Characterization of Non-Toxigenic Clostridioides difficile Strains Isolated from Preterm Neonates and In Vivo Study of Their Protective Effect
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic and Genotypic Characteristics of C. difficile Strains Isolated in Three Cohorts of Preterm Neonates
2.1.1. Description of the Cohorts
2.1.2. C. difficile Isolation and Strain Characterization
2.1.3. PCR-Ribotyping
2.1.4. Multi-Locus Variable-Number Tandem-Reapeat Analysis (MLVA)
2.1.5. Multi-Locus Sequence Typing (MLST)
2.2. In Vivo Study of NTCD Properties to Prevent C. difficile Infection
2.2.1. C. difficile Strain and Spore Preparation
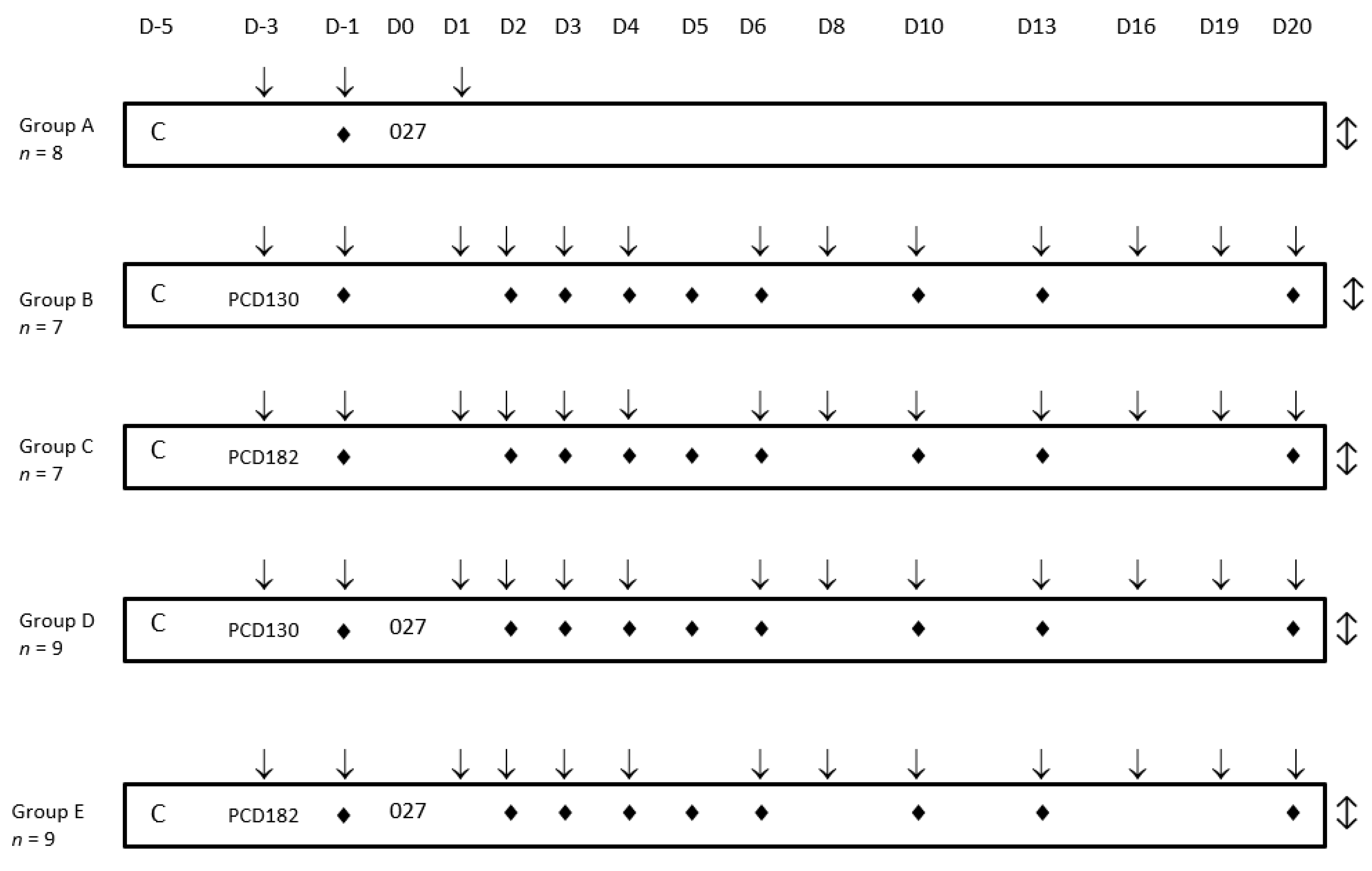
2.2.2. Animal Study Design
2.2.3. Colonization Kinetics and Bacterial Counts
2.3. Statistical Analysis
3. Results
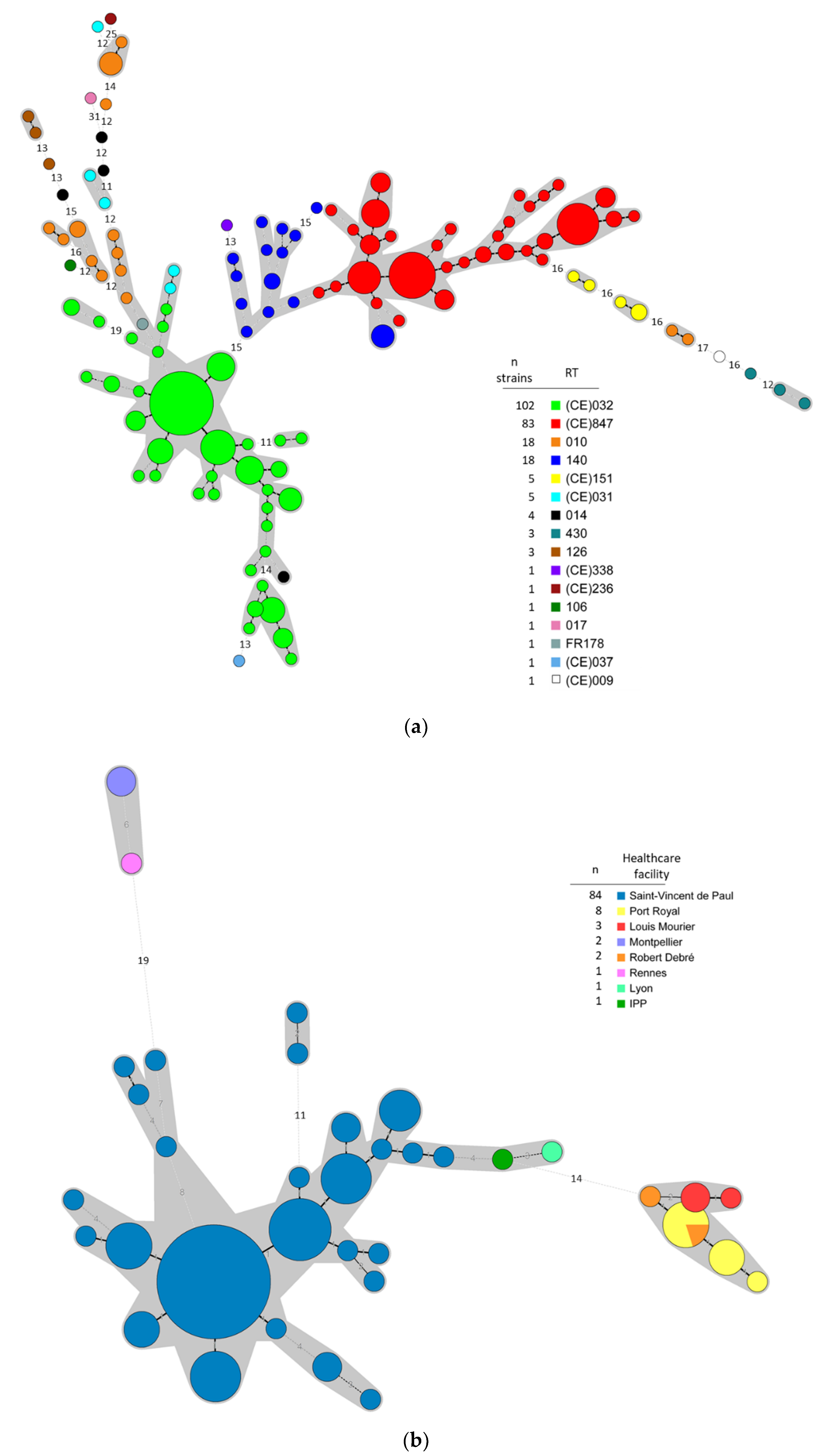
3.1. Typing Data Show that Predominant NTCD in Preterm Neonates Are Genetically Related
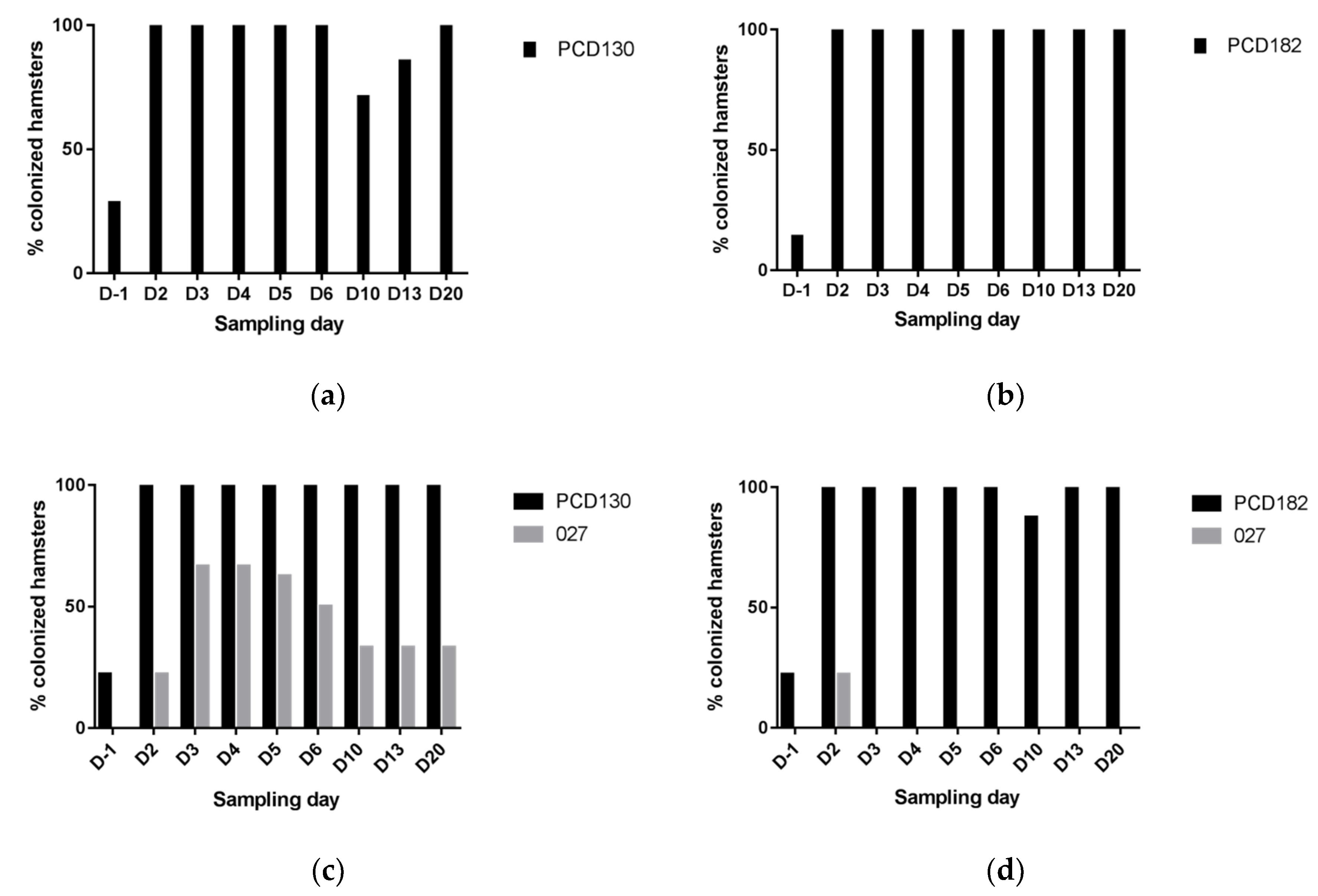
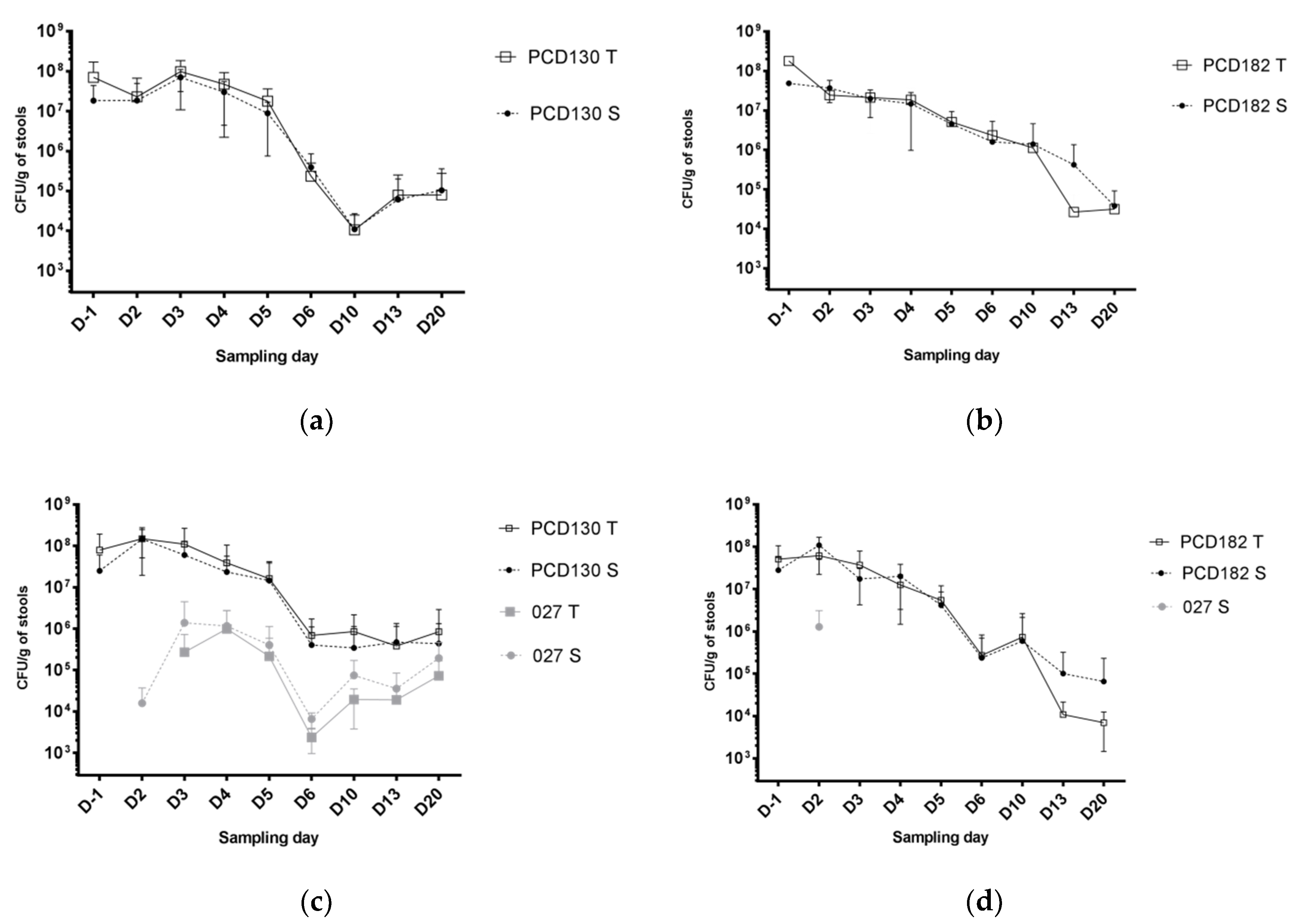
3.2. NTCD Have A Protective Effect against C. difficile Colonization and Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Patents
References
- Bartlett, J.G.; Gerding, D.N. Clinical Recognition and Diagnosis of Clostridium difficile Infection. Clin. Infect. Dis. 2008, 46, S12–S18. [Google Scholar] [CrossRef] [PubMed]
