Improved Isolation and Culture of Urine-Derived Stem Cells (USCs) and Enhanced Production of Immune Cells from the USC-Derived Induced Pluripotent Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Flow Cytometry (FACS) Analysis
2.3. RNA Isolation and Reverse Transcriptase- Polymerase Chain Reaction (RT-PCR)
2.4. RNA Sequencing (RNA-seq) and Gene Ontology (GO)
2.5. Cell Proliferation Assay
2.6. Wound Healing Cell Migration Assay
2.7. Colony Forming Unit Fibroblast (CFU-F) Assay
2.8. Differentiation of USCs
2.9. Generation of iPSCs from USCs and PBMCs
2.10. Kidney Organoid Differentiation
2.11. Immunocytofluorescence Staining
2.12. HPC Differentiation
2.13. HPC Colony-Forming Unit (CFU) Assay
2.14. Statistical Analysis
3. Results
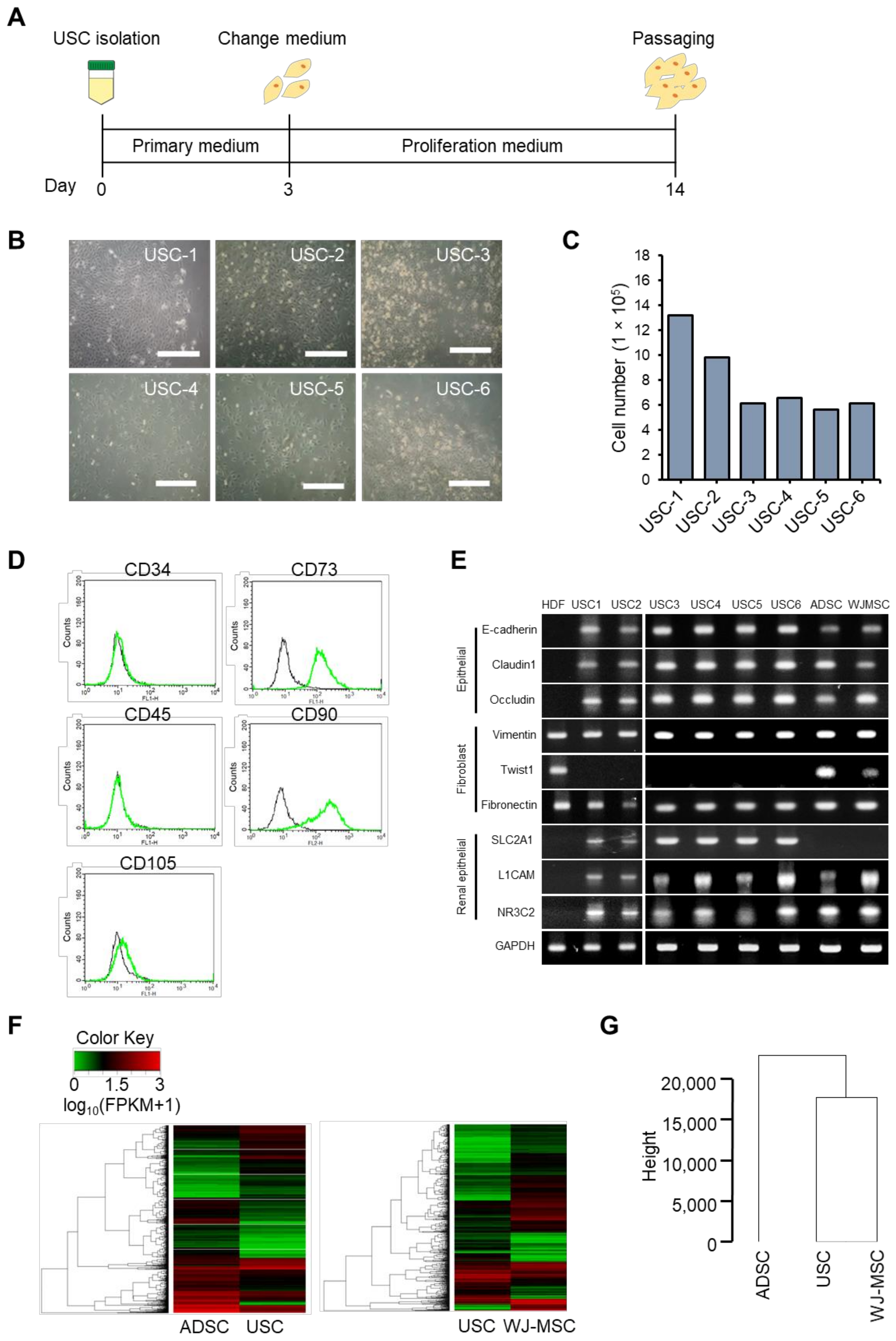
3.1. Isolation and Characterization of USCs
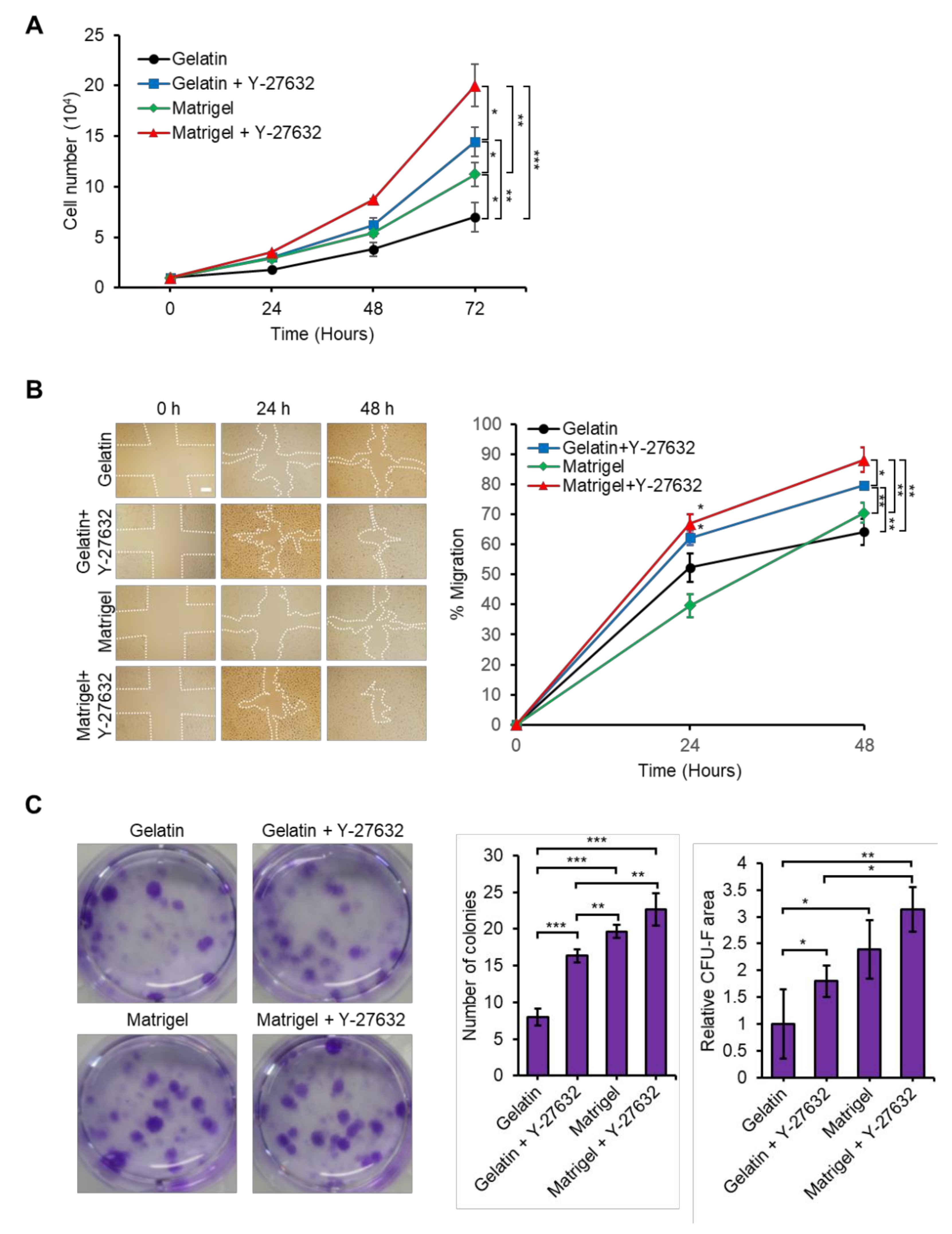
3.2. Y-27632 and Matrigel Enhance USC Isolation Efficiency
3.3. Y-27632 and Matrigel Enhance USCs Properties
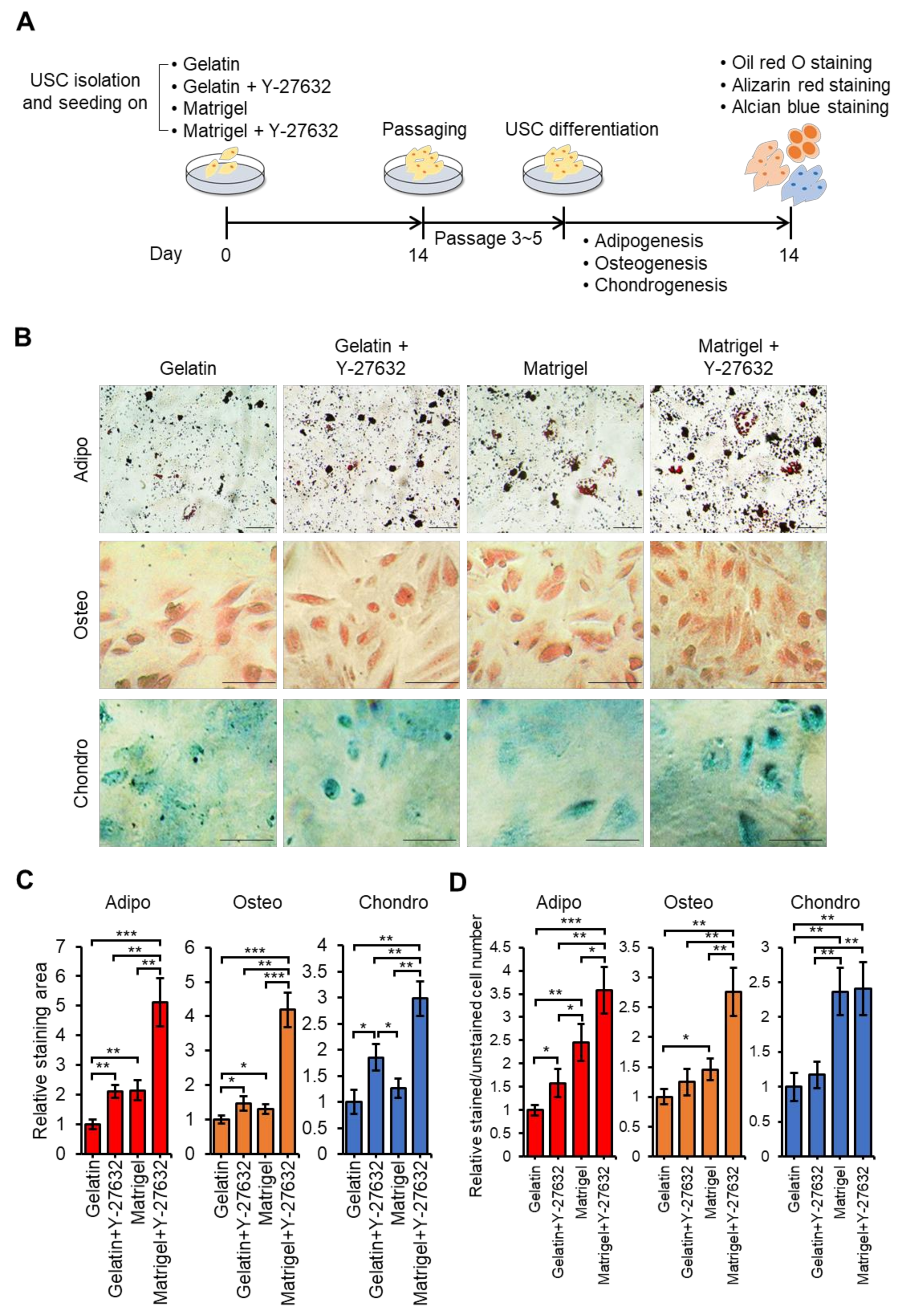
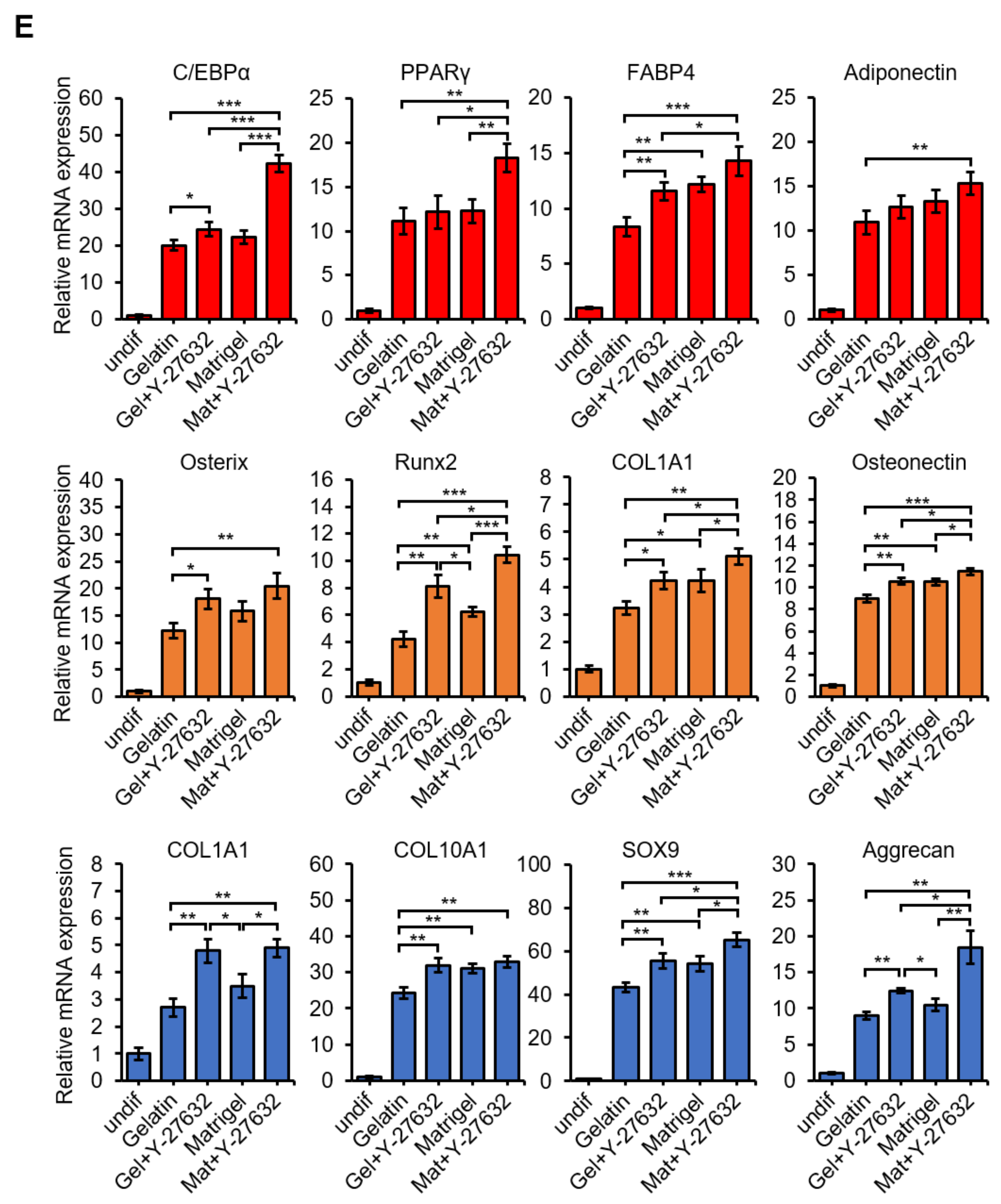
3.4. Y-27632 and Matrigel Enhance Differentiation of USCs
3.5. Flavonoids Increase the Isolation Efficiency of USCs
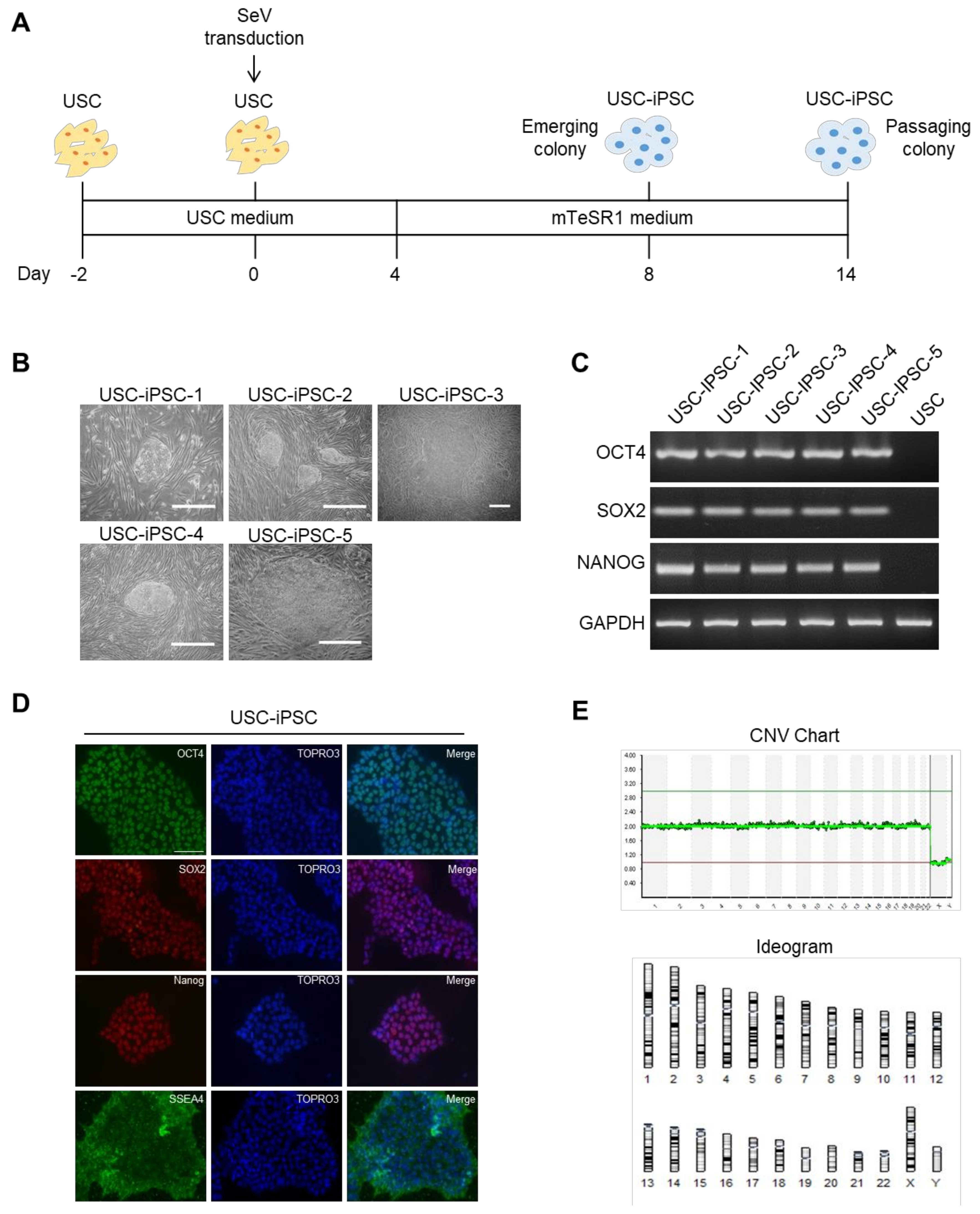
3.6. Generation of USC-iPSCs and Their Characterization
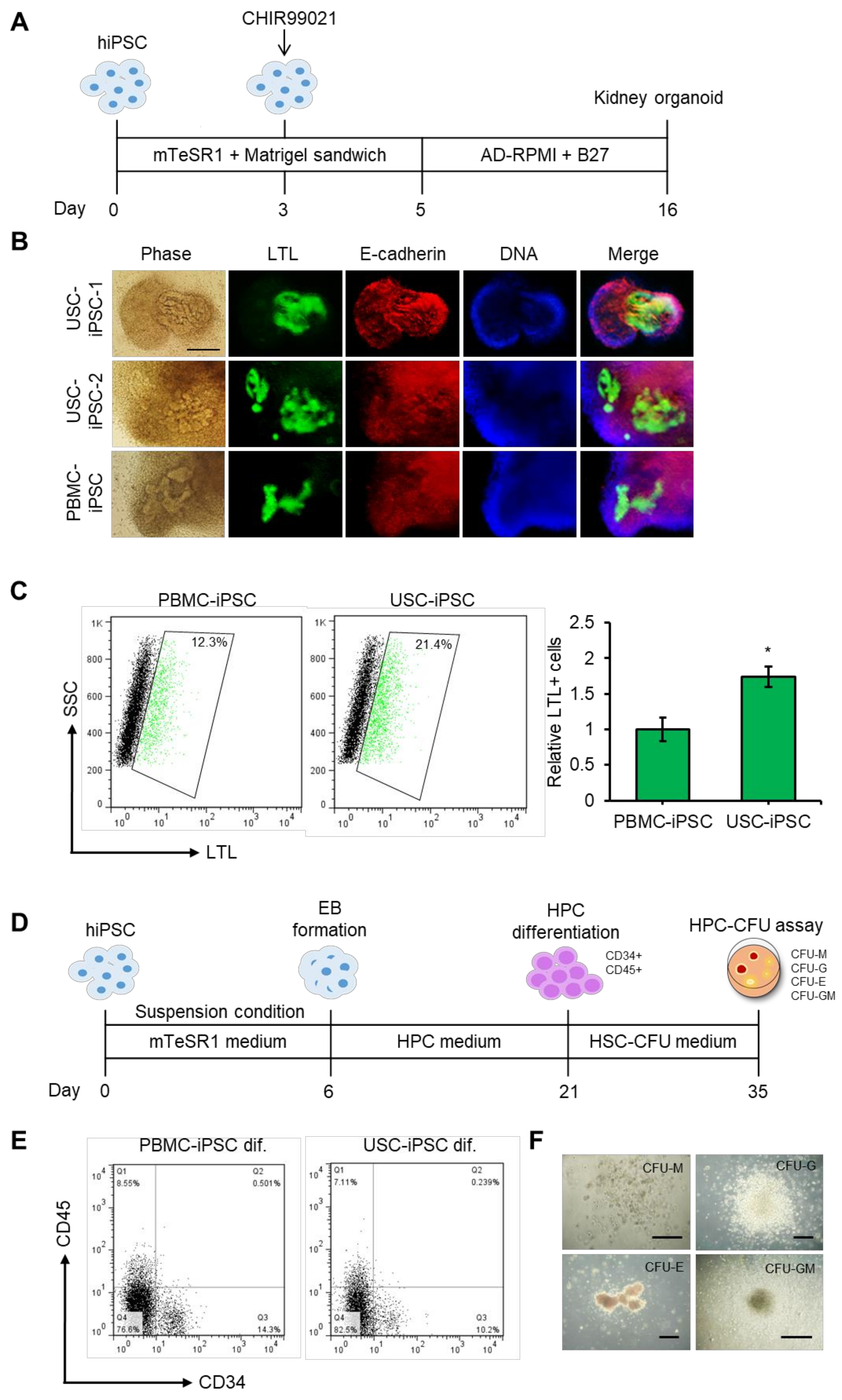
3.7. Differentiation Potential of USC-iPSCs into Kidney Organoids and HPCs.
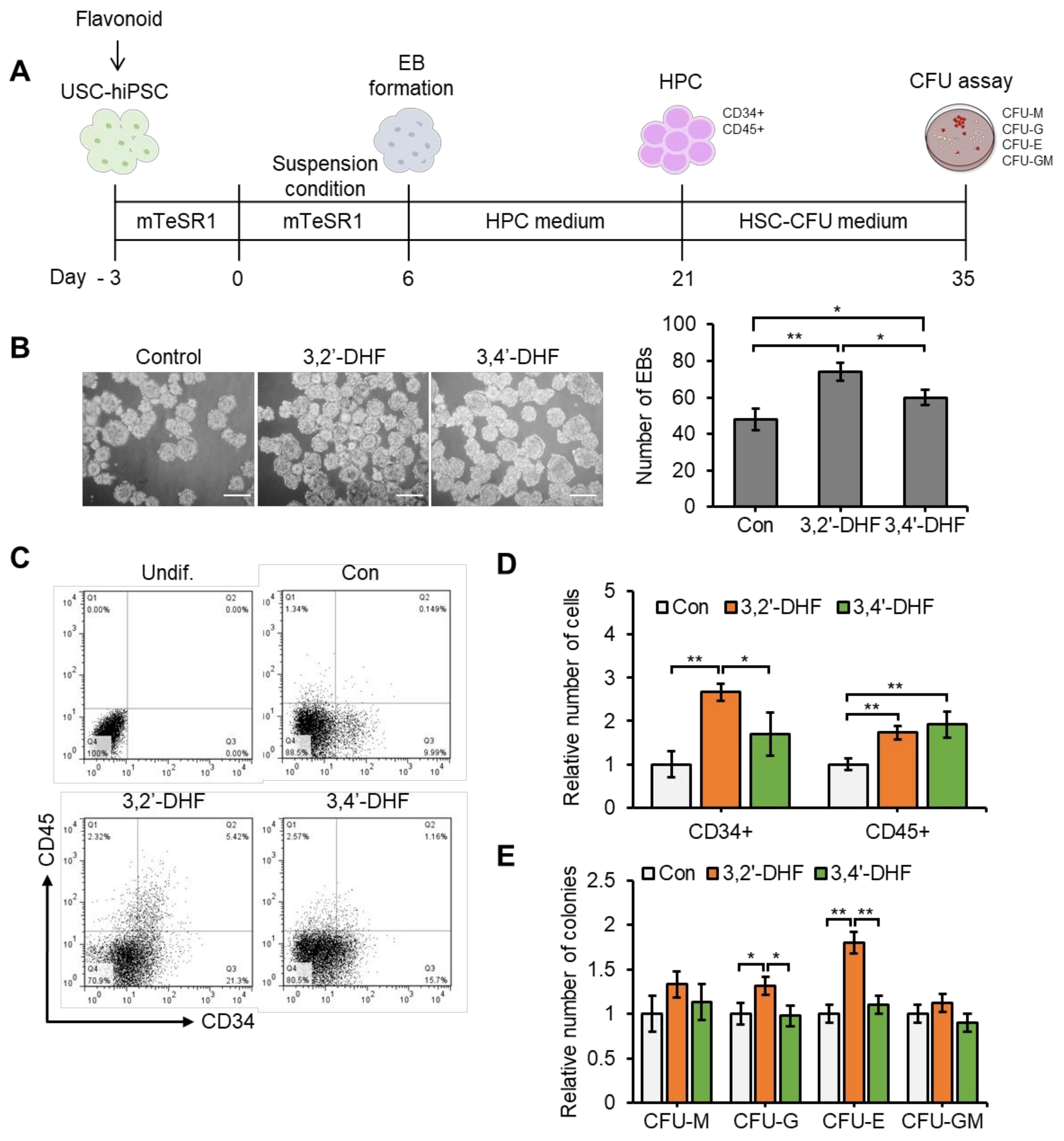
3.8. Flavonoid Treatment of USC-Derived iPSCs Enhances Capacities to Differentiate into HPCs.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gimble, J.M.; Ray, S.P.; Zanata, F.; Wu, X.; Wade, J.; Khoobehi, K.; Ferreira, L.M.; Bunnell, B.A. Adipose derived cells and tissues for regenerative medicine. ACS Biomater. Sci. Eng. 2017, 3, 1477–1482. [Google Scholar] [CrossRef]
- Tabar, V.; Studer, L. Pluripotent stem cells in regenerative medicine: Challenges and recent progress. Nat. Rev. Genet. 2014, 15, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Li, X.; Zhang, Y.; Han, Y.; Chang, F.; Ding, J. Mesenchymal Stem Cells for Regenerative Medicine. Cells 2019, 8, 886. [Google Scholar] [CrossRef] [PubMed]
- Keirstead, H.; Nistor, G.; Bernal, G.; Totoiu, M.; Cloutier, F.; Sharp, K.; Steward, O. Human Embryonic Stem Cell-Derived Oligodendrocyte Progenitor Cell Transplants Remyelinate and Restore Locomotion after Spinal Cord Injury. J. Neurosci. 2005, 25, 4694–4705. [Google Scholar] [CrossRef] [PubMed]
- Millman, J.R.; Xie, C.; Van Dervort, A.; Gürtler, M.; Pagliuca, F.W.; Melton, D.A. Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef]
- Soldner, F.; Hockemeyer, D.; Beard, C.; Gao, Q.; Bell, G.W.; Cook, E.G.; Hargus, G.; Blak, A.; Cooper, O.; Mitalipova, M.J.C. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 2009, 136, 964–977. [Google Scholar] [CrossRef]
