Multi-Platform Characterization of Cerebrospinal Fluid and Serum Metabolome of Patients Affected by Relapsing–Remitting and Primary Progressive Multiple Sclerosis
Abstract
1. Introduction
2. Materials and Methods
2.1. BiocratesAbsoluteIDQ p180 Kit
2.2. NMR and GC-MS Analysis
2.2.1. Sample Preparation
2.2.2. NMR Analysis and Data Processing
2.2.3. GC-MS Analysis and Data Processing
2.3. Statistical Analysis
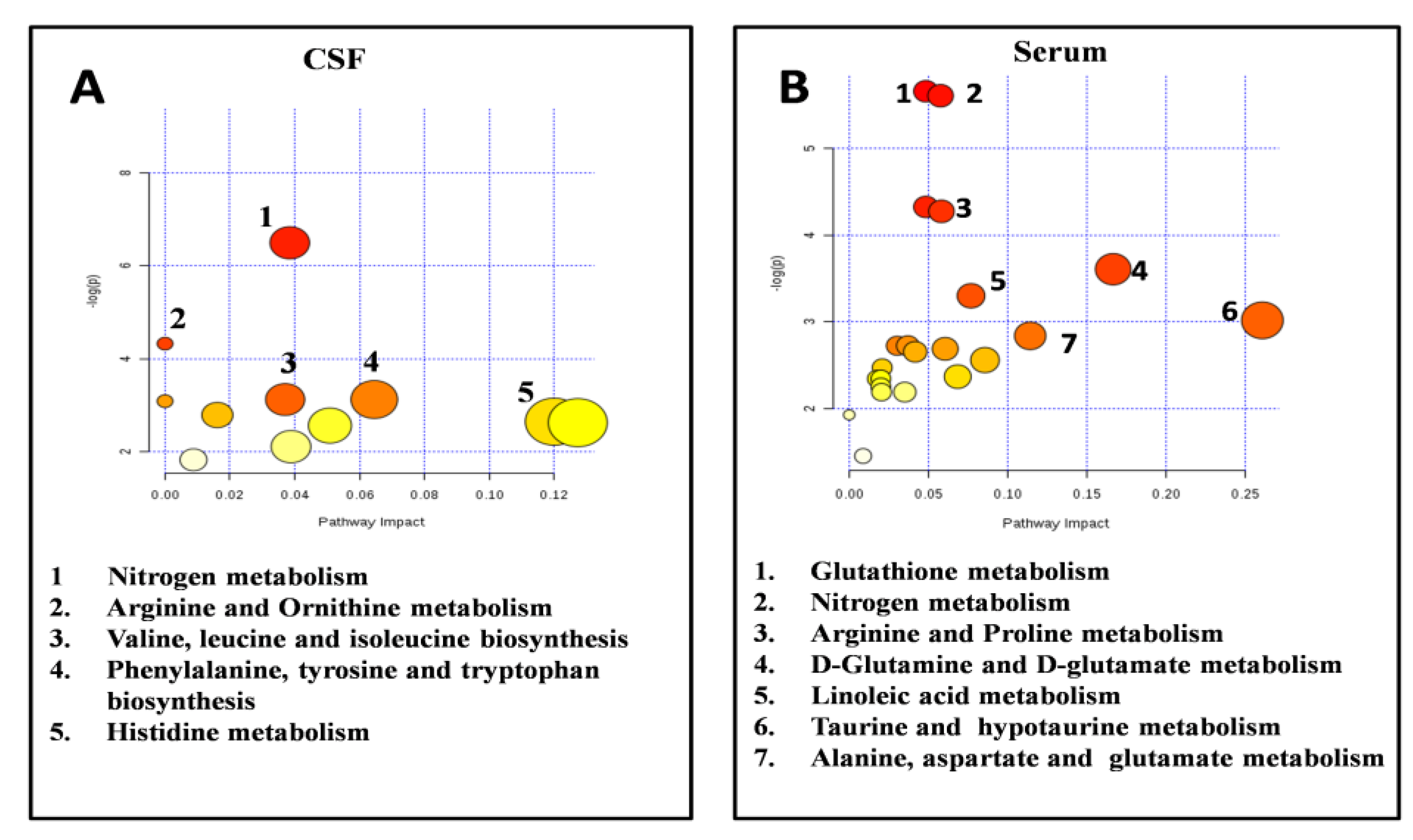
2.4. Pathways Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Haider, L.; Zrzavy, T.; Hametner, S.; Höftberger, R.; Bagnato, F.; Grabner, G.; Trattnig, S.; Pfeifenbring, S.; Brück, W.; Lassmann, H. The topograpy of demyelination and neurodegeneration in the multiple sclerosis brain. Brain 2016, 139, 807–815. [Google Scholar] [CrossRef]
- Ebers, G.C. Environmental factors and multiple sclerosis. Lancet Neurol. 2008, 7, 268–277. [Google Scholar] [CrossRef]
- Lublin, F.D.; Reingold, S.C.; For the National Multiple Sclerosis Society (USA) Advisory Committee on Clinical Trials of New Agents in Multiple Sclerosis. Defining the clinical course of multiple sclerosis: Results of an international survey. Neurology 1996, 46, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Lublin, F.; Reingold, S.C.; Cohen, J.A.; Cutter, G.R.; Sørensen, P.S.; Thompsonm, A.J.; Wolinsky, J.S.; Balcer, L.J.; Banwell, B.; Barkhof, F.; et al. Defining the clinical course of multiple sclerosis. The 2013 revisions. Neurology 2014, 83, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Lucchinetti, C.; Bruck, W.; Parisi, J.; Scheithauer, B.; Rodriguez, M.; Lassmann, H. Heterogeneity of multiple sclerosis lesions: Implications for the pathogenesis of demyelination. Ann. Neurol. 2000, 47, 707–717. [Google Scholar] [CrossRef]
- Leary, S.M.; Stevenson, V.L.; Miller, D.H.; Thompson, A.J. Problems in designing and recruiting to therapeutic trials in primary progressive multiple sclerosis. J. Neurol. 1999, 246, 562–568. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, G.V.; Hawkins, S.A. Clinical study of primary progressive multiple sclerosis in Northern Ireland. J. Neurol. Neurosurg. Psychiatry 1998, 64, 451–454. [Google Scholar] [CrossRef]
- Weinshenker, B. Progressive forms of MS: Classification streamlined or consensus overturned? Lancet 2000, 353, 162–163. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytic techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef]
- Madsen, R.; Lundstedt, T.; Trygg, J. Chemometrics in metabolomics—A review in human disease diagnosis. Anal. Chim. Acta 2010, 659, 23–33. [Google Scholar] [CrossRef]
