochondrial DAMPs Are Released During Cardiopulmonary Bypass Surgery and Are Associated With Postoperative Atrial Fibrillation. Heart Lung Circ. 2018, 27, 122–129. [Google Scholar] [CrossRef]
- Kertai, M.D.; Qi, W.; Li, Y.-J.; Lombard, F.W.; Liu, Y.; Smith, M.P.; Stafford-Smith, M.; Newman, M.F.; Milano, C.A.; Mathew, J.P.; et al. Gene signatures of postoperative atrial fibrillation in atrial tissue after coronary artery bypass grafting surgery in patients receiving β-blockers. J. Mol. Cell. Cardiol. 2016, 92, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Kertai, M.D.; Li, Y.-W.; Li, Y.-J.; Shah, S.H.; Kraus, W.E.; Fontes, M.L.; Stafford-Smith, M.; Newman, M.F.; Podgoreanu, M.V.; Mathew, J.P. G Protein-Coupled Receptor Kinase 5 Gene Polymorphisms Are Associated With Postoperative Atrial Fibrillation After Coronary Artery Bypass Grafting in Patients Receiving -Blockers. Circ. Cardiovasc. Genet. 2014, 7, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Kertai, M.D.; Li, Y.J.; Ji, Y.; Qi, W.; Lombard, F.W.; Shah, S.H.; Kraus, W.E.; Stafford-Smith, M.; Newman, M.F.; Milano, C.A.; et al. Genome-wide association study of new-onset atrial fibrillation after coronary artery bypass grafting surgery. Am. Heart J. 2015, 170, 580.e28–590.e28. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Wang, X.W.; Huang, C.; Qiu, F.; Xiang, X.Y.; Lu, Z.Q. High mobility group box 1 gene polymorphism is associated with the risk of postoperative atrial fibrillation after coronary artery bypass surgery. J. Cardiothorac. Surg. 2015, 10, 88. [Google Scholar] [CrossRef][Green Version]
- Liu, L.; Zhang, L.; Liu, M.; Zhang, Y.; Han, X.; Zhang, Z. GRK5 polymorphisms and Postoperative Atrial Fibrillation following Coronary Artery Bypass Graft Surgery. Sci. Rep. 2015, 5, 12768. [Google Scholar] [CrossRef]
- Virani, S.S.; Brautbar, A.; Lee, V.V.; Elayda, M.; Sami, S.; Nambi, V.; Frazier, L.; Wilson, J.M.; Willerson, J.T.; Boerwinkle, E.; et al. Usefulness of single nucleotide polymorphism in chromosome 4q25 to predict in-hospital and long-term development of atrial fibrillation and survival in patients undergoing coronary artery bypass grafting. Am. J. Cardiol. 2011, 107, 1504–1509. [Google Scholar] [CrossRef]
- Gurpreet, S.; Shea, J.; Najam, F.; Solomon, A.J. Can a Genetic Test Predict the Development of Postoperative Atrial Fibrillation. Int. J. Clin. Cardiol. 2015, 2, 32. [Google Scholar]
- Body, S.C.; Collard, C.D.; Shernan, S.K.; Fox, A.A.; Liu, K.Y.; Ritchie, M.D.; Perry, T.E.; Muehlschlegel, J.D.; Aranki, S.; Donahue, B.S.; et al. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ. Cardiovasc. Genet. 2009, 2, 499–506. [Google Scholar] [CrossRef]
- Luo, X.; Yang, B.; Nattel, S. MicroRNAs and atrial fibrillation: Mechanisms and translational potential. Nat. Rev. Cardiol. 2015, 12, 80–90. [Google Scholar] [CrossRef]
- Arora, P.; Wu, C.; Khan, A.M.; Bloch, D.B.; Davis-Dusenbery, B.N.; Ghorbani, A.; Spagnolli, E.; Martinez, A.; Ryan, A.; Tainsh, L.T.; et al. Atrial natriuretic peptide is negatively regulated by microRNA−425. J. Clin. Investig. 2013, 123, 3378–3382. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Miller, N.; Kerin, M.J. MiRNAs as biomarkers and therapeutic targets in cancer. Curr. Opin. Pharmacol. 2010, 10, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Buchan, R.J.; Cook, S.A. MicroRNA−223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc. Res. 2010, 86, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Mayr, M.; Yusuf, S.; Weir, G.; Chung, Y.-L.; Mayr, U.; Yin, X.; Ladroue, C.; Madhu, B.; Roberts, N.; De Souza, A.; et al. Combined Metabolomic and Proteomic Analysis of Human Atrial Fibrillation. J. Am. Coll. Cardiol. 2008, 51, 585–594. [Google Scholar] [CrossRef]