- Kondo, T.; Asai, M.; Tsukita, K.; Kutoku, Y.; Ohsawa, Y.; Sunada, Y.; Imamura, K.; Egawa, N.; Yahata, N.; Okita, K.; et al. Modeling Alzheimer’s Disease with iPSCs Reveals Stress Phenotypes Associated with Intracellular Aβ and Differential Drug Responsiveness. Cell Stem Cell 2013, 12, 487–496. [Google Scholar] [CrossRef]
- Mazzini, L.; Ferrero, I.; Luparello, V.; Rustichelli, D.; Gunetti, M.; Mareschi, K.; Testa, L.; Stecco, A.; Tarletti, R.; Miglioretti, M.; et al. Mesenchymal stem cell transplantation in amyotrophic lateral sclerosis: A Phase I clinical trial. Exp. Neurol. 2010, 223, 229–237. [Google Scholar] [CrossRef]
- Burd, A.; Ahmed, K.; Lam, S.; Ayyappan, T.; Huang, L. Stem cell strategies in burns care. Burns 2007, 33, 282–291. [Google Scholar] [CrossRef]
- Brusen, R.M.; Cheng, R.K.; Masri, S.C.; Leedy, D.; Sorror, M.L. Moderate or severe valvular heart disease and outcomes in allogeneic stem cell transplantation. Int. J. Cardiol. 2019, 292, 166–170. [Google Scholar] [CrossRef]
- Bang, O.Y.; Lee, J.S.; Lee, P.H.; Lee, G. Autologous mesenchymal stem cell transplantation in stroke patients. Ann. Neurol. 2005, 57, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Knorr, D.A.; Ni, Z.; Hermanson, D.; Hexum, M.K.; Bendzick, L.; Cooper, L.J.; Lee, D.-J.; Kaufman, D.S. Clinical-Scale Derivation of Natural Killer Cells From Human Pluripotent Stem Cells for Cancer Therapy. Stem Cells Transl. Med. 2013, 2, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wu, X.; Liang, Y.; Gu, H.; Song, K.; Zou, X.; Zhou, G. Repair of cartilage defects in osteoarthritis rats with induced pluripotent stem cell derived chondrocytes. BMC Biotechnol. 2016, 16, 78. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.A. Human embryonic stem cell research: Ethical and legal issues. Nat. Rev. Genet. 2001, 2, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Locke, M.; Windsor, J.; Dunbar, P.R. Human adipose-derived stem cells: Isolation, characterization and applications in surgery. ANZ J. Surg. 2009, 79, 235–244. [Google Scholar] [CrossRef] [PubMed]
- García Quiroz, F.; Posada Estefan, O.M.; Gallego Pérez, D.; Higuita Castro, N.; Sarassa Velásquez, C.A.; Hansford, D.J.; Agudelo Florez, P.; López Rojas, L.E. Isolation of human bone marrow mesenchymal stem cells and evaluation of their osteogenic potential. Rev. Ing. Bioméd. 2008, 2, 48–55. [Google Scholar]
- Aggarwal, S.; Pittenger, M.F. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood 2005, 105, 1815–1822. [Google Scholar] [CrossRef]
- Bellini, E.; Grieco, M.P.; Raposio, E. A journey through liposuction and liposculture: Review. Ann. Med. Surg. 2017, 24, 53–60. [Google Scholar] [CrossRef]
- Cantu, C.; Pavlisko, E.N. Liposuction-Induced Fat Embolism Syndrome: A Brief Review and Postmortem Diagnostic Approach. Arch. Pathol. Lab. Med. 2018, 142, 871–875. [Google Scholar] [CrossRef]
