Changes in the Intestinal Microbiota Are Seen Following Treatment with Infliximab in Children with Crohn’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Next-Generation Sequencing (NGS) and Bioinformatic Analysis
2.3. Ethical Considerations
3. Results
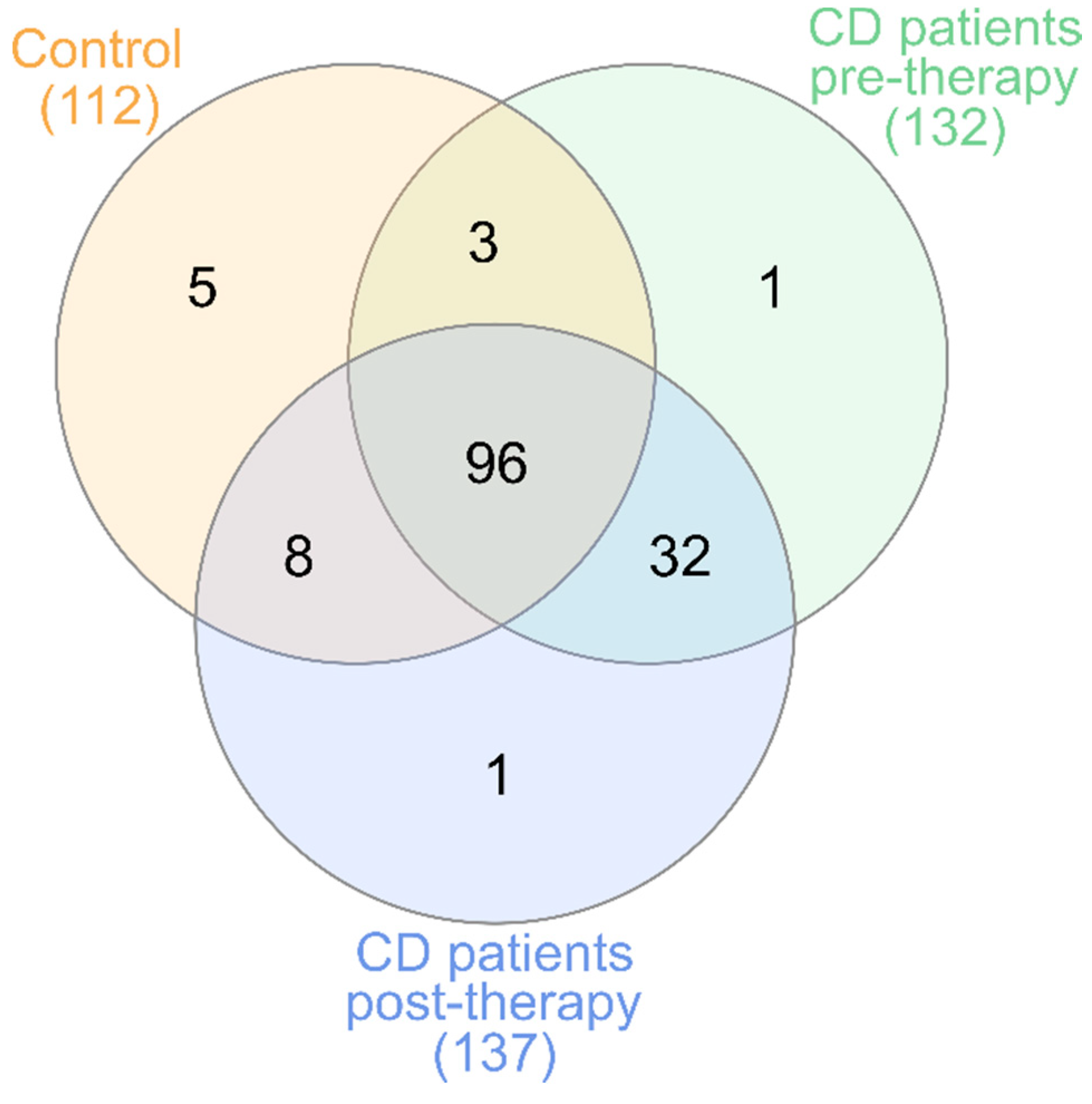
3.1. 16.S rRNA Sequencing Analysis
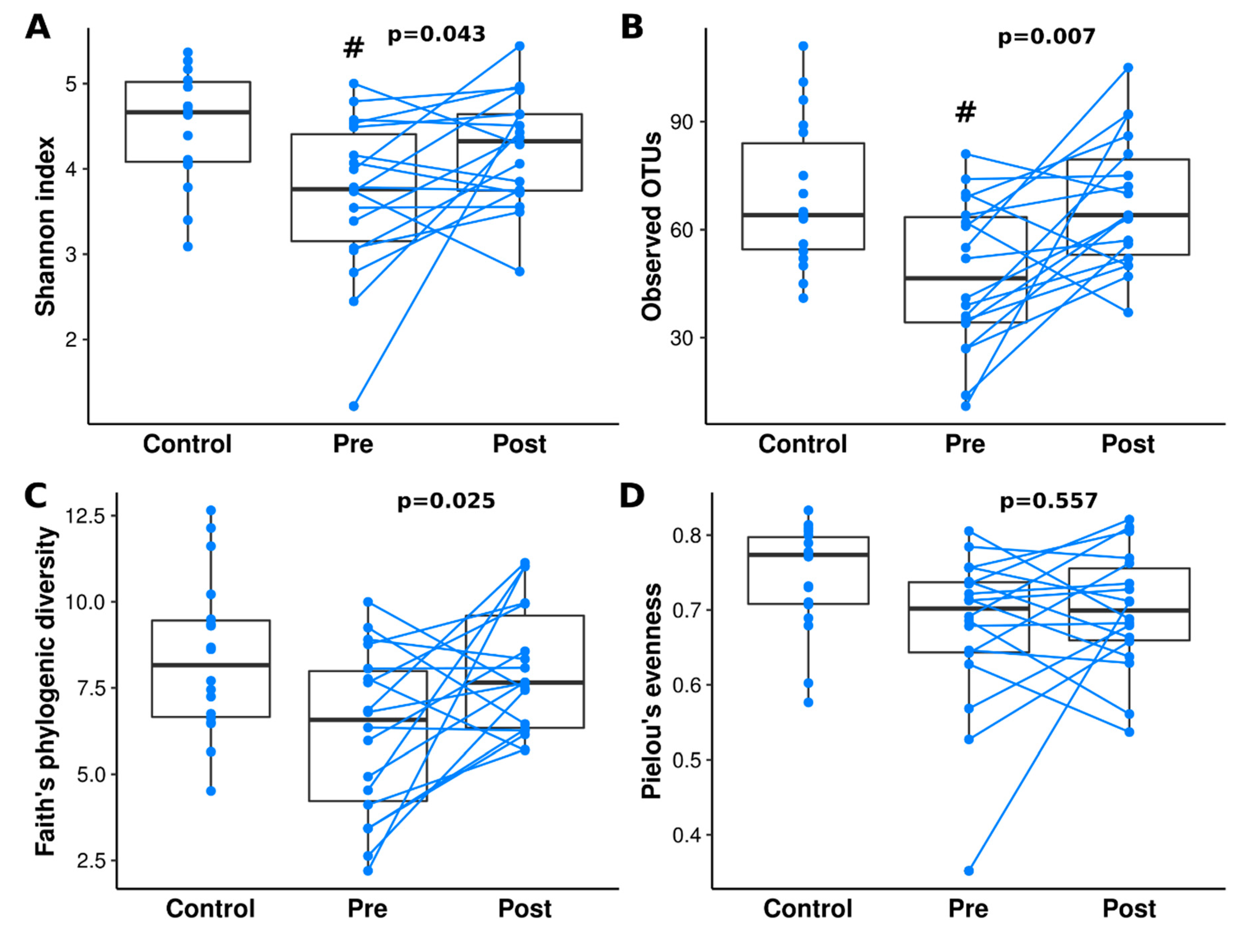
3.2. Diversity Analysis
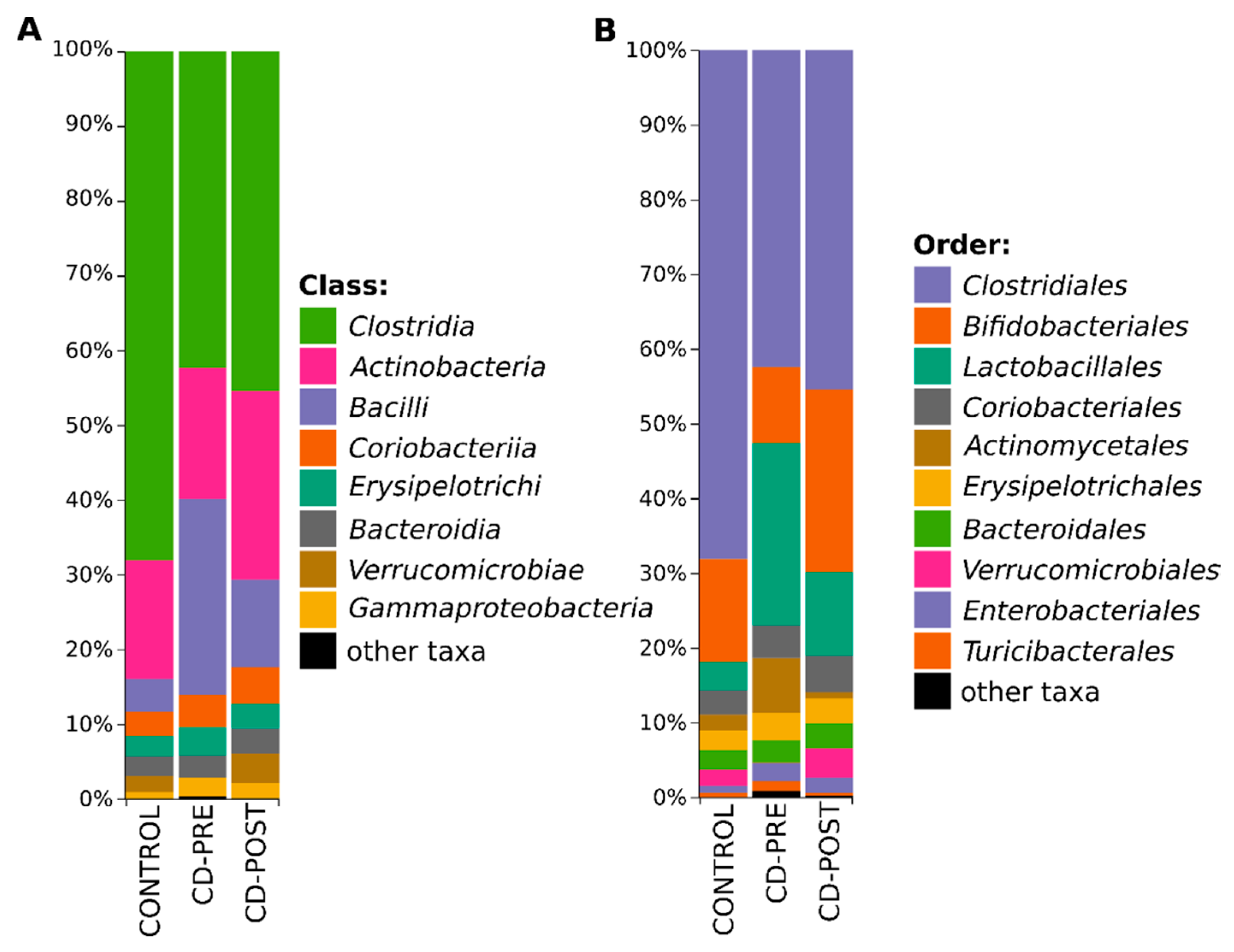
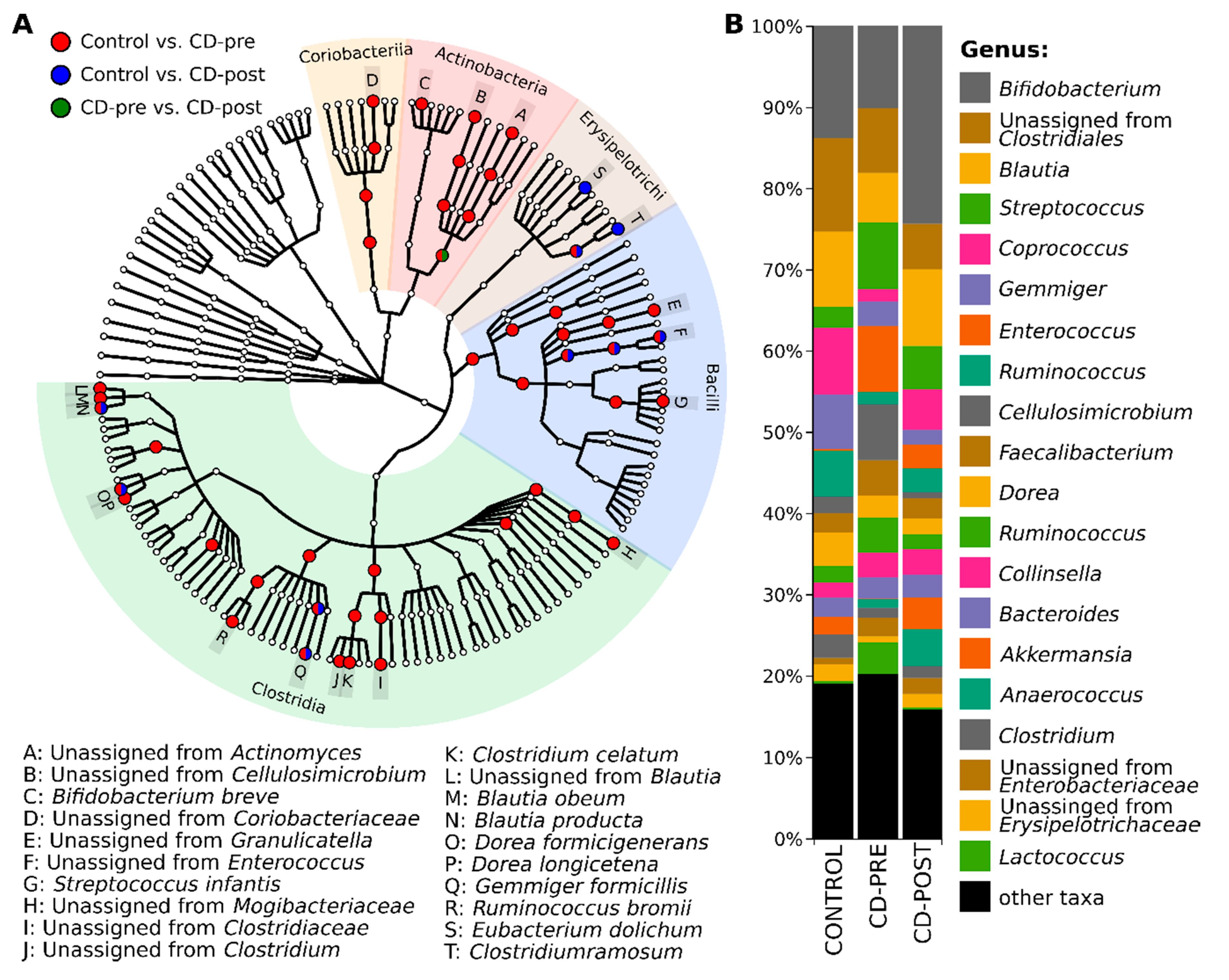
3.3. Bacterial Profile
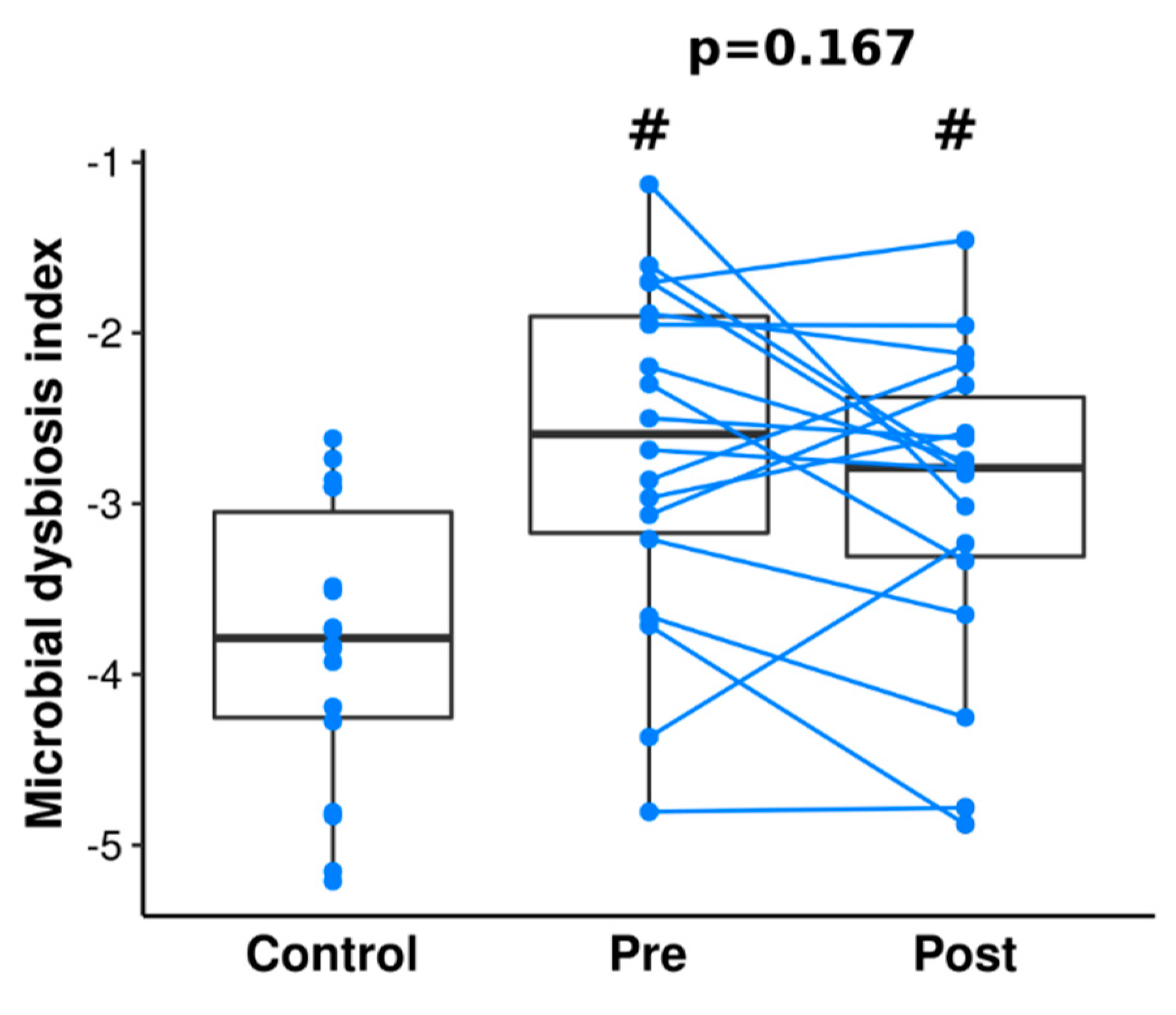
3.4. Gut Microbiome Differences and Dysbiosis between Groups
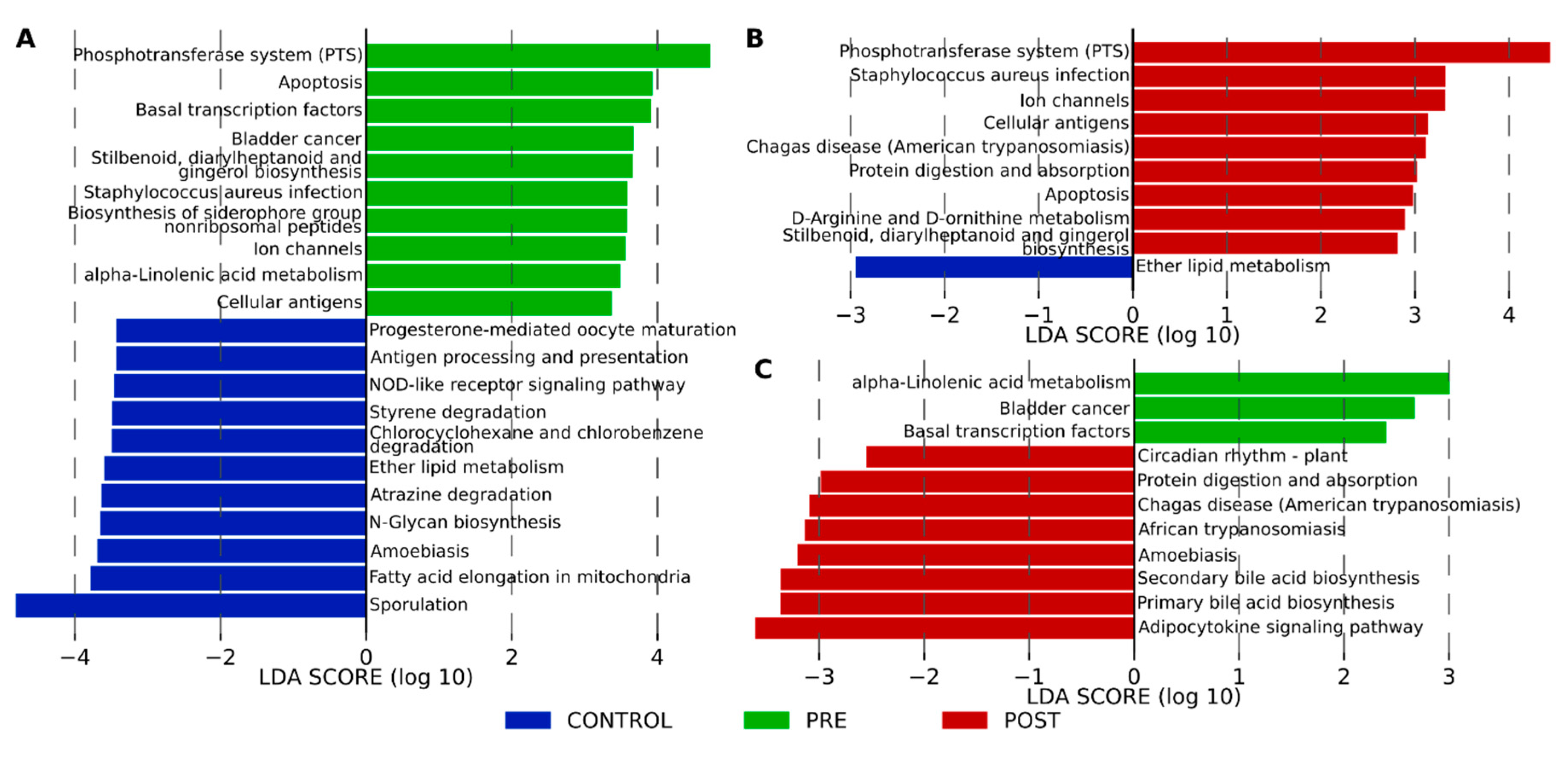
3.5. Functional Profiling with KEGG Pathway Functions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.; Chan, F.K.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Benchimol, E.I.; Fortinsky, K.J.; Gozdyra, P.; Van den Heuvel, M.; Van Limbergen, J.; Griffiths, A.M. Epidemiology of pediatric inflammatory bowel disease: A systematic review of international trends. Inflamm. Bowel Dis. 2011, 17, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Burisch, J.; Jess, T.; Martinato, M.; Lakatos, P.L.; ECCO-EpiCom. The burden of inflammatory bowel disease in Europe. J. Crohn’s Colitis 2013, 7, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Sartor, R.B.; Wu, G.D. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 2017, 152, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.A.; Bertha, M.; Hofmekler, T.; Chopra, P.; Vatanen, T.; Srivatsa, A.; Prince, J.; Kumar, A.; Sauer, C.; Zwick, M.E.; et al. Dysbiosis, inflammation, and response to treatment: A longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 2016, 8, 75. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Integrative HMP iHMP Research Network Consortium. The integrative human microbiome project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef]
