RAD51Bme Levels as a Potential Predictive Biomarker for PD-1 Blockade Response in Non-Small Cell Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Clinical and Pathological Data Collection
2.3. Assessment of PD-L1 Expression by Immunohistochemistry
2.4. Methylation Analysis
2.5. Statistical Analysis
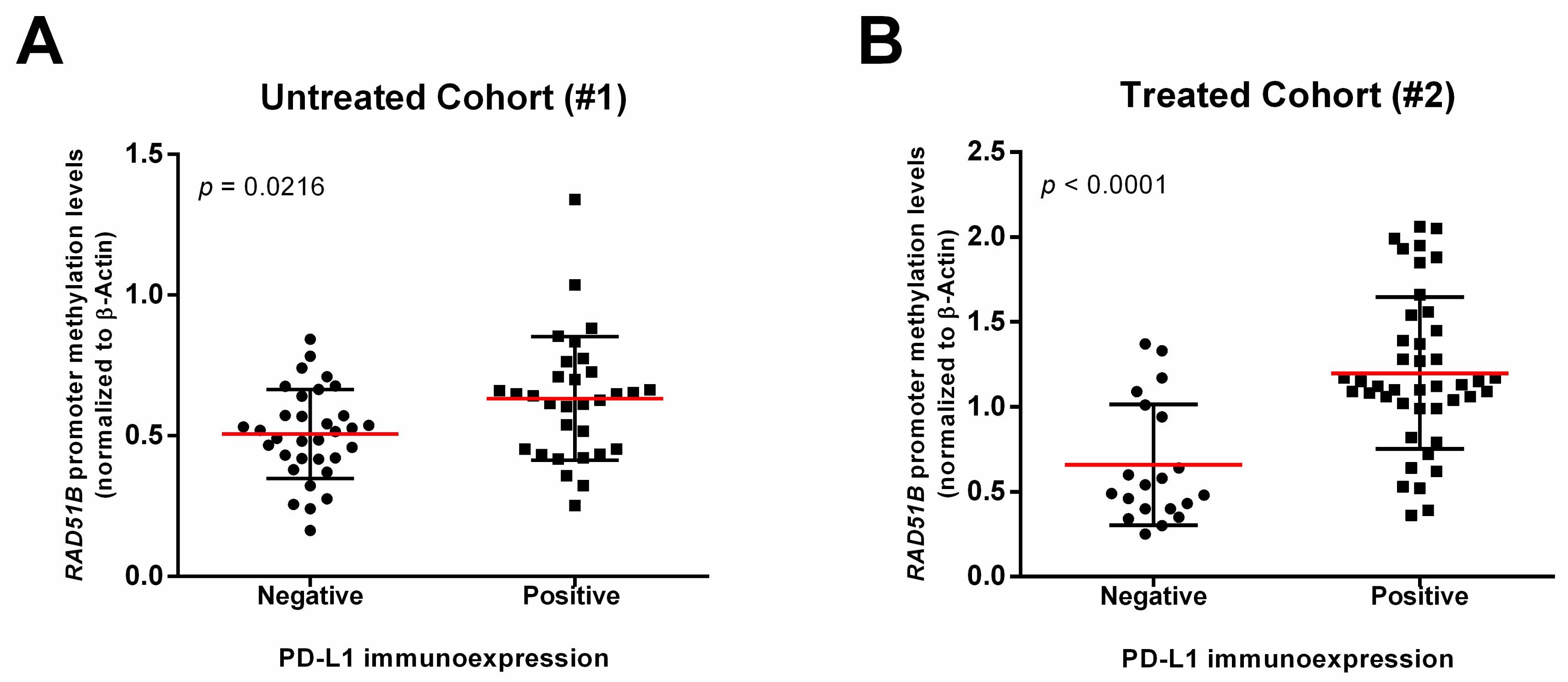
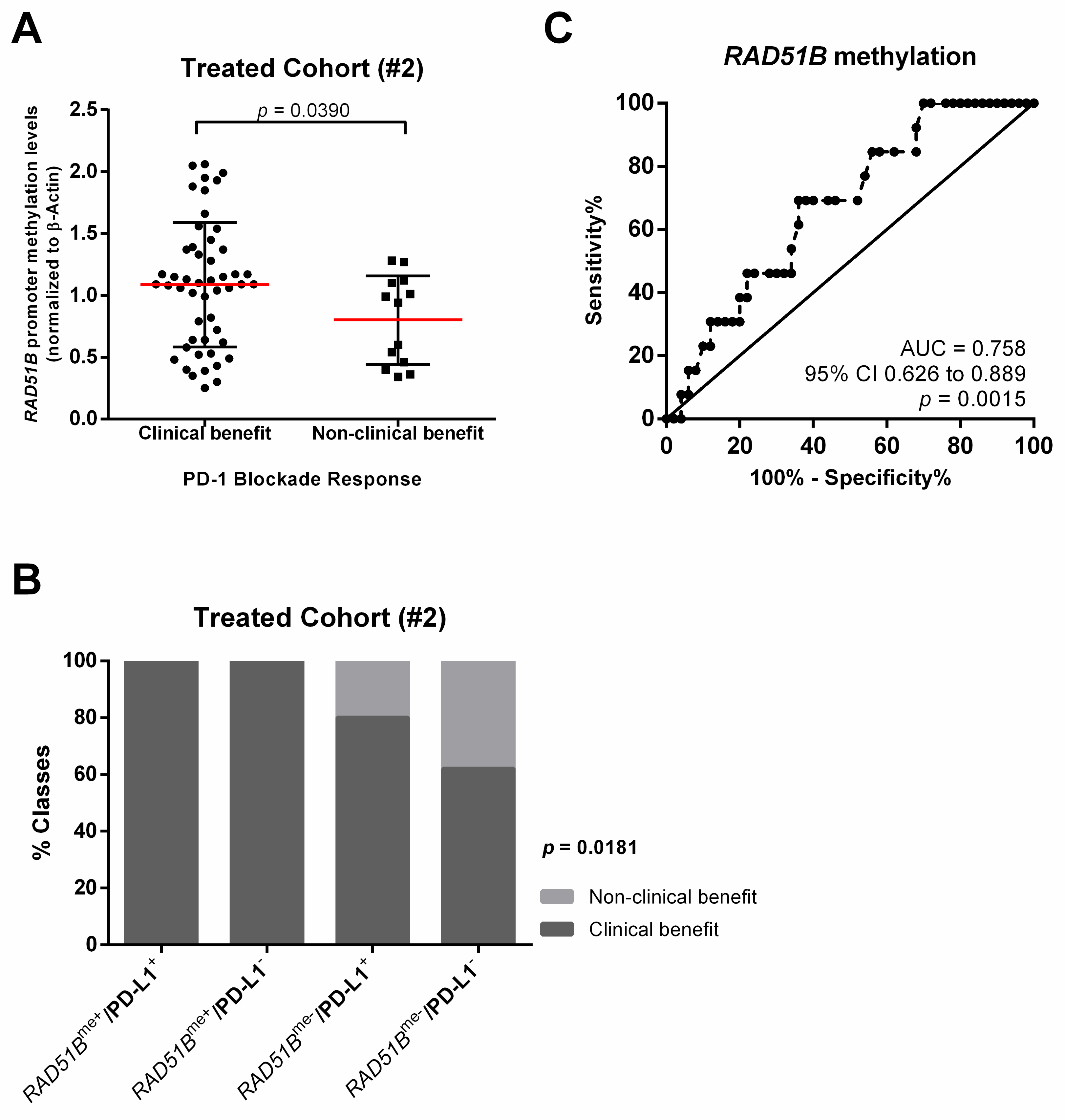
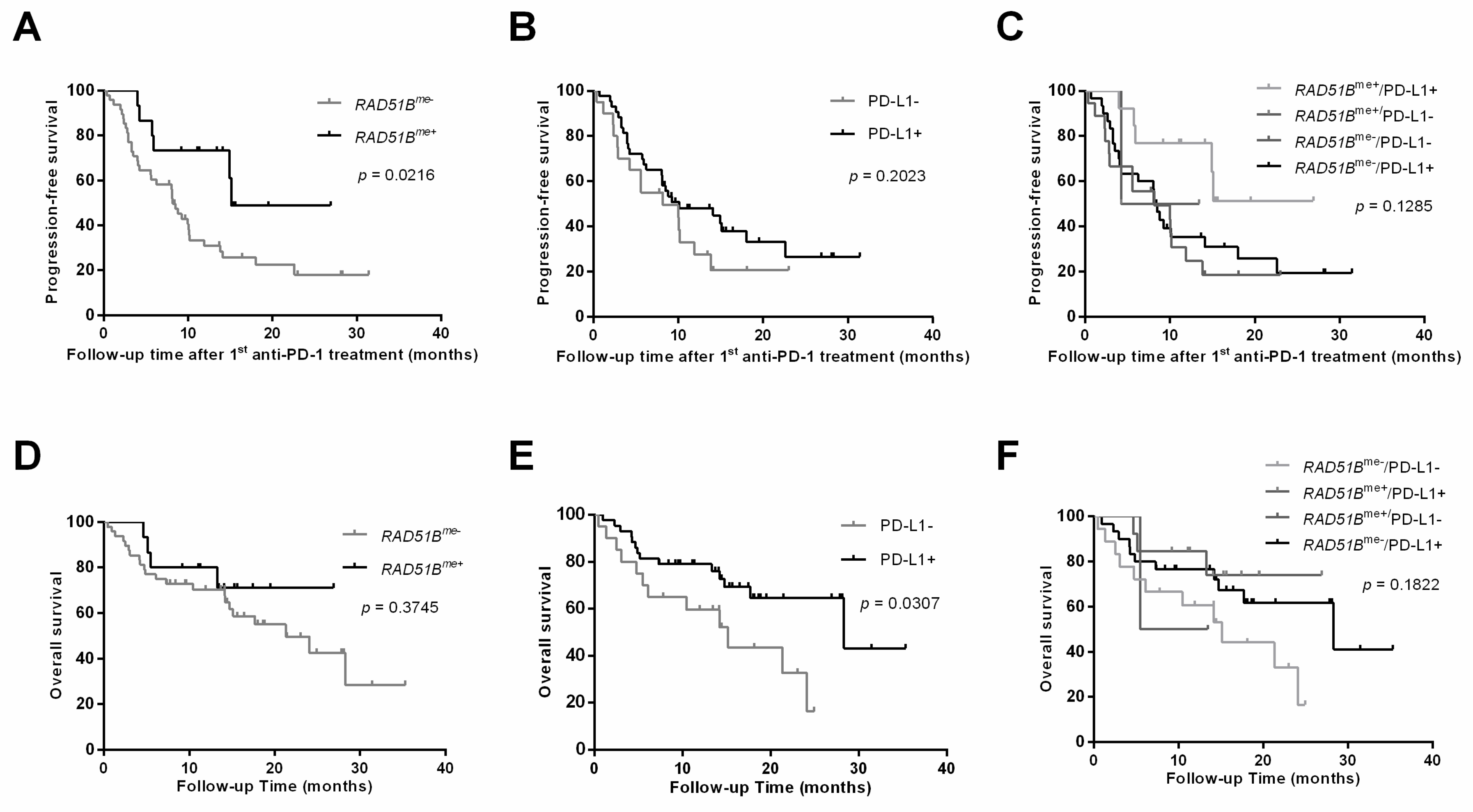
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.-L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Akerley, W.; Bazhenova, L.A.; Borghaei, H.; Camidge, D.R.; Cheney, R.T.; Chirieac, L.R.; D’Amico, T.A.; Dilling, T.J. NCCN guidelines insights: Non–small cell lung cancer, version 4.2016. J. Natl. Compr. Cancer Netw. 2016, 14, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B. The 2015 World Health Organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S. Personalized medicine in lung cancer: What we need to know. Nat. Rev. Clin. Oncol. 2011, 8, 661–668. [Google Scholar] [CrossRef]
- Kerr, K.M.; Bubendorf, L.; Edelman, M.J.; Marchetti, A.; Mok, T.; Novello, S.; O’Byrne, K.; Stahel, R.; Peters, S.; Felip, E. Second ESMO consensus conference on lung cancer: Pathology and molecular biomarkers for non-small-cell lung cancer. Ann. Oncol. 2014, 25, 1681–1690. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L. Pembrolizumab for the treatment of non–small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Petrelli, F.; Maltese, M.; Tomasello, G.; Conti, B.; Borgonovo, K.; Cabiddu, M.; Ghilardi, M.; Ghidini, M.; Passalacqua, R.; Barni, S. Clinical and Molecular Predictors of PD-L1 Expression in Non–Small-Cell Lung Cancer: Systematic Review and Meta-analysis. Clin. Lung Cancer 2018, 19, 315–322. [Google Scholar] [CrossRef]
- Syn, N.L.; Teng, M.W.; Mok, T.S.; Soo, R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017, 18, e731–e741. [Google Scholar] [CrossRef]