- Reinke, D.I.; Broadhurst, B.D.; Sykes, G.B.; Baker, I.; KG Warren, C.; Power, C. Metabolomic profiling in multiple sclerosis: Insights into biomarkers and pathogenesi SN. Mult. Scler. J. 2014, 20, 1396–1400. [Google Scholar] [CrossRef] [PubMed]
- Cocco, E.; Murgia, F.; Lorefice, L.; Barberini, L.; Poddighe, S.; Frau, J.; Fenu, G.; Coghe, G.; Murru, M.R.; Murru, R.; et al. 1H-NMR analysis provides a metabolomic profile of patients with multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e185. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Calabresi, P.A. Metabolomics in multiple sclerosis. Mult. Scler. J. 2016, 22, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Stoessel, D.; Stellmann, J.P.; Willing, A.; Behrens, B.; Rosenkranz, S.C.; Hodecker, S.C.; Sturner, K.H.; Reinhardt, S.; Fleischer, S.; Deuschle, C.; et al. Metabolomic profiles for primary progressive multiple sclerosis stratification and disease course monitoring. Front. Hum. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Dickens, A.M.; Larkin, J.R.; Griffin, J.L.; Cavey, A.; Matthews, L.; Turner, M.R.; Wilcock, G.K.; Davis, B.G.; Claridge, T.C.W.; Palace, J.; et al. A type 2 biomarker separates relapsing-remitting from secondary progressive multiple sclerosis. Neurology 2014, 83, 1492–1499. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.K.; Bilgin, A.; Lovejoy, D.B.; Tan, V.; Bustamante, S.; Taylor, B.V.; Bessede, A.; Brew, B.J.; Guillemin, G.J. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Sci. Rep. 2017, 7, 41473. [Google Scholar] [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carrol, W.M.; Coetzee, T.; Comi, G.; Correale, F.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Kurtzke, J.F. Rating neurologic impairment in multiple sclerosis: An expanded disability status scale (EDSS). Neurology 1983, 33, 1444–1452. [Google Scholar] [CrossRef]
- Ramsay, S.L.; Stoegg, W.M.; Weinberger, K.M.; Graber, A.; Guggenbichler, W. Apparatus and method for analyzing a metabolite profile. CA2608963A1. 11 January 2007. [Google Scholar]
- Murgia, F.; Svegliati, S.; Poddighe, S.; Lussu, M.; Manzin, A.; Spadoni, T.; Fischetti, C.; Gabrielli, A.; Atzori, L. Metabolomic profile of systemic sclerosis patients. Sci. Rep. 2018, 8, 7626. [Google Scholar] [CrossRef]
- Available online: http://www.nist.gov/srd/mslist.cfm (accessed on 20 March 2020).
- Available online: http://fiehnlab.ucdavis.edu/Metabolite-Library-2007 (accessed on 20 March 2020).
- Available online: http://gmd.mpimp-golm.mpg.de (accessed on 20 March 2020).