- Loo, V.G.; Bourgault, A.-M.; Poirier, L.; Lamothe, F.; Michaud, S.; Turgeon, N.; Toye, B.; Beaudoin, A.; Frost, E.H.; Gilca, R.; et al. Host and Pathogen Factors for Clostridium difficile Infection and Colonization. N. Engl. J. Med. 2011, 365, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V.; Mulligan, M.E.; Kwok, R.Y.; Stamm, W.E. Nosocomial acquisition of Clostridium difficile infection. N. Engl. J. Med. 1989, 320, 204–210. [Google Scholar] [CrossRef]
- DePestel, D.D.; Aronoff, D.M. Epidemiology of Clostridium difficile Infection. J. Pharm. Pract. 2013, 26, 464–475. [Google Scholar] [CrossRef]
- Fluit, A.C.; Wolfhagen, M.J.; Verdonk, G.P.; Jansze, M.; Torensma, R.; Verhoef, J. Nontoxigenic strains of Clostridium difficile lack the genes for both toxin A and toxin B. J. Clin. Microbiol. 1991, 29, 2666–2667. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, E.; Kato, H.; Kita, H.; Karasawa, T.; Maegawa, T.; Koino, Y.; Matsumoto, K.; Takada, T.; Nomoto, K.; Tanaka, R.; et al. Clostridium difficile colonization in healthy adults: Transient colonization and correlation with enterococcal colonization. J. Med. Microbiol. 2004, 53, 167–172. [Google Scholar] [CrossRef]
- Galdys, A.L.; Nelson, J.S.; Shutt, K.A.; Schlackman, J.L.; Pakstis, D.L.; Pasculle, A.W.; Marsh, J.W.; Harrison, L.H.; Curry, S.R. Prevalence and Duration of Asymptomatic Clostridium difficile Carriage among Healthy Subjects in Pittsburgh, Pennsylvania. J. Clin. Microbiol. 2014, 52, 2406–2409. [Google Scholar] [CrossRef]