- Shi, Y. Regulatory mechanisms of PD-L1 expression in cancer cells. Cancer Immunol. Immunother. 2018, 67, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Wang, H.; Liu, Y.; Zhao, M.; Zhang, H.; Lu, Z.; Fang, Y.; Chen, X.; Liu, G. The prognostic value of PD-L1 expression for non-small cell lung cancer patients: A meta-analysis. Eur. J. Surg. Oncol. (EJSO) 2015, 41, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Iarovaia, O.V.; Rubtsov, M.; Ioudinkova, E.; Tsfasman, T.; Razin, S.V.; Vassetzky, Y.S. Dynamics of double strand breaks and chromosomal translocations. Mol. Cancer 2014, 13, 249. [Google Scholar] [CrossRef] [PubMed]
- O’Kane, G.M.; Connor, A.A.; Gallinger, S. Characterization, detection, and treatment approaches for homologous recombination deficiency in cancer. Trends Mol. Med. 2017, 23, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Rodrigue, A.; Lafrance, M.; Gauthier, M.C.; McDonald, D.; Hendzel, M.; West, S.C.; Jasin, M.; Masson, J.Y. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 2006, 25, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Gachechiladze, M.; Škarda, J.; Soltermann, A.; Joerger, M. RAD51 as a potential surrogate marker for DNA repair capacity in solid malignancies. Int. J. Cancer 2017, 141, 1286–1294. [Google Scholar] [CrossRef]
- Rieke, D.T.; Ochsenreither, S.; Klinghammer, K.; Seiwert, T.Y.; Klauschen, F.; Tinhofer, I.; Keilholz, U. Methylation of RAD51B, XRCC3 and other homologous recombination genes is associated with expression of immune checkpoints and an inflammatory signature in squamous cell carcinoma of the head and neck, lung and cervix. Oncotarget 2016, 7, 75379–75393. [Google Scholar] [CrossRef]
- Thacker, J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005, 219, 125–135. [Google Scholar] [CrossRef]