- Kaoutzanis, C.; Gupta, V.; Winocour, J.; Layliev, J.; Ramirez, R.; Grotting, J.C.; Higdon, K.K. Cosmetic Liposuction: Preoperative Risk Factors, Major Complication Rates, and Safety of Combined Procedures. Aesthetic Surg. J. 2017, 37, 680–694. [Google Scholar] [CrossRef]
- Baghaei, K.; Hashemi, S.M.; Tokhanbigli, S.; Rad, A.A.; Assadzadeh-Aghdaei, H.; Sharifian, A.; Zali, M.R. Isolation, differentiation, and characterization of mesenchymal stem cells from human bone marrow. Gastroenterol. Gastroenterol. Hepatol. Bed Bench 2017, 10, 208–213. [Google Scholar] [PubMed]
- Kang, H.S.; Choi, S.H.; Kim, B.S.; Choi, J.Y.; Park, G.B.; Kwon, T.G.; Chun, S.Y. Advanced Properties of Urine Derived Stem Cells Compared to Adipose Tissue Derived Stem Cells in Terms of Cell Proliferation, Immune Modulation and Multi Differentiation. J. Korean Med. Sci. 2015, 30, 1764–1776. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wei, G.; Li, P.; Zhou, X.; Zhang, Y. Urine-derived stem cells: A novel and versatile progenitor source for cell-based therapy and regenerative medicine. Genes Dis. 2014, 1, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, G.; Shantaram, B.; Atala, A.; Zhang, Y. 736 Urine Derived Stem Cells with High TelomeRase Activity for Cell Based Therapy in Urology. J. Urol. 2012, 187, e302. [Google Scholar] [CrossRef]
- Qin, D.; Long, T.; Deng, J.; Zhang, Y. Urine-derived stem cells for potential use in bladder repair. Stem Cell Res. Ther. 2014, 5, 69. [Google Scholar] [CrossRef]
- Mullen, P. The use of Matrigel to facilitate the establishment of human cancer cell lines as xenografts. Methods Mol. Med. 2004, 88, 287–292. [Google Scholar]
- Lam, M.T.; Longaker, M.T. Comparison of several attachment methods for human iPS, embryonic and adipose-derived stem cells for tissue engineering. J. Tissue Eng. Regen. Med. 2012, 6, s80–s86. [Google Scholar] [CrossRef]
- Anguiano, M.; Castilla, C.; Maška, M.; Ederra, C.; Peláez, R.; Morales, X.; Muñoz-Arrieta, G.; Mujika, M.; Kozubek, M.; Muñoz-Barrutia, A. Characterization of three-dimensional cancer cell migration in mixed collagen-Matrigel scaffolds using microfluidics and image analysis. PLoS ONE 2017, 12, e0171417. [Google Scholar] [CrossRef]
- Shao, J.; Welch, W.J.; Diamond, M.I. ROCK and PRK-2 mediate the inhibitory effect of Y-27632 on polyglutamine aggregation. FEBS Lett. 2008, 582, 1637–1642. [Google Scholar] [CrossRef]
- Claassen, D.A.; Desler, M.M.; Rizzino, A. ROCK inhibition enhances the recovery and growth of cryopreserved human embryonic stem cells and human induced pluripotent stem cells. Mol. Reprod. Dev. 2009, 76, 722–732. [Google Scholar] [CrossRef]