- Jones-Hall, Y.L.; Nakatsu, C.H. The intersection of TNF, IBD and the microbiome. Gut Microbes 2016, 7, 58–62. [Google Scholar] [CrossRef]
- Himmel, M.E.; Yao, Y.; Orban, P.C.; Steiner, T.S.; Levings, M.K. Regulatory T-cell therapy for inflammatory bowel disease: More questions than answers. Immunology 2012, 136, 115–122. [Google Scholar] [CrossRef]
- Lee, D.; Albenberg, L.; Compher, C.; Baldassano, R.; Piccoli, D.; Lewis, J.D.; Wu, G.D. Diet in the pathogenesis and treatment of inflammatory bowel diseases. Gastroenterology 2015, 148, 1087–1106. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Tlaskalová-Hogenová, H.E.; Tucková, L.; Stepánková, R.; Hudcovic, T.; Palová-Jelínková, L.E.; Kozáková, H.; Rossmann, P.; Sanchez, D.; Cinová, J.; Hrnčíř, T.; et al. Involvement of innate immunity in the development of inflammatory and autoimmune diseases. Ann. N. Y. Acad. Sci. 2005, 1051, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.K.; Kamm, M.A.; Teo, S.M.; Inouye, M.; Wagner, J.; Kirkwood, C.D. Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: A systematic review. Inflamm. Bowel Dis. 2015, 21, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Fyderek, K.; Strus, M.; Kowalska-Duplaga, K.; Gosiewski, T.; Wędrychowicz, A.; Jedynak-Wąsowicz, U.; Sładek, M.; Pieczarkowski, S.; Adamski, P.; Kochan, P.; et al. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J. Gastroenterol. 2009, 15, 5287–5294. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, K.; Kanai, T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef]
