Whole-Genome DNA Methylation Profiling of CD14+ Monocytes Reveals Disease Status and Activity Differences in Crohn’s Disease Patients
Abstract
1. Introduction
2. Experimental Section
2.1. CD14 Cells Isolation
2.2. DNA Isolation and Methylation Analysis
2.3. Monocyte Gene Expression Data
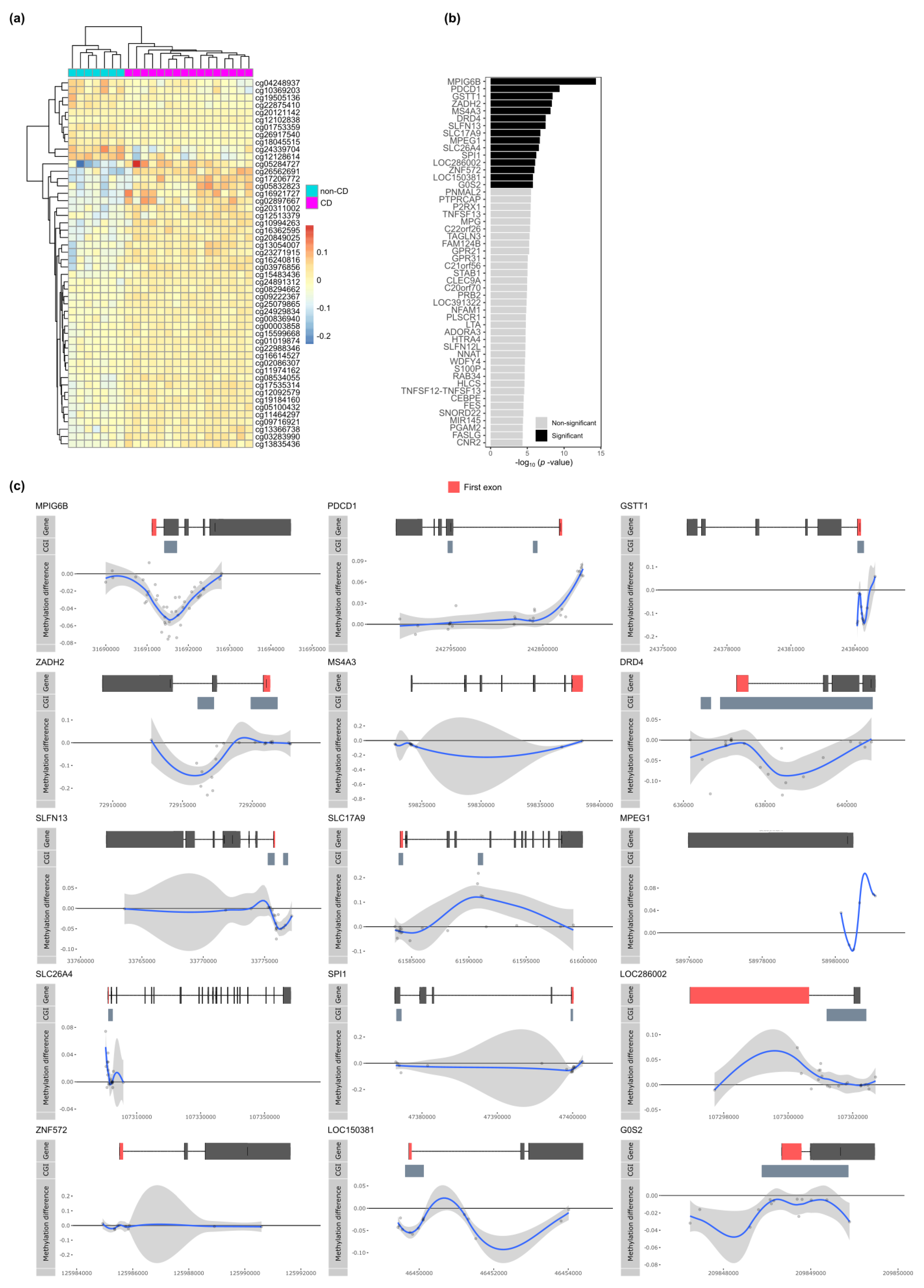
3. Results
3.1. CD-Associated Differential Methylation
3.2. CD-Associated Differential Methylation
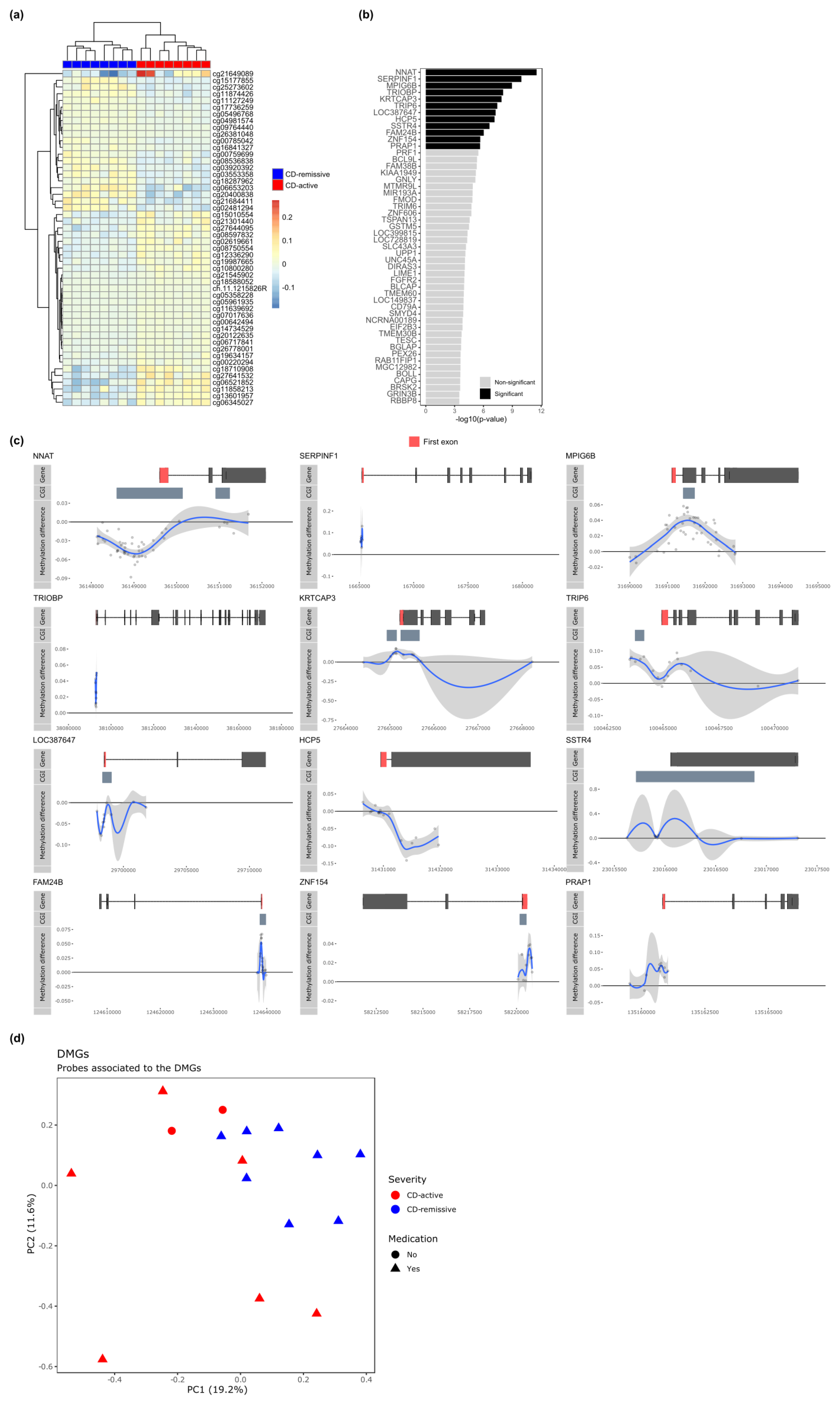
3.3. Differential Methylation Associated with Disease Activity in CD Monocytes
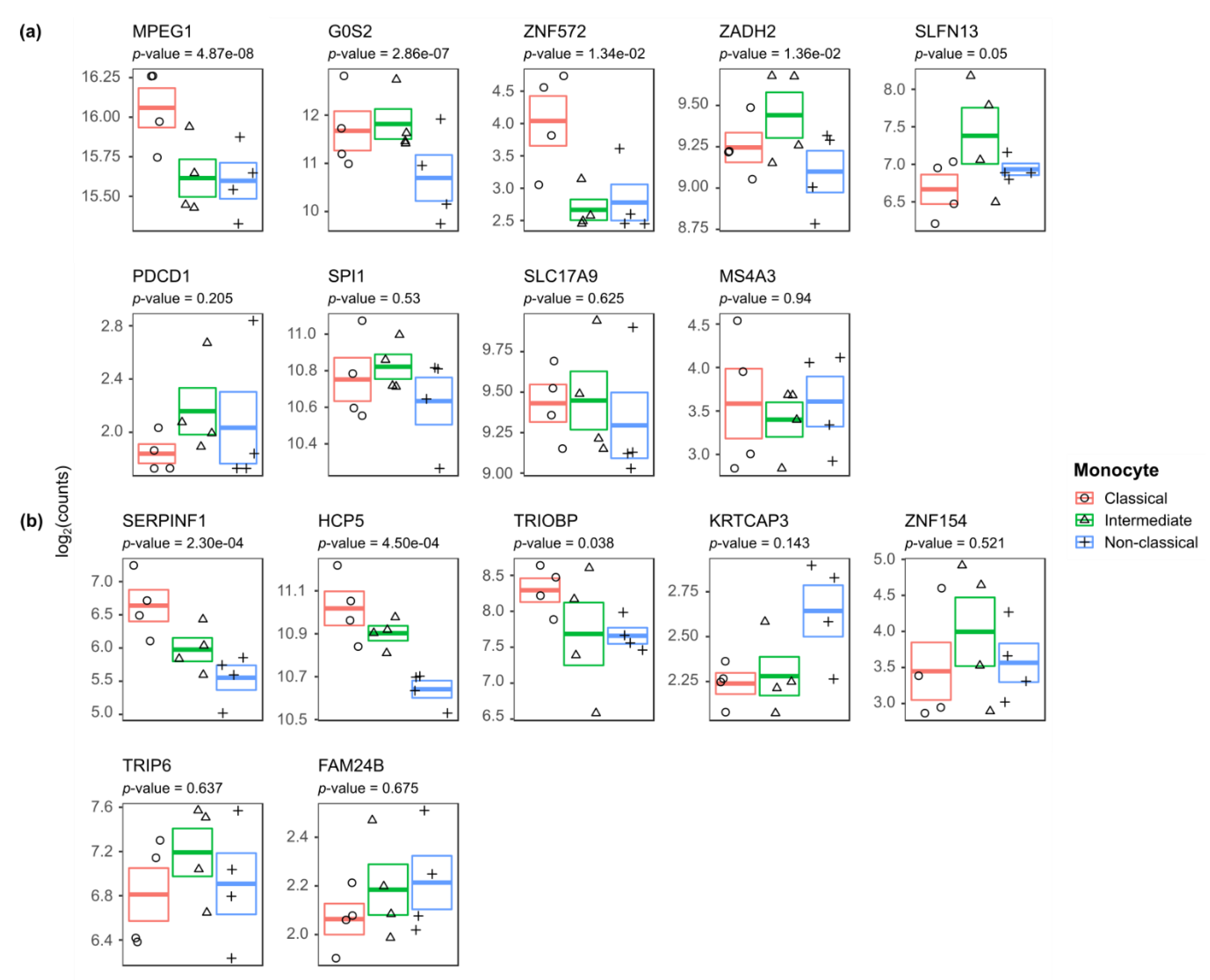
3.4. Differences in Methylation may be Associated with Disease Dynamics in Monocyte Populations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- De Souza, H.S.P.; Fiocchi, C. Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Arseneau, K.O.; Cominelli, F. Targeting leukocyte trafficking for the treatment of inflammatory bowel disease. Clin. Pharmacol. Ther. 2015, 97, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Gordon, H.; Trier Moller, F.; Andersen, V.; Harbord, M. Heritability in Inflammatory Bowel Disease: From the First Twin Study to Genome-Wide Association Studies. Inflamm. Bowel Dis. 2015, 21, 1428–1434. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Anderson, C.A. Genetic studies of Crohn’s disease: Past, present and future. Best Pract. Res. Clin. Gastroenterol. 2014, 28, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Mateos, B.; Palanca-Ballester, C.; Saez-Gonzalez, E.; Moret, I.; Lopez, A.; Sandoval, J. Epigenetics of Inflammatory Bowel Disease: Unraveling Pathogenic Events. Crohn’s Colitis 360 2019, 1. [Google Scholar] [CrossRef]
- Fogel, O.; Richard-Miceli, C.; Tost, J. Epigenetic Changes in Chronic Inflammatory Diseases. In Advances in Protein Chemistry and Structural Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2017; Volume Chromatin Remodelling and Immunity; pp. 139–189. ISBN 9780128123928. [Google Scholar]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Bird, A.P. CpG-Rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. [Google Scholar] [CrossRef]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Razin, A.; Cedar, H. DNA methylation and gene expression. Microbiol. Rev. 1991, 55, 451–458. [Google Scholar] [CrossRef]