- Suwaki, N.; Klare, K.; Tarsounas, M. RAD51 paralogs: Roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin. Cell Dev. Biol. 2011, 22, 898–905. [Google Scholar] [CrossRef]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S. Pembrolizumab versus chemotherapy for PD-L1–positive non–small-cell lung cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E. Nivolumab versus docetaxel in advanced squamous-cell non–small-cell lung cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E. Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, M.D.; Ciuleanu, T.-E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef]
- Rizvi, H.; Sanchez-Vega, F.; La, K.; Chatila, W.; Jonsson, P.; Halpenny, D.; Plodkowski, A.; Long, N.; Sauter, J.L.; Rekhtman, N. Molecular determinants of response to anti–programmed cell death (PD)-1 and anti–programmed death-ligand 1 (PD-L1) blockade in patients with non–small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 2018, 36, 633–641. [Google Scholar] [CrossRef]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Srinivasan, K.; Abdisalaam, S.; Su, F.; Raj, P.; Dozmorov, I.; Mishra, R.; Wakeland, E.K.; Ghose, S.; Mukherjee, S. RAD51 interconnects between DNA replication, DNA repair and immunity. Nucleic Acids Res. 2017, 45, 4590–4605. [Google Scholar] [CrossRef]
- Duruisseaux, M.; Martínez-Cardús, A.; Calleja-Cervantes, M.E.; Moran, S.; de Moura, M.C.; Davalos, V.; Piñeyro, D.; Sanchez-Cespedes, M.; Girard, N.; Brevet, M. Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: A multicentre, retrospective analysis. Lancet Respir. Med. 2018, 6, 771–781. [Google Scholar] [CrossRef]
- Abdel-Rahman, O. Correlation between PD-L1 expression and outcome of NSCLC patients treated with anti-PD-1/PD-L1 agents: A meta-analysis. Crit. Rev. Oncol. Hematol. 2016, 101, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Zhang, Q.; Goradia, A.; Raghunath, P.N.; Liu, X.; Paessler, M.; Wang, H.Y.; Wysocka, M.; Cheng, M.; Ruggeri, B.A. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc. Natl. Acad. Sci. USA 2008, 105, 20852–20857. [Google Scholar] [CrossRef] [PubMed]