- Kang, S.; Denman, S.E.; Morrison, M.; Yu, Z.; Dore, J.; Leclerc, M.; McSweeney, C.S. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm. Bowel Dis. 2010, 16, 2034–2042. [Google Scholar] [CrossRef]
- Kolho, K.L.; Korpela, K.; Jaakkola, T.; Pichai, M.V.; Zoetendal, E.G.; Salonen, A.; De Vos, W.M. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am. J. Gastroenterol. 2015, 110, 921–930. [Google Scholar] [CrossRef]
- Busquets, D.; Mas-de-Xaxars, T.; López-Siles, M.; Martínez-Medina, M.; Bahí, A.; Sàbat, M.; Louvriex, R.; Miquel-Cusachs, J.O.; Garcia-Gil, J.L.; Aldeguer, X. Anti-tumour necrosis factor treatment with adalimumab induces changes in the microbiota of Crohn’s disease. J. Crohn’s Colitis 2015, 9, 899–906. [Google Scholar] [CrossRef]
- Ruemmele, F.M.; Veres, G.; Kolho, K.L.; Griffiths, A.; Levine, A.; Escher, J.C.; Amil Dias, J.; Barabino, A.; Braegger, C.P.; Bronsky, J.; et al. Consensus guidelines of ECCO/ESPGHAN on the medical management of pediatric Crohn’s disease. J. Crohn’s Colitis 2014, 8, 1179–1207. [Google Scholar] [CrossRef]
- Corica, D.; Romano, C. Biological therapy in pediatric inflammatory bowel disease. J. Clin. Gastroenterol. 2017, 51, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Guariso, G.; Gasparetto, M. Treating children with inflammatory bowel disease: Current and new perspectives. World J. Gastroenterol. 