- Ziakas, P.D.; Zacharioudakis, I.M.; Zervou, F.N.; Grigoras, C.; Pliakos, E.E.; Mylonakis, E. Asymptomatic carriers of toxigenic C. difficile in long-term care facilities: A meta-analysis of prevalence and risk factors. PLoS ONE 2015, 10, e0117195. [Google Scholar] [CrossRef]
- Clabots, C.R.; Johnson, S.; Olson, M.M.; Peterson, L.R.; Gerding, D.N. Acquisition of Clostridium difficile by hospitalized patients: Evidence for colonized new admissions as a source of infection. J. Infect. Dis. 1992, 166, 561–567. [Google Scholar] [CrossRef]
- Johnson, S.; Clabots, C.R.; Linn, F.V.; Olson, M.M.; Peterson, L.R.; Gerding, D.N. Nosocomial Clostridium difficile colonisation and disease. Lancet 1990, 336, 97–100. [Google Scholar] [CrossRef]
- Dubberke, E.R.; Reske, K.A.; Seiler, S.; Hink, T.; Kwon, J.H.; Burnham, C.-A.D. Risk Factors for Acquisition and Loss of Clostridium difficile Colonization in Hospitalized Patients. Antimicrob. Agents Chemother. 2015, 59, 4533–4543. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Seo, M.-R.; Kim, J.; Pai, H. Ribotype variability of Clostridioides difficile strains in patients with hospital-acquired C. difficile infections, community-acquired C. difficile infections, and colonization with toxigenic and non-toxigenic strains of C. difficile. Anaerobe 2019, 60, 102086. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.K.; Johnson, S.; Samore, M.H.; Bliss, D.Z.; Gerding, D.N. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 1998, 351, 633–636. [Google Scholar] [CrossRef]