- Akbay, E.A.; Koyama, S.; Carretero, J.; Altabef, A.; Tchaicha, J.H.; Christensen, C.L.; Mikse, O.R.; Cherniack, A.D.; Beauchamp, E.M.; Pugh, T.J. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013, 3, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.-J.; Zhan, P.; Song, Y. PD-L1 over-expression and survival in patients with non-small cell lung cancer: A meta-analysis. Transl. Lung Cancer Res. 2015, 4, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Sheng, Z.; Jiang, L.; Liu, Z.; Bi, Y.; Shen, Y. Overexpression of RAD51B predicts a preferable prognosis for non-small cell lung cancer patients. Oncotarget 2017, 8, 91471–97480. [Google Scholar] [CrossRef] [PubMed]
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| ß-Actin | TGGTGATGGAGGAGGTTTAGTAAGT | AACCAATAAAACCTACTCCTCCCTTAA |
| RAD51Bme | AGATTTTTAGGGTCGAGAGC | CGCCCGACTAATTTTTTTAT |
| Characteristics | Untreated Cohort (#1) n = 64 | Treated Cohort (#2) n = 63 |
|---|---|---|
| Gender, (n, %) Male Female | 51 (79.7) 13 (20.3) | 49 (77.8) 14 (22.2) |
| Age (year), median (IQR) | 62.5 (29.0–84.0) | 62.0 (32.0–77.0) |
| Histologic subtype (n, %) Adenocarcinoma Squamous NOS | 41 (64.1) 22 (34.4) 1 (1.6) | 46 (73.0) 17 (27.0) - |
| Smoking habits (n, %) Never Smoker Previous smoker | 16 (25.0) 20 (31.3) 28 (43.7) | 10 (15.9) 20 (31.7) 33 (53.4) |
| PD-L1 immunoexpression (n, %) Negative Intermediate (1–49%) Strong (≥ 50%) | 33 (51.6) 18 (28.1) 13 (20.3) | 20 (31.7) 14 (22.2) 29 (46.0) |
| Anti-PD-1 agent (n, %) Pembrolizumab Nivolumab | n.a. | 38 (60.3) 25 (39.7) |
| PD-1 blockade (n, %) Clinical benefit Non-clinical benefit | n.a. | 13 (20.6) 50 (79.4) |
| End of PD-1 blockade treatment (n, %) Not applicable Disease progression Toxicity | n.a. | 18 (28.6) 39 (61.9) 6 (9.5) |
| Progression-free survival since PD-1 blockade, months median (IQR) | n.a. | 8.1 (5.1–11.1) |