- Sharma, D.; Li, G.; Xu, G.; Liu, Y.; Xu, Y. Atrial remodeling in atrial fibrillation and some related microRNAs. Cardiology 2011, 120, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, Y.; Wang, N.; Pan, Z.; Gao, X.; Zhang, F.; Zhang, Y.; Shan, H.; Luo, X.; Bai, Y.; et al. MicroRNA−328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 2010, 122, 2378–2387. [Google Scholar] [CrossRef] [PubMed]
- Dawson, K.; Wakili, R.; Ördög, B.; Clauss, S.; Chen, Y.; Iwasaki, Y.; Voigt, N.; Qi, X.Y.; Sinner, M.F.; Dobrev, D.; et al. MicroRNA29: A mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 2013, 127, 1466–1475. [Google Scholar] [CrossRef]
- Mariscalco, G.; Musumeci, F.; Banach, M. Factors influencing post-coronary artery bypass grafting atrial fibrillation episodes. Kardiol. Pol. 2013, 71, 1115–1120. [Google Scholar] [CrossRef]
- Santulli, G.; Iaccarino, G.; De Luca, N.; Trimarco, B.; Condorelli, G. Atrial fibrillation and microRNAs. Front. Physiol. 2014, 5, 15. [Google Scholar] [CrossRef]
- Jalife, J.; Kaur, K. Atrial remodeling, fibrosis, and atrial fibrillation. Trends Cardiovasc. Med. 2015, 25, 475–484. [Google Scholar] [CrossRef]
- Krogstad, L.E.B.; Slagsvold, K.H.; Wahba, A. Remote ischemic preconditioning and incidence of postoperative atrial fibrillation. Scand. Cardiovasc. J. 2015, 49, 117–122. [Google Scholar] [CrossRef]
- Lutter, D.; Marr, C.; Krumsiek, J.; Lang, E.W.; Theis, F.J. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genom. 2010, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Ma, N.; Wang, X.; Hui, Y.; Li, F.; Xiang, Y.; Zhou, J.; Zou, C.; Jin, J.; Lv, G.; et al. MiR−483−5p controls angiogenesis in vitro and targets serum response factor. FEBS Lett. 2011, 585, 3095–3100. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR−29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef]
- Di, Y.; Zhang, D.; Hu, T.; Li, D. miR−23 regulate the pathogenesis of patients with coronary artery disease. Int. J. Clin. Exp. Med. 2015, 8, 11759–11769. [Google Scholar] [PubMed]
- Dong, J.; Bao, J.; Feng, R.; Zhao, Z.; Lu, Q.; Wang, G.; Li, H.; Su, D.; Zhou, J.; Jing, Q.; et al. Circulating microRNAs: A novel potential biomarker for diagnosing acute aortic dissection. Sci. Rep. 2017, 7, 12784. [Google Scholar] [CrossRef] [PubMed]
- Jansen, F.; Schäfer, L.; Wang, H.; Schmitz, T.; Flender, A.; Schueler, R.; Hammerstingl, C.; Nickenig, G.; Sinning, J.-M.; Werner, N. Kinetics of Circulating MicroRNAs in Response to Cardiac Stress in Patients With Coronary Artery Disease. J. Am. Heart Assoc. 2017, 6, e005270. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Pan, Z.; Shan, H.; Xiao, J.; Sun, X.; Wang, N.; Lin, H.; Xiao, L.; Maguy, A.; Qi, X.-Y.; et al. MicroRNA−26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J. Clin. Investig. 2013, 123, 1939–1951. [Google Scholar] [CrossRef]