- Liu, X.S.; Wu, H.; Ji, X.; Stelzer, Y.; Wu, X.; Czauderna, S.; Shu, J.; Dadon, D.; Young, R.A.; Jaenisch, R. Editing DNA Methylation in the Mammalian Genome. Cell 2016, 167, 233.e17–247.e17. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Zhang, X.; Su, J.; Jeong, M.; Gundry, M.C.; Huang, Y.-H.; Zhou, Y.; Li, W.; Goodell, M.A. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat. Commun. 2017, 8, 16026. [Google Scholar] [CrossRef] [PubMed]
- Li Yim, A.Y.F.; Duijvis, N.W.; Zhao, J.; de Jonge, W.J.; D′Haens, G.R.A.M.; Mannens, M.M.A.M.; Mul, A.N.P.M.; te Velde, A.A.; Henneman, P. Peripheral blood methylation profiling of female Crohn’s disease patients. Clin. Epigenetics 2016, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Nimmo, E.R.; Prendergast, J.G.; Aldhous, M.C.; Kennedy, N.A.; Henderson, P.; Drummond, H.E.; Ramsahoye, B.H.; Wilson, D.C.; Semple, C.A.; Satsangi, J. Genome-Wide methylation profiling in Crohn’s disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm. Bowel Dis. 2012, 18, 889–899. [Google Scholar] [CrossRef]
- McDermott, E.; Ryan, E.J.; Tosetto, M.; Gibson, D.; Burrage, J.; Keegan, D.; Byrne, K.; Crowe, E.; Sexton, G.; Malone, K.; et al. DNA Methylation Profiling in Inflammatory Bowel Disease Provides New Insights into Disease Pathogenesis. J. Crohn’s Colitis 2016, 10, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Hume, D.A. The mononuclear phagocyte system. Curr. Opin. Immunol. 2006, 18, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Kitchens, R.L. Role of CD14 in Cellular Recognition of Bacterial Lipopolysaccharides. In CD14 in the Inflammatory Response; KARGER: Basel, Switzerland, 1999; pp. 61–82. [Google Scholar]
- Ziegler-Heitbrock, L.; Hofer, T.P.J. Toward a refined definition of monocyte subsets. Front. Immunol. 2013, 4, 1–5. [Google Scholar] [CrossRef]
- Ziegler-Heitbrock, L.; Ancuta, P.; Crowe, S.; Dalod, M.; Grau, V.; Hart, D.N.; Leenen, P.J.M.; Liu, Y.J.; MacPherson, G.; Randolph, G.J.; et al. Nomenclature of monocytes and dendritic cells in blood. Blood 2010, 116. [Google Scholar] [CrossRef]