- Liggi, S.; Hinz, C.; Hall, Z.; Santoru, M.L.; Poddighe, S.; Fjeldsted, J.; Atzori, L.; Griffin, J.L. KniMet: A pipeline for the processing of chromatography-mass spectrometry metabolomics data. Metabolomics 2018, 14, 52. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikstrom, C. Multi-and Megavariate Data Analysis. Basic Principles and Applications; Umetrics Academy: Malmo, Sweden, 2013; pp. 1–50. [Google Scholar]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, E.; Slupsky, C.M. Targeted profiling: Quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 2006, 78, 4430–4442. [Google Scholar] [CrossRef]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, 486–494. [Google Scholar] [CrossRef]
- Bielekova, B.; Martin, R. Development of biomarkers in multiple sclerosis. Brain 2004, 127, 1463–1478. [Google Scholar] [CrossRef] [PubMed]
- Comabella, M.; Montalban, X. Body fluid biomarkers in multiple sclerosis. Lancet Neurol. 2014, 13, 113–126. [Google Scholar] [CrossRef]
- Kuenz, B.; Lutterotti, A.; Ehling, R.; Gneiss, C.; Haemmerle, M.; Rainer, C.; Deisenhammer, F.; Schocke, M.; Berger, T.; Reindl, M. Cerebrospinal fluid B cells correlate with early brain inflammation in multiple sclerosis. PLoS ONE 2008, 3, e2559. [Google Scholar] [CrossRef]
- Gilgun-Sherki, Y.; Melamed, E.; Offen, D. The role of oxidative stress in the pathogenesis of multiple sclerosis: The need for effective antioxidant therapy. J. Neurol. 2004, 251, 261–268. [Google Scholar]
- Winyard, P.G.; Blake, D.R. Antioxidants, redox-regulated transcription factors and inflammation. Adv. Pharmacol. 1997, 38, 403–421. [Google Scholar]
- Naidoo, R.; Knapp, M.L. Studies of lipid peroxidation products in cerebrospinal fluid and serum in multiple sclerosis and other conditions. Clin. Chem. 1992, 38, 2449–2454. [Google Scholar] [CrossRef]
- Bizzozero, O.; DeJesus, G.; Bixler, H.A.; Pastuszyn, A. Evidence of nitrosative damage in the brain white matter of patients with multiple sclerosis. Neurochem. Res. 2005, 30, 139–149. [Google Scholar] [CrossRef] [PubMed]