- Li, Z.; Han, S.; Wang, X.; Han, F.; Zhu, X.; Zheng, Z.; Wang, H.; Zhou, Q.; Wang, Y.; Su, L.; et al. Rho kinase inhibitor Y-27632 promotes the differentiation of human bone marrow mesenchymal stem cells into keratinocyte-like cells in xeno-free conditioned medium. Stem Cell Res. 2015, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Heng, B.C. Effect of Rho-associated kinase (ROCK) inhibitor Y-27632 on the post-thaw viability of cryopreserved human bone marrow-derived mesenchymal stem cells. Tissue Cell 2009, 41, 376–380. [Google Scholar] [CrossRef] [PubMed]
- George, V.C.; Dellaire, G.; Rupasinghe, H.P.V. Plant flavonoids in cancer chemoprevention: Role in genome stability. J. Nutr. Biochem. 2017, 45, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Amawi, H.; Ashby, C.R., Jr.; Tiwari, A.K. Cancer chemoprevention through dietary flavonoids: What’s limiting? Chin. J. Cancer 2017, 36, 50. [Google Scholar] [CrossRef]
- Kim, J.H.; Song, M.; Kang, G.H.; Lee, E.R.; Choi, H.Y.; Lee, C.; Kim, J.H.; Kim, Y.; Koo, B.N.; Cho, S.G. Combined treatment of 3-hydroxyflavone and imatinib mesylate increases apoptotic cell death of imatinib mesylate-resistant leukemia cells. Leuk. Res. 2012, 36, 1157–1164. [Google Scholar] [CrossRef]
- Lee, E.-R.; Kang, Y.-J.; Kim, J.-H.; Lee, H.T.; Cho, S.-G. Modulation of apoptosis in HaCaT keratinocytes via differential regulation of ERK signaling pathway by flavonoids. J. Biol. Chem. 2005, 280, 31498–31507. [Google Scholar] [CrossRef]
- Lee, K.S.; Kim, E.Y.; Jeon, K.; Cho, S.G.; Han, Y.J.; Yang, B.C.; Lee, S.S.; Ko, M.S.; Riu, K.J.; Lee, H.T.; et al. 3,4-Dihydroxyflavone acts as an antioxidant and antiapoptotic agent to support bovine embryo development in vitro. J. Reprod. Dev. 2011, 57, 127–134. [Google Scholar] [CrossRef]
- Abdal Dayem, A.; Choi, H.Y.; Kim, Y.B.; Cho, S.G. Antiviral effect of methylated flavonol isorhamnetin against influenza. PLoS ONE 2015, 10, e0121610. [Google Scholar] [CrossRef]
- Hossain, M.K.; Choi, H.Y.; Hwang, J.S.; Dayem, A.A.; Kim, J.H.; Kim, Y.B.; Poo, H.; Cho, S.G. Antiviral activity of 3,4’-dihydroxyflavone on influenza a virus. J. Microbiol. 2014, 52, 521–526. [Google Scholar] [CrossRef]
- Kawser Hossain, M.; Abdal Dayem, A.; Han, J.; Yin, Y.; Kim, K.; Kumar Saha, S.; Yang, G.M.; Choi, H.Y.; Cho, S.G. Molecular Mechanisms of the Anti-Obesity and Anti-Diabetic Properties of Flavonoids. Int. J. Mol. Sci. 2016, 17, 569. [Google Scholar] [CrossRef]
- Han, D.; Kim, H.J.; Choi, H.Y.; Kim, B.; Yang, G.; Han, J.; Dayem, A.A.; Lee, H.-R.; Kim, J.H.; Lee, K.-M. 3,2′-dihydroxyflavone-treated pluripotent stem cells show enhanced proliferation, pluripotency marker expression, and neuroprotective properties. Cell Transplant. 2015, 24, 1511–1532. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell 2012, 10, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Dayem, A.A.; Gil, M.; Yang, G.M.; Lee, S.B.; Kwon, O.H.; Choi, S.; Kang, G.H.; Lim, K.M.; Kim, D.; et al. 3,2′-Dihydroxyflavone Improves the Proliferation and Survival of Human Pluripotent Stem Cells and Their Differentiation into Hematopoietic Progenitor Cells. J. Clin. Med. 2020, 9, 669. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Benda, C.; Duzinger, S.; Huang, Y.; Li, X.; Li, Y.; Guo, X.; Cao, G.; Chen, S.; Hao, L. Generation of induced pluripotent stem cells from urine. J. Am. Soc. Nephrol. 2011, 22, 1221–1228. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46. [Google Scholar] [CrossRef]