2017, 23, 5469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, X.; Ghozlane, A.; Hu, H.; Li, X.; Xiao, Y.; Li, D.; Yu, G.; Zhang, T. Characteristics of faecal microbiota in paediatric Crohn’s disease and their dynamic changes during infliximab therapy. J. Crohn’s Colitis 2018, 12, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, M.K.; Strid, H.; Sapnara, M.; Lasson, A.; Bajor, A.; Ung, K.A.; Öhman, L. Anti-TNF therapy response in patients with ulcerative colitis is associated with colonic antimicrobial peptide expression and microbiota composition. J. Crohn’s Colitis 2016, 10, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Varum, F.; Bravo, R.; Furrer, E.; Bojic, D.; Basit, A.W. Inflammatory bowel disease: Exploring gut pathophysiology for novel therapeutic targets. Transl. Res. 2016, 176, 38–68. [Google Scholar] [CrossRef]
- Levine, A.; Koletzko, S.; Turner, D.; Escher, J.C.; Cucchiara, S.; De Ridder, L.; Kolho, K.L.; Veres, G.; Russell, R.K.; Paerregaard, A.; et al. ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J. Pediatr. Gastroenterol. Nutr. 2014, 58, 795–806. [Google Scholar] [CrossRef]
- Levine, A.; Griffiths, A.; Markowitz, J.; Wilson, D.C.; Turner, D.; Russell, R.K.; Fell, J.; Ruemmele, F.M.; Walters, T.; Sherlock, M.; et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: The Paris classification. Inflamm. Bowel. Dis. 2011, 17, 1314–1321. [Google Scholar] [CrossRef]
- Daperno, M.; D’Haens, G.; Van Assche, G.; Baert, F.; Bulois, P.; Maunoury, V.; Sostegni, R.; Rocca, R.; Pera, A.; Gevers, A.; et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: The SES-CD. Gastrointest. Endosc. 2004, 60, 505–512. [Google Scholar] [CrossRef]