- Passlick, B.; Flieger, D.; Ziegler-Heitbrock, H. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 1989, 74, 2527–2534. [Google Scholar] [CrossRef]
- Gren, S.T.; Rasmussen, T.B.; Janciauskiene, S.; Håkansson, K.; Gerwien, J.G.; Grip, O. A Single-Cell Gene-Expression Profile Reveals Inter-Cellular Heterogeneity within Human Monocyte Subsets. PLoS ONE 2015, 10, e0144351. [Google Scholar] [CrossRef]
- Loon Wong, K.; Jing-Yi Tai, J.; Wong, W.-C.; Han, H.; Sem, X.; Yeap, W.-H.; Kourilsky, P.; Wong, S.-C. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets New official nomenclature subdivides human monocytes into 3 subsets: The classical (CD14 CD16 ), intermediate (CD14 CD16 ), and nonclassical (CD14 CD16 ) monocytes. Blood J. Am. Soc. Hematol. 2011. [Google Scholar] [CrossRef]
- Kapellos, T.S.; Bonaguro, L.; Gemünd, I.; Reusch, N.; Saglam, A.; Hinkley, E.R.; Schultze, J.L. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, E.C.; Plevy, S.E. The role of macrophages and dendritic cells in the initiation of inflammation in IBD. Inflamm. Bowel Dis. 2014. [CrossRef] [PubMed]
- Na, Y.R.; Stakenborg, M.; Seok, S.H.; Matteoli, G. Macrophages in intestinal inflammation and resolution: A potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 531–543. [Google Scholar] [CrossRef]
- Kühl, A.A.; Erben, U.; Kredel, L.I.; Siegmund, B.; Kuhl, A.A.; Erben, U.; Kredel, L.I.; Siegmund, B. Diversity of intestinal macrophages in inflammatory bowel diseases. Front. Immunol. 2015, 6, 1–7. [Google Scholar] [CrossRef]
- Thiesen, S.; Janciauskiene, S.; Uronen-Hansson, H.; Agace, W.; Hogerkorp, C.-M.; Spee, P.; Hakansson, K.; Grip, O. CD14hiHLA-DRdim macrophages, with a resemblance to classical blood monocytes, dominate inflamed mucosa in Crohn’s disease. J. Leukoc. Biol. 2014, 95, 531–541. [Google Scholar] [CrossRef]
- Gren, S.T.; Grip, O. Role of Monocytes and Intestinal Macrophages in Crohn’s Disease and Ulcerative Colitis. Inflamm. Bowel Dis. 2016, 22, 1. [Google Scholar] [CrossRef]