- Kukreja, R.C.; Yin, C.; Salloum, F.N. MicroRNAs: New players in cardiac injury and protection. Mol. Pharmacol. 2011, 80, 558–564. [Google Scholar] [CrossRef]
- Kilic, U.; Gok, O.; Bacaksiz, A.; Izmirli, M.; Elibol-Can, B.; Uysal, O. SIRT1 gene polymorphisms affect the protein expression in cardiovascular diseases. PLoS ONE 2014, 9, e90428. [Google Scholar] [CrossRef]
- Choi, S.E.; Kemper, J.K. Regulation of SIRT1 by microRNAs. Mol. Cells 2013, 36, 385–392. [Google Scholar] [CrossRef]
- Yamac, A.H.; Huyut, M.A.; Yilmaz, E.; Celikkale, I.; Bacaksiz, A.; Demir, Y.; Demir, A.R.; Erturk, M.; Bakhshaliyev, N.; Ozdemir, R.; et al. MicroRNA 199a is downregulated in patients after coronary artery bypass graft surgery and is associated with increased levels of sirtuin 1 (SIRT 1) protein and major adverse cardiovascular events at 3-year follow-up. Med. Sci. Monit. 2018, 24, 6245–6254. [Google Scholar] [CrossRef]
- Rane, S.; He, M.; Sayed, D.; Vashistha, H.; Malhotra, A.; Sadoshima, J.; Vatner, D.E.; Vatner, S.F.; Abdellatif, M. Downregulation of MiR−199a derepresses hypoxia-inducible factor−1α and sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 2009, 104, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.L.; Bu, P.L.; Liu, J.N.; Wang, X.; Wu, X.N.; Zhao, L.X. Expression of SIRT1 in right auricle tissues and the relationship with oxidative stress in patients with atrial fibrillation. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi Chin. J. Cell. Mol. Immunol. 2012, 28, 972–974. [Google Scholar]
- Vegter, E.L.; Ovchinnikova, E.S.; van Veldhuisen, D.J.; Jaarsma, T.; Berezikov, E.; van der Meer, P.; Voors, A.A. Low circulating microRNA levels in heart failure patients are associated with atherosclerotic disease and cardiovascular-related rehospitalizations. Clin. Res. Cardiol. 2017, 106, 598–609. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Clauss, S.; Sinner, M.F.; Kääb, S.; Wakili, R. The role of MicroRNAs in antiarrhythmic therapy for atrial fibrillation. Arrhythmia Electrophysiol. Rev. 2015, 4, 146–155. [Google Scholar] [CrossRef]
- Girmatsion, Z.; Biliczki, P.; Bonauer, A.; Wimmer-Greinecker, G.; Scherer, M.; Moritz, A.; Bukowska, A.; Goette, A.; Nattel, S.; Hohnloser, S.H.; et al. Changes in microRNA−1 expression and IK1 up-regulation in human atrial fibrillation. Heart Rhythm 2009, 6, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Ide, T.; Tsutsui, H.; Hayashidani, S.; Kang, D.; Suematsu, N.; Nakamura, K.; Utsumi, H.; Hamasaki, N.; Takeshita, A. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ. Res. 2001, 88, 529–535. [Google Scholar] [CrossRef]
- Shokolenko, I.; Venediktova, N.; Bochkareva, A.; Wilson, G.L.; Alexeyev, M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009, 37, 2539–2548. [Google Scholar] [CrossRef]
- Phan, K.; Khuong, J.N.; Xu, J.; Kanagaratnam, A.; Yan, T.D. Obesity and postoperative atrial fibrillation in patients undergoing cardiac surgery: Systematic review and meta-analysis. Int. J. Cardiol. 2016, 217, 49–57. [Google Scholar] [CrossRef]
- Turagam, M.K.; Mirza, M.; Werner, P.H.; Sra, J.; Kress, D.C.; Tajik, A.J.; Jahangir, A. Circulating biomarkers predictive of postoperative atrial fibrillation. Cardiol. Rev. 2016, 24, 76–87. [Google Scholar] [CrossRef]