- Qian, S.-W.; Li, X.; Zhang, Y.-Y.; Huang, H.-Y.; Liu, Y.; Sun, X.; Tang, Q. Characterization of adipocyte differentiation from human mesenchymal stem cells in bone marrow. BMC Dev. Boil. 2010, 10, 47. [Google Scholar] [CrossRef]
- Solchaga, L.A.; Penick, K.J.; Welter, J.F. Chondrogenic Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells: Tips and Tricks. In Mesenchymal Stem Cell Assays and Applications; Springer: Berlin/Heidelberg, Germany, 2011; pp. 253–278. [Google Scholar]
- Jaiswal, N.; Haynesworth, S.E.; Caplan, A.I.; Bruder, S.P. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J. Cell. Biochem. 1997, 64, 295–312. [Google Scholar] [CrossRef]
- Donzelli, E.; Salvade, A.; Mimo, P.; Viganò, M.; Morrone, M.; Papagna, R.; Carini, F.; Zaopo, A.; Miloso, M.; Baldoni, M.; et al. Mesenchymal stem cells cultured on a collagen scaffold: In vitro osteogenic differentiation. Arch. Oral Boil. 2007, 52, 64–73. [Google Scholar] [CrossRef]
- Freedman, B.S.; Brooks, C.R.; Lam, A.Q.; Fu, H.; Morizane, R.; Agrawal, V.; Saad, A.F.; Li, M.K.; Hughes, M.R.; Vander Werff, R. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 2015, 6, 8715. [Google Scholar] [CrossRef]
- Galić, Z.; Kitchen, S.G.; Subramanian, A.; Bristol, G.; Marsden, M.D.; Balamurugan, A.; Kacena, A.; Yang, O.; Zack, J.A. Generation of T lineage cells from human embryonic stem cells in a feeder free system. Stem Cells 2009, 27, 100–107. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rönn, R.E.; Guibentif, C.; Moraghebi, R.; Chaves, P.; Saxena, S.; Garcia, B.; Woods, N.B. Retinoic acid regulates hematopoietic development from human pluripotent stem cells. Stem Cell Rep. 2015, 4, 269–281. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, Y.; Han, X.; Quintana Bustamante, O.; Bessa de Castro, R.; Zhang, K.; Zhang, P.; Li, Y.; Liu, Z.; Liu, X.; Ferrari, M. Highly efficient genome editing of human hematopoietic stem cells via a nano-silicon-blade delivery approach. Integr. Biol. 2017, 9, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Pacini, S.; Barachini, S.; Montali, M.; Carnicelli, V.; Fazzi, R.; Parchi, P.; Petrini, M. Mesangiogenic progenitor cells derived from one novel CD64brightCD31brightCD14neg population in human adult bone marrow. Stem Cells Dev. 2016, 25, 661–673. [Google Scholar] [CrossRef]
- Bharadwaj, S.; Liu, G.; Shi, Y.; Markert, C.; Andersson, K.-E.; Atala, A.; Zhang, Y. Characterization of urine-derived stem cells obtained from upper urinary tract for use in cell-based urological tissue engineering. Tissue Eng. Part A 2011, 17, 2123–2132. [Google Scholar] [CrossRef]