| Overall survival since PD-1 blockade, months median (IQR) | n.a. | 21.3 (13.7–28.9) |
| RAD51Bme levels (normalized to β-actin), median (IQR) | 0.54 (0.16–1.34) | 1.08 (0.25–2.06) |
| Predictive Biomarkers of PD-1 Blockade Response | |||
|---|---|---|---|
| RAD51Bme+ | PD-L1+ | RAD51Bme+/PD-L1+ | |
| Sensitivity | 38% | 74% | 68% |
| Specificity | 85% | 54% | 85% |
| Accuracy | 48% | 70% | 71% |
| PPV | 90% | 86% | 94% |
| NPV | 26% | 35% | 41% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerreiro, I.M.; Barros-Silva, D.; Lopes, P.; Cantante, M.; Cunha, A.L.; Lobo, J.; Antunes, L.; Rodrigues, A.; Soares, M.; Henrique, R.; et al. RAD51Bme Levels as a Potential Predictive Biomarker for PD-1 Blockade Response in Non-Small Cell Lung Cancer. J. Clin. Med. 2020, 9, 1000. https://doi.org/10.3390/jcm9041000
Guerreiro IM, Barros-Silva D, Lopes P, Cantante M, Cunha AL, Lobo J, Antunes L, Rodrigues A, Soares M, Henrique R, et al. RAD51Bme Levels as a Potential Predictive Biomarker for PD-1 Blockade Response in Non-Small Cell Lung Cancer. Journal of Clinical Medicine. 2020; 9(4):1000. https://doi.org/10.3390/jcm9041000
Chicago/Turabian StyleGuerreiro, Inês Maria, Daniela Barros-Silva, Paula Lopes, Mariana Cantante, Ana Luísa Cunha, João Lobo, Luís Antunes, Ana Rodrigues, Marta Soares, Rui Henrique, and et al. 2020. "RAD51Bme Levels as a Potential Predictive Biomarker for PD-1 Blockade Response in Non-Small Cell Lung Cancer" Journal of Clinical Medicine 9, no. 4: 1000. https://doi.org/10.3390/jcm9041000
APA StyleGuerreiro, I. M., Barros-Silva, D., Lopes, P., Cantante, M., Cunha, A. L., Lobo, J., Antunes, L., Rodrigues, A., Soares, M., Henrique, R., & Jerónimo, C. (2020). RAD51Bme Levels as a Potential Predictive Biomarker for PD-1 Blockade Response in Non-Small Cell Lung Cancer. Journal of Clinical Medicine, 9(4), 1000. https://doi.org/10.3390/jcm9041000