- Perrier, S.; Meyer, N.; Hoang Minh, T.; Announe, T.; Bentz, J.; Billaud, P.; Mommerot, A.; Mazzucotelli, J.-P.; Kindo, M. Predictors of Atrial Fibrillation After Coronary Artery Bypass Grafting: A Bayesian Analysis. Ann. Thorac. Surg. 2017, 103, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Philipp, M.; Berger, I.M.; Just, S.; Caron, M.G. Overlapping and opposing functions of G protein-coupled receptor kinase 2 (GRK2) and Grk5 during heart development. J. Biol. Chem. 2014, 289, 26119–26130. [Google Scholar] [CrossRef] [PubMed]
- Liggett, S.B.; Cresci, S.; Kelly, R.J.; Syed, F.M.; Matkovich, S.J.; Hahn, H.S.; Diwan, A.; Martini, J.S.; Sparks, L.; Parekh, R.R.; et al. A GRK5 polymorphism that inhibits β-adrenergic receptor signaling is protective in heart failure. Nat. Med. 2008, 14, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Matkovich, S.J.; Duan, X.; Gold, J.I.; Koch, W.J.; Dorn, G.W. Nuclear effects of G-protein receptor kinase 5 on histone deacetylase 5-regulated gene transcription in heart failure. Circ. Heart Fail. 2011, 4, 659–668. [Google Scholar] [CrossRef]
- Gold, J.I.; Martini, J.S.; Hullmann, J.; Gao, E.; Chuprun, J.K.; Lee, L.; Tilley, D.G.; Rabinowitz, J.E.; Bossuyt, J.; Bers, D.M.; et al. Nuclear Translocation of Cardiac G Protein-Coupled Receptor Kinase 5 Downstream of Select Gq-Activating Hypertrophic Ligands Is a Calmodulin-Dependent Process. PLoS ONE 2013, 8, e57324. [Google Scholar] [CrossRef]
- Crystal, E.; Garfinkle, M.S.; Connolly, S.; Ginger, T.; Sleik, K.; Yusuf, S. Interventions for preventing post-operative atrial fibrillation in patients undergoing heart surgery. In Cochrane Database of Systematic Reviews; Crystal, E., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2004; p. CD003611. [Google Scholar]
- Kääb, S.; Darbar, D.; Van Noord, C.; Dupuis, J.; Pfeufer, A.; Newton-Cheh, C.; Schnabel, R.; Makino, S.; Sinner, M.F.; Kannankeril, P.J.; et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur. Heart J. 2009, 30, 813–819. [Google Scholar] [CrossRef]
- Anselmi, C.V.; Novelli, V.; Roncarati, R.; Malovini, A.; Bellazzi, R.; Bronzini, R.; Marchese, G.; Condorelli, G.; Montenero, A.S.; Puca, A.A. Association of rs2200733 at 4q25 with atrial flutter/fibrillation diseases in an Italian population. Heart 2008, 94, 1394–1396. [Google Scholar] [CrossRef]
- Chen, F.; Yang, Y.; Zhang, R.; Zhang, S.; Dong, Y.; Yin, X.; Chang, D.; Yang, Z.; Wang, K.; Gao, L.; et al. Polymorphism rs2200733 at chromosome 4q25 is associated with atrial fibrillation recurrence after radiofrequency catheter ablation in the Chinese Han population. Am. J. Transl. Res. 2016, 8, 688–697. [Google Scholar]
- Lubitz, S.A.; Sinner, M.F.; Lunetta, K.L.; Makino, S.; Pfeufer, A.; Rahman, R.; Veltman, C.E.; Barnard, J.; Bis, J.C.; Danik, S.P.; et al. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation 2010, 122, 976–984. [Google Scholar] [CrossRef]
- Oral, H. Post-Operative Atrial Fibrillation and Oxidative Stress. J. Am. Coll. Cardiol. 2008, 51, 75–76. [Google Scholar] [CrossRef]
- Hsu, J.; Hanna, P.; Van Wagoner, D.R.; Barnard, J.; Serre, D.; Chung, M.K.; Smith, J.D. Whole Genome Expression Differences in Human Left and Right Atria Ascertained by RNA Sequencing. Circ. Cardiovasc. Genet. 2012, 5, 327–335. [Google Scholar] [CrossRef] [PubMed]