- Guh, A.Y.; Kutty, P.K. Clostridioides difficile Infection. Ann. Intern. Med. 2018, 169, ITC49-64. [Google Scholar] [CrossRef] [PubMed]
- Gerding, D.N.; Meyer, T.; Lee, C.; Cohen, S.H.; Murthy, U.K.; Poirier, A.; Van Schooneveld, T.C.; Pardi, D.S.; Ramos, R.A.; Barron, M.A.; et al. Administration of Spores of Nontoxigenic Clostridium difficile Strain M3 for Prevention of Recurrent C difficile Infection: A Randomized Clinical Trial. JAMA 2015, 313, 1719. [Google Scholar] [CrossRef]
- Enoch, D.A.; Butler, M.J.; Pai, S.; Aliyu, S.H.; Karas, J.A. Clostridium difficile in children: Colonisation and disease. J. Infect. 2011, 63, 105–113. [Google Scholar] [CrossRef]
- Schutze, G.E.; Willoughby, R.E.; Committee on Infectious Diseases. American Academy of Pediatrics. Clostridium difficile infection in infants and children. Pediatrics 2013, 131, 196–200. [Google Scholar]
- Ferraris, L.; Couturier, J.; Eckert, C.; Delannoy, J.; Barbut, F.; Butel, M.J.; Aires, J. Carriage and colonization of C. difficile in preterm neonates: A longitudinal prospective study. PLoS ONE 2019, 14, e0212568. [Google Scholar] [CrossRef]
- Schönherr-Hellec, S.; Klein, G.L.; Delannoy, J.; Ferraris, L.; Rozé, J.C.; Butel, M.J.; Aires, J. Clostridial Strain-Specific Characteristics Associated with Necrotizing Enterocolitis. Appl. Environ. Microbiol. 2018, 84, e02428-17. [Google Scholar]