- Cho, K.-A.; Park, M.; Kim, Y.-H.; Woo, S.-Y.; Ryu, K.-H. RNA sequencing reveals a transcriptomic portrait of human mesenchymal stem cells from bone marrow, adipose tissue, and palatine tonsils. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Christodoulou, I.; Kolisis, F.; Papaevangeliou, D.; Zoumpourlis, V. Comparative evaluation of human mesenchymal stem cells of fetal (Wharton’s jelly) and adult (adipose tissue) origin during prolonged in vitro expansion: Considerations for cytotherapy. Stem Cells Int. 2013, 2013. [Google Scholar] [CrossRef]
- Chen, M.-Y.; Lie, P.-C.; Li, Z.-L.; Wei, X. Endothelial differentiation of Wharton’s jelly–derived mesenchymal stem cells in comparison with bone marrow–derived mesenchymal stem cells. Exp. Hematol. 2009, 37, 629–640. [Google Scholar] [CrossRef]
- Bharadwaj, S.; Liu, G.; Shi, Y.; Wu, R.; Yang, B.; He, T.; Fan, Y.; Lu, X.; Zhou, X.; Liu, H. Multipotential differentiation of human urine-derived stem cells: Potential for therapeutic applications in urology. Stem Cells 2013, 31, 1840–1856. [Google Scholar] [CrossRef]
- Watanabe, K.; Ueno, M.; Kamiya, D.; Nishiyama, A.; Matsumura, M.; Wataya, T.; Takahashi, J.B.; Nishikawa, S.; Nishikawa, S.-i.; Muguruma, K. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007, 25, 681–686. [Google Scholar] [CrossRef]
- Sohni, A.; Verfaillie, C.M. Mesenchymal stem cells migration homing and tracking. Stem Cells Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Liu, G.; Halim, A.; Ju, Y.; Luo, Q.; Song, A.G. Mesenchymal Stem Cell Migration and Tissue Repair. Cells 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jiang, J. Regulatory factors of mesenchymal stem cell migration into injured tissues and their signal transduction mechanisms. Front. Med. 2011, 5, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Pochampally, R. Colony forming unit assays for MSCs. In Mesenchymal Stem Cells; Springer: Berlin/Heidelberg, Germany, 2008; pp. 83–91. [Google Scholar]
- Han, J.; Choi, H.Y.; Dayem, A.A.; Kim, K.; Yang, G.; Won, J.; Do, S.H.; Kim, J.H.; Jeong, K.S.; Cho, S.G. Regulation of adipogenesis through differential modulation of ROS and kinase signaling pathways by 3, 4′-dihydroxyflavone treatment. J. Cell. Biochem. 2017, 118, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, C.C.; Fontes, A.; Ravinder, N.; Kuninger, D.; Kaur, J.; Bailey, M.; Taliana, A.; Vemuri, M.C.; Lieu, P.T. Generation of human-induced pluripotent stem cells by a nonintegrating RNA Sendai virus vector in feeder-free or xeno-free conditions. Stem Cells Int. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jiang, W.-J.; Sun, C.; Hou, C.-Z.; Yang, X.-M.; Gao, J.-G. Induced pluripotent stem cells: Origins, applications, and future perspectives. J. Zhejiang Univ. Sci. B 2013, 14, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Kern, S.; Eichler, H.; Stoeve, J.; Klüter, H.; Bieback, K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells 2006, 24, 1294–1301. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef]