- Kowalska-Duplaga, K.; Gosiewski, T.; Kapusta, P.; Sroka-Oleksiak, A.; Wędrychowicz, A.; Pieczarkowski, S.; Ludwig-Słomczyńska, A.H.; Wołkow, P.P.; Fyderek, K. Differences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn’s disease. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2, High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Janssen, S.; McDonald, D.; Gonzalez, A.; Navas-Molina, J.A.; Jiang, L.; Xu, Z.Z.; Winker, K.; Kado, D.M.; Orwoll, E.; Manary, M.; et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. Msystems 2018, 3, e00021-18. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 2011, 12, 2825–2830. Available online: http://scikit-learn.sourceforge.net (accessed on 12 January 2020).
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Imhann, F.; Vila, A.V.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; Musfeldt, M.; Wenderoth, D.F.; Hampe, J.; Brant, O.; Fölsch, U.R.; Timmis, K.N.; Schreiber, S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004, 53, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Joossens, M.; Huys, G.; Cnockaert, M.; De Preter, V.; Verbeke, K.; Rutgeerts, P.; Vandamme, P.; Vermeire, S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 2011, 60, 631–637. [Google Scholar] [CrossRef]
- Martinez-Medina, M.; Aldeguer, X.; Gonzalez-Huix, F.; Acero, D.; Garcia-Gil, J.L. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm. Bowel Dis. 2006, 12, 1136–1145. [Google Scholar] [CrossRef]
- Andoh, A.; Kuzuoka, H.; Tsujikawa, T.; Nakamura, S.; Hirai, F.; Suzuki, Y.; Matsui, T.; Fujiyama, Y.; Matsumoto, T. Multicenter analysis of fecal microbiota profiles in Japanese patients with Crohn’s disease. J. Gastroenterol. 2012, 47, 1298–1307. [Google Scholar] [CrossRef]
- Barrett, J.C.; Hansoul, S.; Nicolae, D.L.; Cho, J.H.; Duerr, R.H.; Rioux, J.D.; Brant, S.R.; Silverberg, M.S.; Taylor, K.D.; Barmada, M.M.; et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008, 40, 955–962. [Google Scholar] [CrossRef]