- Martin, J.C.; Boschetti, G.; Chang, C.; Ungaro, R.; Giri, M.; Chuang, L.-S.; Nayar, S.; Greenstein, A.; Dubinsky, M.; Walker, L.; et al. Single-Cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti TNF therapy. bioRxiv 2018, 357, 503102. [Google Scholar] [CrossRef]
- Koch, S.; Kucharzik, T.; Heidemann, J.; Nusrat, A.; Luegering, A. Investigating the role of proinflammatory CD16+ monocytes in the pathogenesis of inflammatory bowel disease. Clin. Exp. Immunol. 2010, 161, 332–341. [Google Scholar] [CrossRef]
- Gettler, K.; Giri, M.; Kenigsberg, E.; Martin, J.; Chuang, L.S.; Hsu, N.Y.; Denson, L.A.; Hyams, J.S.; Griffiths, A.; Noe, J.D.; et al. Prioritizing Crohn’s disease genes by integrating association signals with gene expression implicates monocyte subsets. Genes Immun. 2019, 20, 577–588. [Google Scholar] [CrossRef]
- Andus, T.; Gross, V.; Casar, I.; Krumm, D.; Hosp, J.; David, M.; Scholmerich, J. Activation of monocytes during inflammatory bowel disease. In Proceedings of the Pathobiology; Karger: Basel, Switzerland, 1991; Volume 59, pp. 166–170. [Google Scholar]
- Schwarzmaier, D.; Foell, D.; Weinhage, T.; Varga, G.; Däbritz, J. Peripheral Monocyte Functions and Activation in Patients with Quiescent Crohn’s Disease. PLoS ONE 2013, 8, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, F.; Jung, S.; Littman, D.R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 2003, 19, 71–82. [Google Scholar] [CrossRef]
- Bain, C.C.; Schridde, A. Origin, Differentiation, and Function of Intestinal Macrophages. Front. Immunol. 2018, 9, 2733. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Hisamatsu, T.; Okamoto, S.; Chinen, H.; Kobayashi, T.; Sato, T.; Sakuraba, A.; Kitazume, M.T.; Sugita, A.; Koganei, K.; et al. Unique CD14+intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-γ axis. J. Clin. Investig. 2008, 118, 2269–2280. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; ISBN 3-900051-07-0. [Google Scholar]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Van Iterson, M.; Tobi, E.W.; Slieker, R.C.; den Hollander, W.; Luijk, R.; Slagboom, P.E.; Heijmans, B.T. MethylAid: Visual and interactive quality control of large Illumina 450k datasets. Bioinformatics 2014, 30, 3435–3437. [Google Scholar] [CrossRef]