- LeVine, S.M. The role of reactive oxygen species in the pathogenesis of multiple sclerosis. Med. Hypotheses 1992, 39, 271–274. [Google Scholar] [CrossRef]
- Bhargava, P.; Fitzgerald, K.C.; Calabresi, P.A.; Mowry, E.M. Metabolic alterations in multiple sclerosis and the impact of vitamin D supplementation. JCI Insight 2017, 2, e95302. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, V.; Scapagnini, G.; Ravagna, A.; Bella, R.; Foresti, R.; Bates, T.E.; Giuffrida Stella, A.M.; Pennisi, G. Nitric oxide synthase is present in the cerebrospinal fluid of patients with active multiple sclerosis and is associated with increases in cerebrospinal fluid protein nitrotyrosine and S-nitrosothiols and with changes in glutathione levels. J. Neurosci. Res. 2002, 70, 580–587. [Google Scholar] [CrossRef]
- Koch, M.; Ramsaransing, G.S.; Arutjunyan, A.V.; Stepanov, M.; Teelken, A.; Heersema, D.J.; De Keyser, J. Oxidative stress in serum and peripheral blood leukocytes in patients with different disease courses of multiple sclerosis. J. Neurol. 2006, 253, 483–487. [Google Scholar] [CrossRef]
- Gonsette, R.E. Neurodegeneration in multiple sclerosis: The role of oxidative stress and excitotoxicity. J. Neurol. Sci. 2008, 274, 48–53. [Google Scholar] [CrossRef]
- Groom, A.J.; Smith, T.; Lursky, L. Multiple sclerosis and glutamate. Ann. N. Y. Acad. Sci. 2003, 993, 229–275. [Google Scholar] [CrossRef]
- Pitt, D.; Nagelmeier, I.E.; Wilson, H.C.; Raine, C.S. Glutamate uptake by oligodendrocytes Implications for excitotoxicity in multiple sclerosis. Neurology 2003, 61, 1113–1120. [Google Scholar] [CrossRef]
- Pieragostino, D.; D’Alessandro, M.; di Ioia, M.; Rossi, C.; Zucchelli, M.; Urbani, A.; Di Ilio, C.; Lugaresi, A.; Sacchetta, P.; Del Boccio, P. An integrated metabolomics approach for the research of new cerebrospinal fluid biomarkers of multiple sclerosis. Mol. BioSyst. 2015, 11, 1563–1572. [Google Scholar] [CrossRef]
- Villoslada, P.; Alonso, C.; Agirrezabal, I.; Kotelnikova, E.; Zubizarreta, I.; Pulido-Valdeolivas, I.; Saiz, A.; Comabella, M.; Montalban, X.; Villar, L.; et al. Metabolomic signatures associated with disease severity in multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2017, 4, e321. [Google Scholar] [CrossRef]