- Hampe, J.; Grebe, J.; Nikolaus, S.; Solberg, C.; Croucher, P.J.; Mascheretti, S.; Jahnsen, J.; Moum, B.; Klump, B.; Krawczak, M.; et al. Association of NOD2 (CARD 15) genotype with clinical course of Crohn’s disease: A cohort study. Lancet 2002, 359, 1661–1665. [Google Scholar] [CrossRef]
- Frank, D.N.; Robertson, C.E.; Hamm, C.M.; Kpadeh, Z.; Zhang, T.; Chen, H.; Zhu, W.; Sartor, R.B.; Boedeker, E.C.; Harpaz, N.; et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 2011, 17, 179–184. [Google Scholar] [CrossRef]
- Aden, K.; Rehman, A.; Waschina, S.; Pan, W.H.; Walker, A.; Lucio, M.; Nunez, A.M.; Bharti, R.; Zimmerman, J.; Bethge, J.; et al. Metabolic functions of gut microbes associate with efficacy of tumor necrosis factor antagonists in patients with inflammatory bowel diseases. Gastroenterology 2019, 157, 1279–1292. [Google Scholar] [CrossRef]
- Moustafa, A.; Li, W.; Anderson, E.L.; Wong, E.H.; Dulai, P.S.; Sandborn, W.J.; Biggs, W.; Yooseph, S.; Jones, M.B.; Venter, J.C.; et al. Genetic risk, dysbiosis, and treatment stratification using host genome and gut microbiome in inflammatory bowel disease. Clin. Transl. Gastroenterol. 2018, 9, e132. [Google Scholar] [CrossRef] [PubMed]
- Olbjørn, C.; Småstuen, M.C.; Thiis-Evensen, E.; Nakstad, B.; Vatn, M.H.; Jahnsen, J.; Ricanek, P.; Vatn, S.; Moen, A.E.; Tannæs, T.M.; et al. Fecal microbiota profiles in treatment-naïve pediatric inflammatory bowel disease—Associations with disease phenotype, treatment, and outcome. Clin. Exp. Gastroenterol. 