- Fortin, J.P.; Fertig, E.; Hansen, K. shinyMethyl: Interactive quality control of Illumina 450k DNA methylation arrays in R. F1000Research 2014, 3. [Google Scholar] [CrossRef]
- Du, P.; Zhang, X.; Huang, C.-C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-Value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-Sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Peters, T.J.; Buckley, M.J.; Statham, A.L.; Pidsley, R.; Samaras, K.; Lord, R.V.; Clark, S.J.; Molloy, P.L. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 2015, 8, 6. [Google Scholar] [CrossRef]
- Brown, M.B. A Method for Combining Non-Independent, One-Sided Tests of Significance. Biometrics 1975, 31, 987. [Google Scholar] [CrossRef]
- Poole, W.; Gibbs, D.L. EmpiricalBrownsMethod: Uses Brown’s Method to Combine p-Values from Dependent Tests. 2015. Available online: http://bioconductor.org/packages/release/bioc/html/EmpiricalBrownsMethod.html (accessed on 25 March 2020).
- Koziol, J.A.; Perlman, M.D. Combining independent chi-squared tests. J. Am. Stat. Assoc. 1978, 73, 753–763. [Google Scholar] [CrossRef]
- Zhang, W.; Spector, T.D.; Deloukas, P.; Bell, J.T.; Engelhardt, B.E. Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements. Genome Biol. 2015, 16, 14. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer New York: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- Yin, T.; Cook, D.; Lawrence, M. ggbio: An R package for extending the grammar of graphics for genomic data. Genome Biol. 2012, 13, R77. [Google Scholar] [CrossRef]
- Carlson, M. TxDb.Hsapiens.UCSC.hg19.knownGene: Annotation Package for TxDb Object(s). 2015. Available online: http://bioconductor.riken.jp/packages/3.3/data/annotation/manuals/TxDb.Hsapiens.UCSC.hg38.knownGene/man/TxDb.Hsapiens.UCSC.hg38.knownGene.pdf (accessed on 25 March 2020).
- Morgan, M. AnnotationHub: Client to Access AnnotationHub Resources. 2019. Available online: http://bioconductor.jp/packages/devel/bioc/manuals/AnnotationHub/man/AnnotationHub.pdf (accessed on 25 March 2020).