- Rice, D.P.; Aberg, T.; Chan, Y.; Tang, Z.; Kettunen, P.J.; Pakarinen, L.; Maxson, R.E.; Thesleff, I. Integration of FGF and TWIST in calvarial bone and suture development. Development 2000, 127, 1845–1855. [Google Scholar]
- Johnson, D.; Iseki, S.; Wilkie, A.O.; Morriss-Kay, G.M. Expression patterns of Twist and Fgfr1, -2 and -3 in the developing mouse coronal suture suggest a key role for twist in suture initiation and biogenesis. Mech. Dev 2000, 91, 341–345. [Google Scholar] [CrossRef]
- Quarto, N.; Senarath-Yapa, K.; Renda, A.; Longaker, M.T. TWIST1 silencing enhances in vitro and in vivo osteogenic differentiation of human adipose-derived stem cells by triggering activation of BMP-ERK/FGF signaling and TAZ upregulation. Stem Cells 2015, 33, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Rowland, T.J.; Miller, L.M.; Blaschke, A.J.; Doss, E.L.; Bonham, A.J.; Hikita, S.T.; Johnson, L.V.; Clegg, D.O. Roles of integrins in human induced pluripotent stem cell growth on Matrigel and vitronectin. Stem Cells Dev. 2009, 19, 1231–1240. [Google Scholar] [CrossRef] [PubMed]
- Baharvand, H.; Salekdeh, G.H.; Taei, A.; Mollamohammadi, S. An efficient and easy-to-use cryopreservation protocol for human ES and iPS cells. Nat. Protoc. 2010, 5, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Martin-Ibanez, R.; Unger, C.; Strömberg, A.; Baker, D.; Canals, J.; Hovatta, O. Novel cryopreservation method for dissociated human embryonic stem cells in the presence of a ROCK inhibitor. Hum. Reprod. 2008, 23, 2744–2754. [Google Scholar] [CrossRef] [PubMed]
- Emre, N.; Vidal, J.G.; Elia, J.; O’Connor, E.D.; Paramban, R.I.; Hefferan, M.P.; Navarro, R.; Goldberg, D.S.; Varki, N.M.; Marsala, M. The ROCK inhibitor Y-27632 improves recovery of human embryonic stem cells after fluorescence-activated cell sorting with multiple cell surface markers. PLoS ONE 2010, 5, e12148. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Yoshimura, A.; Kaneko, T.; Sato, K.; Hara, Y. ROCK inhibitor Y-27632 maintains the proliferation of confluent human mesenchymal stem cells. J. Periodontal Res. 2014, 49, 363–370. [Google Scholar] [CrossRef]
- Mellott, A.J.; Godsey, M.E.; Shinogle, H.E.; Moore, D.S.; Forrest, M.L.; Detamore, M.S. Improving viability and transfection efficiency with human umbilical cord wharton’s jelly cells through use of a ROCK inhibitor. Cell. Reprogram. 2014, 16, 91–97. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Yan, X.; Liu, H.; Zhang, L.; Yao, A.; Guo, C.; Liu, X.; Xu, T. The Rho kinase inhibitor Y-27632 facilitates the differentiation of bone marrow mesenchymal stem cells. J. Mol. Histol. 2014, 45, 707–714. [Google Scholar] [CrossRef]