2019, 12, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Rivero, M.; Alonso, J.; Ramón, M.F.; Gonzales, N.; Pozo, A.; Marín, I.; Navascués, A.; Juanbeltz, R. Infections due to Cellulosimicrobium species: Case report and literature review. BMC Infect. Dis. 2019, 19, 816. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wang, H.; Liao, W.D.; Peng, C.; Shu, X.; Zhu, X.; Zhu, Z.H. Characteristics of mucosa-associated gut microbiota during treatment in Crohn’s disease. World J. Gastroenterol. 2019, 25, 2204–2216. [Google Scholar] [CrossRef]
- Balfour Sartor, R.; Muehlbauer, M. Microbial host interactions in IBD: Implications for pathogenesis and therapy. Curr. Gastroenterol. Rep. 2007, 9, 497–507. [Google Scholar] [CrossRef]
- Knox, N.C.; Forbes, J.D.; Van Domselaar, G.; Bernstein, C.N. The gut microbiome as a target for IBD treatment: Are we there yet? Curr. Treat Options Gastroenterol. 2019, 17, 115–126. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Luo, C.; Yajnik, V.; Khalili, H.; Garber, J.J.; Stevens, B.W.; Cleland, T.; Xavier, R.J. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe 2017, 21, 603–610. [Google Scholar] [CrossRef]
- Doherty, M.K.; Ding, T.; Koumpouras, C.; Telesco, S.E.; Monast, C.; Das, A.; Brodmerkel, C.; Schloss, P.D. Fecal microbiota signatures are associated with response to ustekinumab therapy among Crohn’s Disease patients. MBio 2018, 9, e02120-17. [Google Scholar] [CrossRef]
- Takahashi, M.K.; Tan, X.; Dy, A.J.; Braff, D.; Akana, R.T.; Furuta, Y.; Donghia, N.; Ananthakrishnan, A.; Collins, J.J. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]

| Characteristics | Biological Therapy—IFX (n = 18) | Control Group (n = 18) |
|---|---|---|