- Gardiner-Garden, M.; Frommer, M. CpG Islands in vertebrate genomes. J. Mol. Biol. 1987. [Google Scholar] [CrossRef]
- Edgar, R. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Monaco, G.; Lee, B.; Xu, W.; Mustafah, S.; Hwang, Y.Y.; Carré, C.; Burdin, N.; Visan, L.; Ceccarelli, M.; Poidinger, M.; et al. RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Rep. 2019, 26, 1627.e7–1640.e7. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database, C. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-And-Vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Anders, S.; Huber, W. Differential analysis of count data—The DESeq2 package. Genome Biol. 2014, 15, 12. [Google Scholar]
- Walsh, A.J.; Bryant, R.V.; Travis, S.P.L. Current best practice for disease activity assessment in IBD. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Li Yim, A.Y.F.; de Bruyn, J.R.; Duijvis, N.W.; Sharp, C.; Ferrero, E.; de Jonge, W.J.; Wildenberg, M.E.; Mannens, M.M.A.M.; Buskens, C.J.; D’Haens, G.R.; et al. A distinct epigenetic profile distinguishes stenotic from non-inflamed fibroblasts in the ileal mucosa of Crohn’s disease patients. PLoS ONE 2018. [Google Scholar] [CrossRef] [PubMed]
- Taman, H.; Fenton, C.G.; Hensel, I.V.; Anderssen, E.; Florholmen, J.; Paulssen, R.H. Genome-Wide DNA Methylation in Treatment-naïve Ulcerative Colitis. J. Crohn’s Colitis 2018, 12, 1338–1347. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Song, Z.; Lv, Y.; Huang, X.; Mao, B. Glutathione S-Transferase T1 Null Genotype is Associated with Susceptibility to Inflammatory Bowel Disease. Cell. Physiol. Biochem. 2017, 41, 2545–2552. [Google Scholar] [CrossRef]
- Hamm, C.M.; Reimers, M.A.; McCullough, C.K.; Gorbe, E.B.; Lu, J.; Gu, C.C.; Li, E.; Dieckgraefe, B.K.; Gong, Q.; Stappenbeck, T.S.; et al. NOD2 status and human ileal gene expression. Inflamm. Bowel Dis. 2010, 16, 1649–1657. [Google Scholar] [CrossRef][Green Version]
- Kim, H.; Zhao, Q.; Zheng, H.; Li, X.; Zhang, T.; Ma, X. A novel crosstalk between TLR4- and NOD2-Mediated signaling in the regulation of intestinal inflammation. Sci. Rep. 2015, 5, 1–17. [Google Scholar] [CrossRef]
- Arijs, I.; Quintens, R.; Lommel, L.; Van Steen, K.; De Hertogh, G.; Lemaire, K.; Schraenen, A.; Perrier, C.; Van Assche, G.; Vermeire, S.; et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm. Bowel Dis. 2010. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-Wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef]
- Mandelboum, S.; Manber, Z.; Elroy-Stein, O.; Elkon, R. Recurrent functional misinterpretation of RNA-seq data caused by sample-specific gene length bias. PLOS Biol. 2019, 17, e3000481. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, Y.; Lou, J.; Li, J.; Bo, L.; Zhu, K.; Wan, X.; Deng, X.; Cai, Z. PD-L1 blockade improves survival in experimental sepsis by inhibiting lymphocyte apoptosis and reversing monocyte dysfunction. Crit. Care 2010, 14, R220. [Google Scholar] [CrossRef]
- Scott, E.W.; Simon, M.C.; Anastasi, J.; Singh, H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science (80-.) 1994, 265, 1573–1577. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Nishiyama, C.; Nakano, N.; Nishiyama, M.; Usui, Y.; Takeda, K.; Kanada, S.; Fukuyama, K.; Akiba, H.; Tokura, T.; et al. Roles of PU.1 in monocyte- and mast cell-specific gene regulation: PU.1 transactivates CIITA pIV in cooperation with IFN-gamma. Int. Immunol. 2009, 21, 803–816. [Google Scholar] [CrossRef][Green Version]
- Albert, S.; Blons, H.; Jonard, L.; Feldmann, D.; Chauvin, P.; Loundon, N.; Sergent-Allaoui, A.; Houang, M.; Joannard, A.; Schmerber, S.; et al. SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. Eur. J. Hum. Genet. 2006, 14, 773–779. [Google Scholar] [CrossRef] [PubMed]