- Rozé, J.-C.; Ancel, P.-Y.; Lepage, P.; Martin-Marchand, L.; Al Nabhani, Z.; Delannoy, J.; Picaud, J.C.; Lapillonne, A.; Aires, J.; Durox, M.; et al. Nutritional strategies and gut microbiota composition as risk factors for necrotizing enterocolitis in very-preterm infants. Am. J. Clin. Nutr. 2017, 106, 821–830. [Google Scholar] [CrossRef]
- Lemee, L.; Dhalluin, A.; Testelin, S.; Mattrat, M.A.; Maillard, K.; Lemeland, J.F.; Pons, J.L. Multiplex PCR Targeting tpi (Triose Phosphate Isomerase), tcdA (Toxin A), and tcdB (Toxin B) Genes for Toxigenic Culture of Clostridium difficile. J. Clin. Microbiol. 2004, 42, 5710–5714. [Google Scholar] [CrossRef] [PubMed]
- Bidet, P.; Barbut, F.; Lalande, V.; Burghoffer, B.; Petit, J.-C. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol. Lett. 1999, 175, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Eckert, C.; Barbut, F. Spatio-temporal variability of the epidemic 027 Clostridium difficile strains in France based on MLVA typing. Anaerobe 2017, 48, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, D.; Fawley, W.; Kachrimanidou, M.; Bowden, R.; Crook, D.W.; Fung, R.; Golubchik, T.; Harding, R.M.; Jeffery, K.J.M.; Jolley, K.A.; et al. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 2010, 48, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Feil, E.J.; Chan, M.S.; Maiden, M.C. Sequence type analysis and recombinational tests (START). Bioinformatics 2001, 17, 1230–1231. [Google Scholar] [CrossRef]