- Sarchielli, P.; Greco, L.; Floridi, A.; Floridi, A.; Gallai, V. Excitatory amino acids and multiple sclerosis: Evidence from cerebrospinal fluid. Arch. Neurol. 2003, 60, 1082–1088. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, I.; Vojinovic, S.; Ljubisavljevic, S.; Pavlovic, R.; Basic, J.; Pavlovic, D.; Ilic, A.; Cvetkovic, T.; Stukalov, M. INF-β1b therapy modulates L-arginine and nitric oxide metabolism in patients with relapse remittent multiple sclerosis. J. Neurol. Sci. 2012, 323, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Poddighe, S.; Murgia, F.; Lorefice, L.; Liggi, S.; Cocco, E.; Marrosu, M.G.; Atzori, L. Metabolomic analysis identifies altered metabolic pathways in Multiple Sclerosis. Int. J. Biochem. Cell Biol. 2017, 93, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Mangalam, A.K.; Poisson, L.M.; Nemutlu, E.; Datta, I.; Denic, A.; Dzeja, P.; Rodriguez, M.; Rattan, R.; Giri, S. Profile of Circulatory Metabolites in a Relapsing-remitting Animal Model of Multiple Sclerosis using Global Metabolomics. J. Clin. Cell. Immunol. 2013, 4. [Google Scholar] [CrossRef]
- Pritzker, L.B.; Joshi, S.; Gowan, J.J.; Harauz, G.; Moscarello, M.A. Deimination of myelin basic protein I. Effect of deimination of arginyl residues of myelin basic protein on its structure and susceptibility to digestion by cathespsin D. Biochemistry 2000, 39, 5324–5381. [Google Scholar] [CrossRef]
- Moscarello, M.A.; Mastronardi, F.G.; Wood, D.D. The role of citrullinated proteins suggests a novel mechanism in the pathogenesis of multiple sclerosis. Neurochem. Res. 2007, 32, 251–256. [Google Scholar] [CrossRef]
- Finch, P.R.; Wood, D.D.; Moscarello, M.A. The presence of citrulline in a myelin protein fraction. FEBS Lett. 1971, 15, 145–148. [Google Scholar] [CrossRef]
- Fernstrom, J.D. Branched-chain amino acids and brain. J. Nutr. 2005, 135, 1539–1546. [Google Scholar] [CrossRef]