| RNAs and Gene Expression | Protein/Gene/Loci | CABG | Source | All Patients | Study Type | With POAF | p Value | AUC | Sensitivity | Specificity | Technique | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA−483−5p | On-pump | Blood | 34 | Prospective | 12 (35.3%) | 0.046 | 0.78 | 77.78 | 77.27 | qPCR | [34] | |
| miRNA−26a | On-pump | Serum | 48 | Prospective | 24 (50.0%) | 0.010 | 0.66 | - | - | qPCR | [36] | |
| miRNA−23a | On-pump | Serum | 48 | Prospective | 24 (50.0%) | 0.020 | 0.63 | - | - | qPCR | [36] | |
| miRNA−199a | SIRT1 | On-pump | RAA | 63 | Prospective | 20 (31.7%) | 0.022 | - | - | - | qPCR | [16] |
| miRNA−1 and miRNA−133a | On/Off-pump | RAA | 42 | Prospective | 14 (33.3%) | <0.05 | - | - | - | qPCR | [37] | |
| circRNA−025016 | Off-pump | Plasma | 284 | Prospective | 68 (23.9%) | <0.01 | - | 73.52 | 77.83 | qPCR | [38] | |
| mtDNA | Off-pump | Blood | 485 | Prospective | 101 (20.8%) | <0.01 | 0.81 | 70.3 | 80.2 | qPCR | [39] | |
| mtDNA | On-pump | Plasma | 16 | Prospective | 6 (37.5%) | <0.01 | - | - | - | qPCR | [40] | |
| SNP (VOPP1) | Off-pump | RAA | 45 | Prospective | 13 (28.9%) | <0.01 | - | - | - | eQTL | [41] | |
| SNP (rs3740563) | On-pump | Blood | 245 | Prospective | 42 (17.1%) | 0.011 | - | - | - | OMNI1-Quad BeadChip | [42] | |
| SNP (rs2249825) | HMGB1 | On-pump | Blood | 128 | Prospective | 37 (29.9%) | <0.001 | - | - | - | qPCR | [44] |
| SNP (rs4572292 and rs11198893) | GRK5 | On/Off-pump | Blood | 1348 | Reterospective | 405 (30.0%) | <0.01 | - | - | - | qPCR | [45] |
| SNP (rs2200733 and rs10033464) # | 4q25 | On/Off-pump | Buccal swabs | 143 | Prospective | 23 (16.1%) | NS | - | 16 | 71 | deCODE | [47] |
| SNP (rs2200733 and rs10033464) | 4q25 | On/Off-pump | Blood | 1166 | Reterospective | 425 (36.4%) | 0.048 | - | - | - | qPCR | [46] |
| SNP (rs2200733 and rs13143308) | 4q25 | On-pump | Blood | 959 * 494 ** | Prospective | 289 (30.1%) * 151 (30.6%) ** | <0.01 | 0.72 | - | - | deCODE | [48] |
| SNP (rs10504554) | LY96 | On-pump | Blood | 877 * 304 ** | Prospective | 257 (29.3%) 84 (27.6%) | <0.01 | - | - | - | OMNI1-Quad BeadChip | [43] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.S.; Yamashita, K.; Sharma, V.; Ranjan, R.; Dosdall, D.J. RNAs and Gene Expression Predicting Postoperative Atrial Fibrillation in Cardiac Surgery Patients Undergoing Coronary Artery Bypass Grafting. J. Clin. Med. 2020, 9, 1139. https://doi.org/10.3390/jcm9041139
Khan MS, Yamashita K, Sharma V, Ranjan R, Dosdall DJ. RNAs and Gene Expression Predicting Postoperative Atrial Fibrillation in Cardiac Surgery Patients Undergoing Coronary Artery Bypass Grafting. Journal of Clinical Medicine. 2020; 9(4):1139. https://doi.org/10.3390/jcm9041139
Chicago/Turabian StyleKhan, Muhammad Shuja, Kennosuke Yamashita, Vikas Sharma, Ravi Ranjan, and Derek James Dosdall. 2020. "RNAs and Gene Expression Predicting Postoperative Atrial Fibrillation in Cardiac Surgery Patients Undergoing Coronary Artery Bypass Grafting" Journal of Clinical Medicine 9, no. 4: 1139. https://doi.org/10.3390/jcm9041139
APA StyleKhan, M. S., Yamashita, K., Sharma, V., Ranjan, R., & Dosdall, D. J. (2020). RNAs and Gene Expression Predicting Postoperative Atrial Fibrillation in Cardiac Surgery Patients Undergoing Coronary Artery Bypass Grafting. Journal of Clinical Medicine, 9(4), 1139. https://doi.org/10.3390/jcm9041139