- Marvaud, J.-C.; Quevedo-Torres, S.; Eckert, C.; Janoir, C.; Barbut, F. Virulence of new variant strains of Clostridium difficile producing only toxin A or binary toxin in the hamster model. New Microbes New Infect. 2019, 32, 100590. [Google Scholar] [CrossRef]
- Nagaro, K.J.; Phillips, S.T.; Cheknis, A.K.; Sambol, S.P.; Zukowski, W.E.; Johnson, S.; Gerding, D.N. Nontoxigenic Clostridium difficile protects hamsters against challenge with historic and epidemic strains of toxigenic BI/NAP1/027 C. difficile. Antimicrob. Agents Chemother. 2013, 57, 5266–5270. [Google Scholar] [CrossRef]
- Jangi, S.; Lamont, J.T. Asymptomatic colonization by Clostridium difficile in infants: Implications for disease in later life. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 2–7. [Google Scholar] [CrossRef]
- Rousseau, C.; Lemée, L.; Le Monnier, A.; Poilane, I.; Pons, J.-L.; Collignon, A. Prevalence and diversity of Clostridium difficile strains in infants. J. Med. Microbiol. 2011, 60, 1112–1118. [Google Scholar] [CrossRef]
- Stoesser, N.; Eyre, D.W.; Quan, T.P.; Godwin, H.; Pill, G.; Mbuvi, E.; Vaughan, A.; Griffiths, D.; Martin, J.; Fawley, W.; et al. Epidemiology of Clostridium difficile in infants in Oxfordshire, UK: Risk factors for colonization and carriage, and genetic overlap with regional C. difficile infection strains. PLoS ONE 2017, 12, e0182307. [Google Scholar] [CrossRef]