- Hutson, S.M.; Sweatt, A.J.; LaNoue, K.F. Branched Chain Amino Acids (BCAAs) in Brain. In Handbook of Neurochemistry and Molecular Neurobiology: Amino Acids and Peptides in the Nervous System; Springer Science: New York, NY, USA, 2007; pp. 117–131. [Google Scholar]
- Li, P.; Yin, Y.L.; Li, D.; Kim, S.W.; Wu, G. Amino acids and immune function. Br. J. Nutr. 2007, 98, 237–252. [Google Scholar] [CrossRef]
- Schwarcz, R. The kynurenine pathway of tryptophan degradation as a drug target. Curr. Opin. Pharmacol. 2004, 4, 12–17. [Google Scholar] [CrossRef]
- Watzlawik, J.O.; Wootla, B.; Rodriguez, M. Tryptophan catabolites and their impact on multiple sclerosis progression. Curr. Pharm. Des. 2016, 22, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Rajda, C.; Bergquist, J.; Vécsei, L. Kynurenines, redox disturbances and neurodegeneration in multiple sclerosis. Neuropsychiatric Disorders an Integrative Approach. J. Neural Transm. 2007, 72, 323–329. [Google Scholar]
- Lovelace, M.D.; Varney, B.; Sundaram, G.; Franco, N.F.; Ng, M.L.; Pai, S.; Lim, C.K.; Guillemin, G.L.; Brew, B.J. Current evidence for a role of the Kynurenine pathway of tryptophan metabolism in multiple sclerosis. Front. Immunol. 2016, 7, 246. [Google Scholar] [CrossRef] [PubMed]
- Lorefice, L.; Murgia, F.; Fenu, G.; Frau, J.; Coghe, G.; Murru, M.R.; Tranquilli, S.; Visconti, A.; Marrosu, M.G.; Atzori, L.; et al. Assessing the metabolomic profile of multiple sclerosis patients treated with interferon beta 1a by 1H-NMR spectroscopy. Neurotherapeutics 2019, 16, 797–807. [Google Scholar] [CrossRef] [PubMed]

| MS Patients (34) | Relapsing (22) | Progressive (12) | p-Value | |
|---|---|---|---|---|
| Male Gender | 14 (41.2%) | 6 (27.2%) | 8 (66.6%) | ns |
| Age (mean ± SD) years | 37.3 ± 12.8 | 32±8.4 | 47.1 ± 12.8 | <0.05 |
| MS Disease Duration (mean ± SD) years | 2.1 ± 1.5 | 1.2 ± 1.4 | 3.7 ± 1.2 | <0.05 |
| Expanded Disability Status Scale (EDSS) score | 2.1 ± 1.1 | 1.1 ± 1.5 | 3.9 ± 1.7 | <0.05 |
| CSF | SERUM | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2X | R2Y | Q2 | p-Value | Permutation Test: Intercept R2\Q2 | R2X | R2Y | Q2 | p-Value | Permutation Test: Intercept R2\Q2 | |