| Male:Female, n (ratio) | 11:7 (1.57) | 8:10 (0.8) |
| Age at Diagnosis, months | 147.18 ± 45.56 | N/A |
| Age at Inclusion into the Study; months | 160.28 ± 44.56 * | 138.28 ± 35.16 †,* |
| Weight kg | 44.25 ± 14.23 | 41.75 ± 17.37 |
| Height, cm | 152.16 ± 18.67 | 146.67 ± 20.52 |
| BMI, kg/m2 | 18.54 ± 3.17 | 18.3 ± 3.49 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalska-Duplaga, K.; Kapusta, P.; Gosiewski, T.; Sroka-Oleksiak, A.; Ludwig-Słomczyńska, A.H.; Wołkow, P.P.; Fyderek, K. Changes in the Intestinal Microbiota Are Seen Following Treatment with Infliximab in Children with Crohn’s Disease. J. Clin. Med. 2020, 9, 687. https://doi.org/10.3390/jcm9030687
Kowalska-Duplaga K, Kapusta P, Gosiewski T, Sroka-Oleksiak A, Ludwig-Słomczyńska AH, Wołkow PP, Fyderek K. Changes in the Intestinal Microbiota Are Seen Following Treatment with Infliximab in Children with Crohn’s Disease. Journal of Clinical Medicine. 2020; 9(3):687. https://doi.org/10.3390/jcm9030687
Chicago/Turabian StyleKowalska-Duplaga, Kinga, Przemysław Kapusta, Tomasz Gosiewski, Agnieszka Sroka-Oleksiak, Agnieszka H. Ludwig-Słomczyńska, Paweł P. Wołkow, and Krzysztof Fyderek. 2020. "Changes in the Intestinal Microbiota Are Seen Following Treatment with Infliximab in Children with Crohn’s Disease" Journal of Clinical Medicine 9, no. 3: 687. https://doi.org/10.3390/jcm9030687
APA StyleKowalska-Duplaga, K., Kapusta, P., Gosiewski, T., Sroka-Oleksiak, A., Ludwig-Słomczyńska, A. H., Wołkow, P. P., & Fyderek, K. (2020). Changes in the Intestinal Microbiota Are Seen Following Treatment with Infliximab in Children with Crohn’s Disease. Journal of Clinical Medicine, 9(3), 687. https://doi.org/10.3390/jcm9030687