- Xu, C.; Inokuma, M.S.; Denham, J.; Golds, K.; Kundu, P.; Gold, J.D.; Carpenter, M.K. Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 2001, 19, 971–974. [Google Scholar] [CrossRef]
- Zachar, L.; Bačenková, D.; Rosocha, J. Activation, homing, and role of the mesenchymal stem cells in the inflammatory environment. J. Inflamm. Res. 2016, 9, 231. [Google Scholar] [CrossRef]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.; Ji, H.; Ehrlich, L. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-R.; Kim, J.-H.; Kang, Y.-J.; Cho, S.-G. The anti-apoptotic and anti-oxidant effect of eriodictyol on UV-induced apoptosis in keratinocytes. Biol. Pharm. Bull. 2007, 30, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-R.; Kim, J.-Y.; Kang, Y.-J.; Ahn, J.-Y.; Kim, J.-H.; Kim, B.-W.; Choi, H.-Y.; Jeong, M.-Y.; Cho, S.-G. Interplay between PI3K/Akt and MAPK signaling pathways in DNA-damaging drug-induced apoptosis. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2006, 1763, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-R.; Kim, J.-H.; Choi, H.Y.; Jeon, K.; Cho, S.-G. Cytoprotective effect of eriodictyol in UV-irradiated keratinocytes via phosphatase-dependent modulation of both the p38 MAPK and Akt signaling pathways. Cell. Physiol. Biochem. 2011, 27, 513–524. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Gil, M.; Dayem, A.A.; Choi, S.; Kang, G.-H.; Yang, G.-M.; Cho, S.; Jeong, Y.; Kim, S.J.; Seok, J.; et al. Improved Isolation and Culture of Urine-Derived Stem Cells (USCs) and Enhanced Production of Immune Cells from the USC-Derived Induced Pluripotent Stem Cells. J. Clin. Med. 2020, 9, 827. https://doi.org/10.3390/jcm9030827
Kim K, Gil M, Dayem AA, Choi S, Kang G-H, Yang G-M, Cho S, Jeong Y, Kim SJ, Seok J, et al. Improved Isolation and Culture of Urine-Derived Stem Cells (USCs) and Enhanced Production of Immune Cells from the USC-Derived Induced Pluripotent Stem Cells. Journal of Clinical Medicine. 2020; 9(3):827. https://doi.org/10.3390/jcm9030827
Chicago/Turabian StyleKim, Kyeongseok, Minchan Gil, Ahmed Abdal Dayem, Sangbaek Choi, Geun-Ho Kang, Gwang-Mo Yang, Sungha Cho, Yeojin Jeong, Se Jong Kim, Jaekwon Seok, and et al. 2020. "Improved Isolation and Culture of Urine-Derived Stem Cells (USCs) and Enhanced Production of Immune Cells from the USC-Derived Induced Pluripotent Stem Cells" Journal of Clinical Medicine 9, no. 3: 827. https://doi.org/10.3390/jcm9030827
APA StyleKim, K., Gil, M., Dayem, A. A., Choi, S., Kang, G.-H., Yang, G.-M., Cho, S., Jeong, Y., Kim, S. J., Seok, J., Kwak, H. J., Kumar Saha, S., Kim, A., & Cho, S.-G. (2020). Improved Isolation and Culture of Urine-Derived Stem Cells (USCs) and Enhanced Production of Immune Cells from the USC-Derived Induced Pluripotent Stem Cells. Journal of Clinical Medicine, 9(3), 827. https://doi.org/10.3390/jcm9030827