| FIA-MS/MS | 0.272 | 0.862 | 0.634 | 2,6e-05 | 0.59/−0.23 | 0.523 | 0.666 | 0.512 | 0.0004 | 0.33/−0.19 |
| LC-MS | 0.395 | 0.697 | 0.496 | 0.002 | 0.29/−0.28 | 0.224 | 0.846 | 0.514 | 0.0002 | 0.35/−0.26 |
| CSF | ||||||||
|---|---|---|---|---|---|---|---|---|
| METABOLITES | RR vs. PP | p-Value | Holm–Bonf. Correction | ROC-CURVE | ||||
| AUC | Std. Error | CI | p-Value | |||||
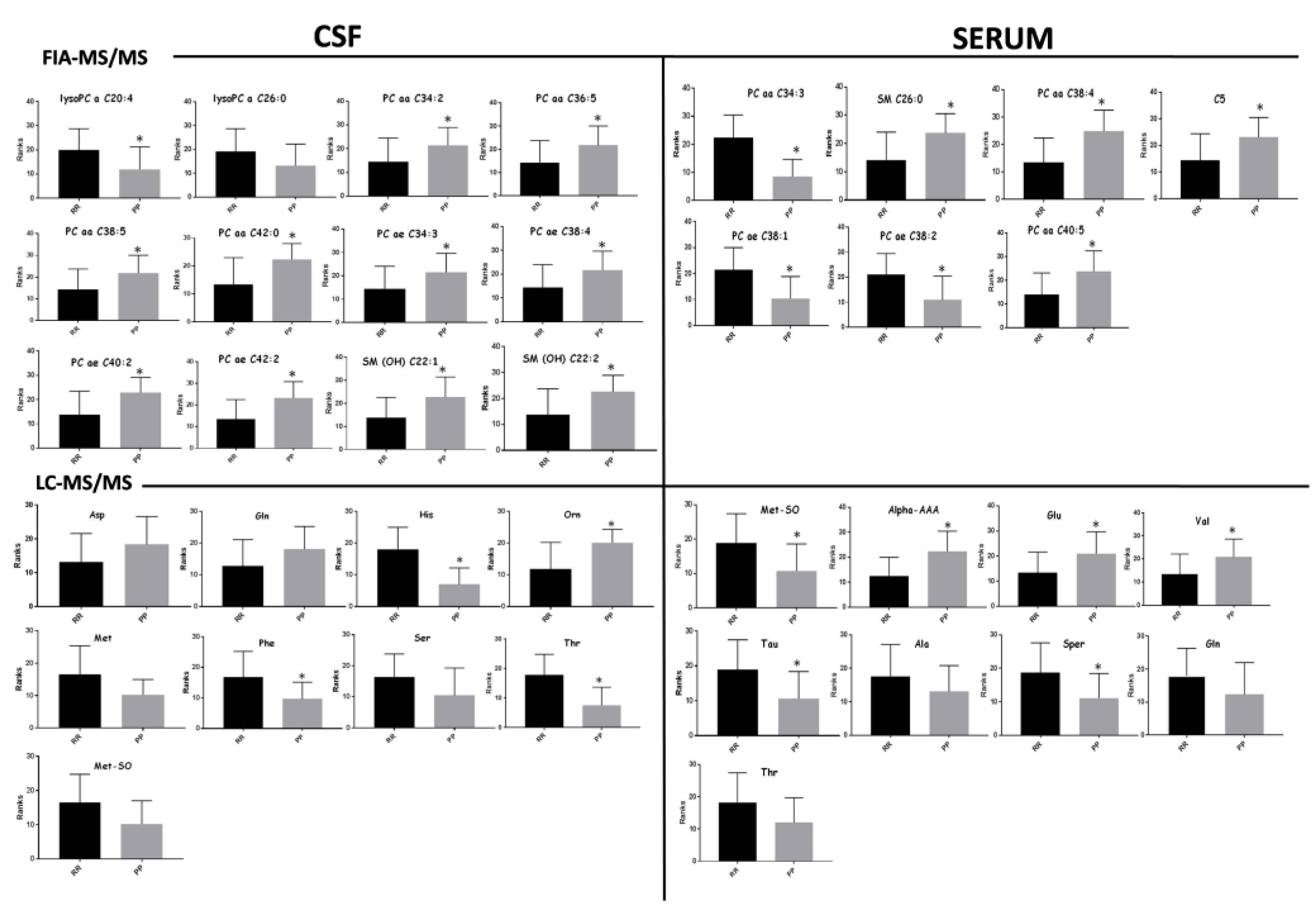
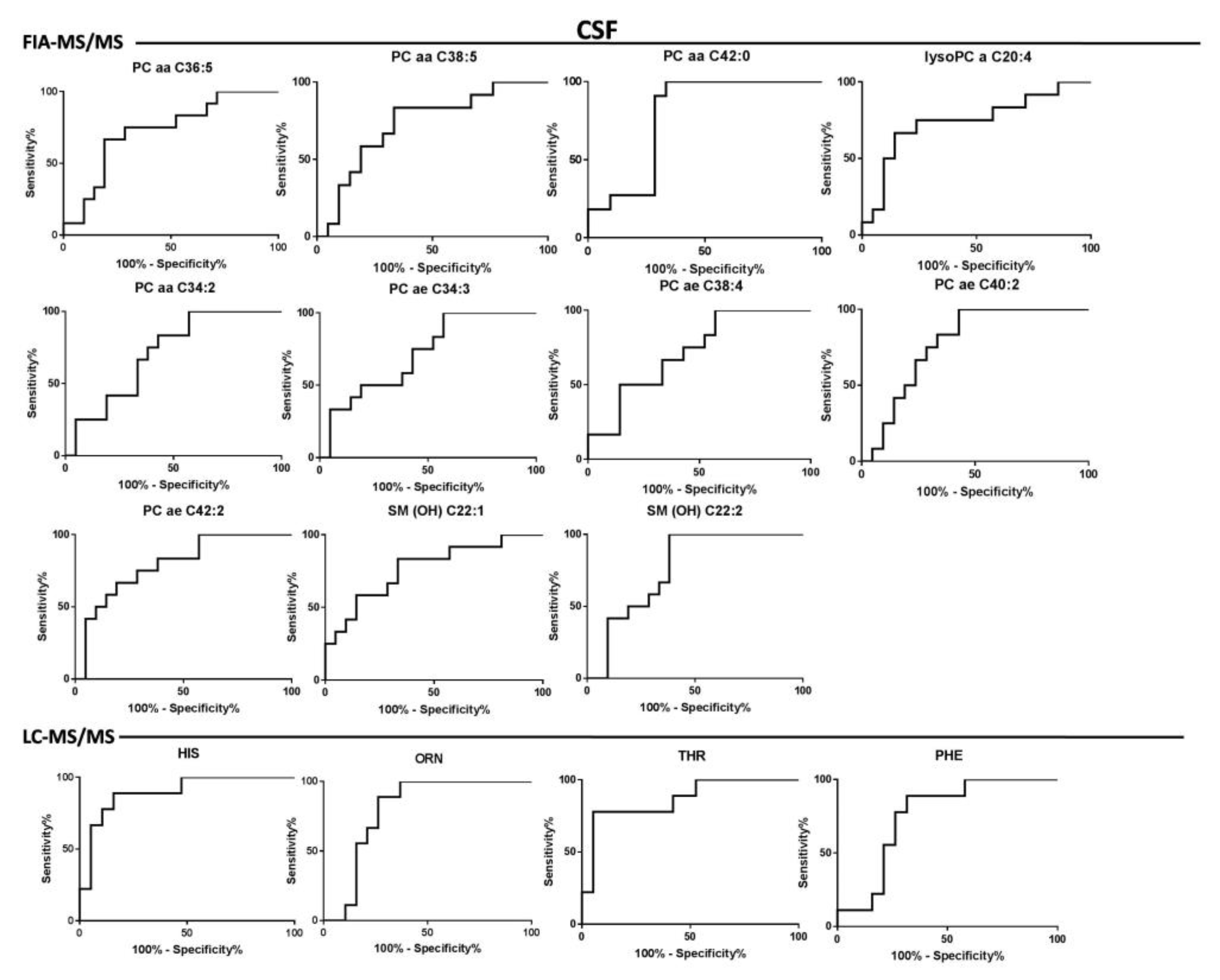
| FIA-MS/MS | -lysoPC a C20:4 | + | 0.02 | ns | 0.74 | 0.09 | 0.55–0.93 | 0.02 |
| -PC aa C34:2 | - | 0.04 | ns | 0.71 | 0.08 | 0.53–0.88 | 0.04 | |
| -PC aa C36:5 | - | 0.03 | ns | 0.72 | 0.09 | 0.54–0.90 | 0.03 | |
| -PC aa C38:5 | - | 0.02 | ns | 0.73 | 0.09 | 0.55–0.90 | 0.03 | |
| -PC aa C42:0 | - | 0.009 | ns | 0.78 | 0.08 | 0.61–0.94 | 0.01 | |
| -PC ae C34:3 | - | 0.04 | ns | 0.71 | 0.09 | 0.53–0.89 | 0.04 | |
| -PC ae C38:4 | - | 0.03 | ns | 0.72 | 0.08 | 0.55–0.89 | 0.03 | |
| -PC ae C40:2 | - | 0.007 | ns | 0.78 | 0.08 | 0.62–0.93 | 0.008 | |
| PC ae C42:2 | - | 0.004 | 0.04 | 0.79 | 0.07 | 0.64–0.95 | 0.005 | |
| SM(OH) C 22:1 | - | 0.010 | ns | 0.77 | 0.08 | 0.6–0.93 | 0.01 | |
| SM(OH) C 22:2 | - | 0.01 | ns | 0.76 | 0.08 | 0.6–0.92 | 0.01 | |
| LC-MS/MS | HIS | + | 0.0004 | 0.001 | 0.89 | 0.06 | 0.77–1 | 0.0009 |
| ORN | - | 0.01 | 0.010 | 0.79 | 0.08 | 0.63–0.96 | 0.03 | |
| PHE | + | 0.03 | 0.010 | 0.75 | 0.09 | 0.57–0.93 | 0.03 | |
| THR | + | 0.001 | 0.002 | 0.86 | 0.07 | 0.71–1 | 0.002 | |
| SERUM | ||||||||
|---|---|---|---|---|---|---|---|---|
| METABOLITES | RR vs. PP | p-Value | Holm––Bonf. Correction | ROC-CURVE | ||||
| AUC | Std. Error | CI | p-Value | |||||
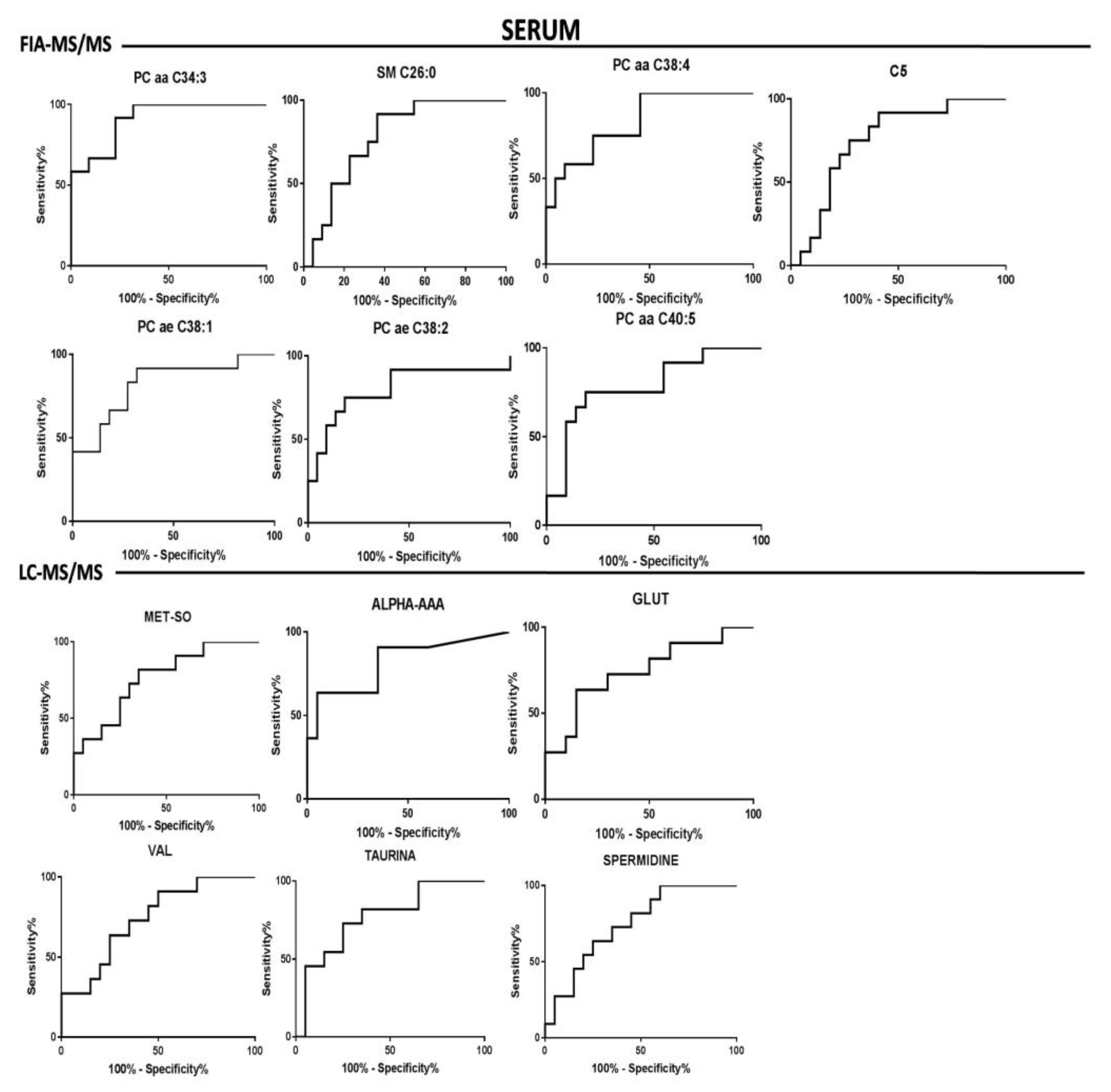
| FIA-MS/MS | PC aa C34:3 | + | <0.0001 | 0.001 | 0.91 | 0.05 | 0.81–1.00 | <0.0001 |
| PC aa C38:4 | - | 0.0010 | 0.005 | 0.83 | 0.07 | 0.70–0.97 | 0.001 | |
| PC ae C38:1 | + | 0.0016 | 0.006 | 0.82 | 0.08 | 0.67–0.97 | 0.002 | |
| PC ae C38:2 | + | 0.0036 | 0.011 | 0.80 | 0.08 | 0.62–0.97 | 0.004 | |