- McCormack, R.M.; de Armas, L.R.; Shiratsuchi, M.; Fiorentino, D.G.; Olsson, M.L.; Lichtenheld, M.G.; Morales, A.; Lyapichev, K.; Gonzalez, L.E.; Strbo, N.; et al. Perforin-2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Fields, K.A.; McCormack, R.; de Armas, L.R.; Podack, E.R. Perforin-2 restricts growth of chlamydia trachomatis in macrophages. Infect. Immun. 2013, 81, 3045–3054. [Google Scholar] [CrossRef]
- Newland, S.A.; Macaulay, I.C.; Floto, R.A.; De Vet, E.C.; Ouwehand, W.H.; Watkins, N.A.; Lyons, P.A.; Campbell, R.D. The novel inhibitory receptor G6B is expressed on the surface of platelets and attenuates platelet function in vitro. Blood 2007, 109, 4806–4809. [Google Scholar] [CrossRef]
- Harries, A.D.; Fitzsimons, E.; Fifield, R.; Dew, M.J.; Rhoades, J. Platelet count: A simple measure of activity in Crohn’s disease. Br. Med. J. 1983, 286, 1476. [Google Scholar] [CrossRef]
- Yan, S.L.S.; Russell, J.; Harris, N.R.; Senchenkova, E.Y.; Yildirim, A.; Granger, D.N. Platelet abnormalities during colonic inflammation. Inflamm. Bowel Dis. 2013, 19, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Bin, L.H.; Li, F.; Liu, Y.; Chen, D.; Zhai, Z.; Shu, H.B. TRIP6 is a RIP2-associated common signaling component of multiple NF-κB activation pathways. J. Cell Sci. 2005, 118, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Kinio, A.; Saleh, M. Functions of NOD-Like Receptors in Human Diseases. Front. Immunol. 2013, 4, 333. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; Yeretssian, G. NOD-Like receptors: Master regulators of inflammation and cancer. Front. Immunol. 2014, 5, 327. [Google Scholar] [CrossRef]
- Van Op den bosch, J.; Torfs, P.; De Winter, B.Y.; De Man, J.G.; Pelckmans, P.A.; Van Marck, E.; Grundy, D.; Van Nassauw, L.; Timmermans, J.P. Effect of genetic SSTR4 ablation on inflammatory peptide and receptor expression in the non-inflamed and inflamed murine intestine. J. Cell. Mol. Med. 2009, 13, 3283–3295. [Google Scholar] [CrossRef]
- Sawada, K.; Echigo, N.; Juge, N.; Miyaji, T.; Otsuka, M.; Omote, H.; Yamamoto, A.; Moriyama, Y. Identification of a vesicular nucleotide transporter. Proc. Natl. Acad. Sci. USA 2008, 105, 5683–5686. [Google Scholar] [CrossRef]
- Sakaki, H.; Tsukimoto, M.; Harada, H.; Moriyama, Y.; Kojima, S. Autocrine Regulation of Macrophage Activation via Exocytosis of ATP and Activation of P2Y11 Receptor. PLoS ONE 2013, 8, e59778. [Google Scholar] [CrossRef]
- Mello, F.V.; Alves, L.R.; Land, M.G.P.; Teodósio, C.; Sanchez, M.-L.; Bárcena, P.; Peres, R.T.; Pedreira, C.E.; Costa, E.S.; Orfao, A. Maturation-associated gene expression profiles along normal human bone marrow monopoiesis. Br. J. Haematol. 2017, 176, 464–474. [Google Scholar] [CrossRef]
- Zawada, A.M.; Schneider, J.S.; Michel, A.I.; Rogacev, K.S.; Hummel, B.; Krezdorn, N.; Müller, S.; Rotter, B.; Winter, P.; Obeid, R.; et al. DNA methylation profiling reveals differences in the 3 human monocyte subsets and identifies uremia to induce DNA methylation changes during differentiation. Epigenetics 2016, 11, 259–272. [Google Scholar] [CrossRef][Green Version]

| Characteristics | Non-CD (n = 7) | CD (n = 16) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Active (n = 8) | Remissive (n = 8) | p-Value | |||||||||||
| Female (n) | 7 | 8 | 8 | 1 | |||||||||
| Age (mean years ± sd) | 31.4 ± 8.34 | 35.7 ± 12.0 | 39.8 ± 4.25 | 0.21 | |||||||||
| CD duration (mean years ± sd) | - | 9.42 ± 10.1 | 16.2 ± 8.54 | 0.169 | |||||||||
| C-reactive protein (mean mg/L ± sd) | - | 22.9 ± 12.0 | 0.825 ± 0.79 | 0.00994 ** | |||||||||
| Harvey Bradshaw Index (mean ± sd) | - | 6.8 ± 2.77 | 1.29 ± 1.8 | 0.00182 ** | |||||||||
| Montreal Classification (n) | |||||||||||||