| PC aa C40:5 | - | 0.0059 | 0.012 | 0.78 | 0.08 | 0.61–0.95 | 0.007 | |
| SM C26:0 | - | 0.006 | 0.012 | 0.79 | 0.08 | 0.63–0.93 | 0.008 | |
| C5 | - | 0.0149 | 0.012 | 0.75 | 0.08 | 0.6–0.92 | 0.015 | |
| LC-MS/MS | MET-SO | + | 0.010 | 0.040 | 0.76 | 0.08 | 0.59–0.94 | 0.01 |
| ALPHA-AAA | - | 0.002 | 0.010 | 0.81 | 0.08 | 0.65–0.98 | 0.003 | |
| GLU | - | 0.02 | 0.040 | 0.74 | 0.09 | 0.56–0.93 | 0.02 | |
| VAL | - | 0.02 | 0.040 | 0.74 | 0.09 | 0.56–0.92 | 0.02 | |
| TAU | - | 0.01 | 0.040 | 0.77 | 0.08 | 0.59–0.94 | 0.01 | |
| SPER | - | 0.02 | 0.040 | 0.75 | 0.08 | 0.57–0.92 | 0.02 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murgia, F.; Lorefice, L.; Poddighe, S.; Fenu, G.; Secci, M.A.; Marrosu, M.G.; Cocco, E.; Atzori, L. Multi-Platform Characterization of Cerebrospinal Fluid and Serum Metabolome of Patients Affected by Relapsing–Remitting and Primary Progressive Multiple Sclerosis. J. Clin. Med. 2020, 9, 863. https://doi.org/10.3390/jcm9030863
Murgia F, Lorefice L, Poddighe S, Fenu G, Secci MA, Marrosu MG, Cocco E, Atzori L. Multi-Platform Characterization of Cerebrospinal Fluid and Serum Metabolome of Patients Affected by Relapsing–Remitting and Primary Progressive Multiple Sclerosis. Journal of Clinical Medicine. 2020; 9(3):863. https://doi.org/10.3390/jcm9030863
Chicago/Turabian StyleMurgia, Federica, Lorena Lorefice, Simone Poddighe, Giuseppe Fenu, Maria Antonietta Secci, Maria Giovanna Marrosu, Eleonora Cocco, and Luigi Atzori. 2020. "Multi-Platform Characterization of Cerebrospinal Fluid and Serum Metabolome of Patients Affected by Relapsing–Remitting and Primary Progressive Multiple Sclerosis" Journal of Clinical Medicine 9, no. 3: 863. https://doi.org/10.3390/jcm9030863
APA StyleMurgia, F., Lorefice, L., Poddighe, S., Fenu, G., Secci, M. A., Marrosu, M. G., Cocco, E., & Atzori, L. (2020). Multi-Platform Characterization of Cerebrospinal Fluid and Serum Metabolome of Patients Affected by Relapsing–Remitting and Primary Progressive Multiple Sclerosis. Journal of Clinical Medicine, 9(3), 863. https://doi.org/10.3390/jcm9030863