| A1 | A2 | - | 2 | 6 | 1 | 7 | 1 | ||||||
| L1 | L2 | L3 | L2+4 | 3 | 2 | 3 | 0 | 2 | 3 | 2 | 1 | 1 | |
| B1 | B2 | B3 | 6 | 2 | 0 | 6 | 1 | 1 | 1 | ||||
| P | 1 | 3 | 0.5692 | ||||||||||
| Any concomitant medication (n) | - | 6 | 8 | 0.4667 | |||||||||
| Anti-TNF | - | 2 | 6 | 0.1319 | |||||||||
| Corticosteroid | - | 2 | 0 | 0.4667 | |||||||||
| Thiopurine | - | 0 | 3 | 0.2 | |||||||||
| Questran | - | 1 | 0 | 1 | |||||||||
| Celcoxib | - | 1 | 0 | 1 | |||||||||
| Pantoprazole | - | 1 | 0 | 1 | |||||||||
| Mercaptopurine | - | 0 | 1 | 1 | |||||||||
| CD vs. Non-CD | CD-Active vs. CD-Remissive | |||
|---|---|---|---|---|
| Differentially Methylated Gene | p-Value | BH-Adjusted p-Value | p-Value | BH-Adjusted p-Value |
| MPIG6B (C6orf25) | 4.63 × 10−15 | 9.19 × 10−11 | 1.08 × 10−9 | 2.15 × 10−5 |
| PDCD1 | 4.05 × 10−10 | 8.04 × 10−6 | 0.905923 | 1 |
| GSTT1 | 3.60 × 10−9 | 7.16 × 10−5 | 0.317294 | 1 |
| ZADH2 | 4.54 × 10−9 | 9.02 × 10−5 | 0.028386 | 1 |
| MS4A3 | 6.90 × 10−9 | 0.000136924 | 0.873469 | 1 |
| DRD4 | 3.15 × 10−8 | 0.000625727 | 0.283934 | 1 |
| SLFN13 | 3.27 × 10−8 | 0.000649811 | 0.010163 | 1 |
| SLC17A9 | 1.67 × 10−7 | 0.003305914 | 0.14758 | 1 |
| MPEG1 | 2.00 × 10−7 | 0.003965532 | 0.498594 | 1 |
| SLC26A4 | 2.66 × 10−7 | 0.005275024 | 0.612185 | 1 |
| SPI1 | 5.95 × 10−7 | 0.011817048 | 0.007133 | 1 |
| LOC286002 | 8.54 × 10−7 | 0.016951922 | 0.578701 | 1 |
| ZNF572 | 1.09 × 10−6 | 0.02165659 | 0.3022 | 1 |
| LOC150381 | 1.72 × 10−6 | 0.034101222 | 0.935613 | 1 |
| G0S2 | 1.74 × 10−6 | 0.03460972 | 0.560081 | 1 |
| NNAT | 1.88 × 10−5 | 0.372484 | 2.98 × 10−12 | 5.91 × 10−8 |
| SERPINF1 | 0.204451 | 1 | 1.13 × 10−10 | 2.24 × 10−6 |
| TRIOBP | 0.634584 | 1 | 9.00 × 10−9 | 1.79 × 10−4 |
| KRTCAP3 | 0.992178 | 1 | 1.31 × 10−8 | 2.61 × 10−4 |
| TRIP6 | 0.036528 | 1 | 3.64 × 10−8 | 7.22 × 10−4 |
| LOC387647 | 0.998103 | 1 | 5.43 × 10−8 | 1.08 × 10−3 |
| HCP5 | 0.809063 | 1 | 7.04 × 10−8 | 1.40 × 10−3 |
| SSTR4 | 0.873458 | 1 | 2.34 × 10−7 | 4.64 × 10−3 |
| FAM24B | 0.071767 | 1 | 9.33 × 10−7 | 0.018513 |
| ZNF154 | 0.003772 | 1 | 2.16 × 10−6 | 0.042834 |
| PRAP1 | 0.901922 | 1 | 2.24 × 10−6 | 0.044415 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li Yim, A.Y.F.; Duijvis, N.W.; Ghiboub, M.; Sharp, C.; Ferrero, E.; Mannens, M.M.A.M.; D’Haens, G.R.; de Jonge, W.J.; te Velde, A.A.; Henneman, P. Whole-Genome DNA Methylation Profiling of CD14+ Monocytes Reveals Disease Status and Activity Differences in Crohn’s Disease Patients. J. Clin. Med. 2020, 9, 1055. https://doi.org/10.3390/jcm9041055
Li Yim AYF, Duijvis NW, Ghiboub M, Sharp C, Ferrero E, Mannens MMAM, D’Haens GR, de Jonge WJ, te Velde AA, Henneman P. Whole-Genome DNA Methylation Profiling of CD14+ Monocytes Reveals Disease Status and Activity Differences in Crohn’s Disease Patients. Journal of Clinical Medicine. 2020; 9(4):1055. https://doi.org/10.3390/jcm9041055
Chicago/Turabian StyleLi Yim, Andrew Y.F., Nicolette W. Duijvis, Mohammed Ghiboub, Catriona Sharp, Enrico Ferrero, Marcel M.A.M. Mannens, Geert R. D’Haens, Wouter J. de Jonge, Anje A. te Velde, and Peter Henneman. 2020. "Whole-Genome DNA Methylation Profiling of CD14+ Monocytes Reveals Disease Status and Activity Differences in Crohn’s Disease Patients" Journal of Clinical Medicine 9, no. 4: 1055. https://doi.org/10.3390/jcm9041055
APA StyleLi Yim, A. Y. F., Duijvis, N. W., Ghiboub, M., Sharp, C., Ferrero, E., Mannens, M. M. A. M., D’Haens, G. R., de Jonge, W. J., te Velde, A. A., & Henneman, P. (2020). Whole-Genome DNA Methylation Profiling of CD14+ Monocytes Reveals Disease Status and Activity Differences in Crohn’s Disease Patients. Journal of Clinical Medicine, 9(4), 1055. https://doi.org/10.3390/jcm9041055