- Romano, V.; Pasquale, V.; Lemee, L.; El Meouche, I.; Pestel-Caron, M.; Capuano, F.; Buono, P.; Dumontet, S. Clostridioides difficile in the environment, food, animals and humans in southern Italy: Occurrence and genetic relatedness. Comp. Immunol. Microbiol. Infect. Dis. 2018, 59, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Camorlinga, M.; Sanchez-Rojas, M.; Torres, J.; Romo-Castillo, M. Phenotypic Characterization of Non-toxigenic Clostridioides difficile Strains Isolated from Patients in Mexico. Front. Microbiol. 2019, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- van der Lugt, N.M.; Steggerda, S.J.; Walther, F.J. Use of rifampin in persistent coagulase negative staphylococcal bacteremia in neonates. BMC Pediatr. 2010, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Borriello, S.P.; Barclay, F.E. Protection of hamsters against Clostridium difficile ileocaecitis by prior colonisation with non-pathogenic strains. J. Med. Microbiol. 1985, 19, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.H.; Sheagren, J.N. Antagonism of toxigenic Clostridium difficile by nontoxigenic C. difficile. J. Infect. Dis. 1983, 147, 733–736. [Google Scholar] [CrossRef]
- Sambol, S.P.; Merrigan, M.M.; Tang, J.K.; Johnson, S.; Gerding, D.N. Colonization for the prevention of Clostridium difficile disease in hamsters. J. Infect. Dis. 2002, 186, 1781–1789. [Google Scholar] [CrossRef]
- Abt, M.C.; McKenney, P.T.; Pamer, E.G. Clostridium difficile colitis: Pathogenesis and host defence. Nat. Rev. Microbiol. 2016, 14, 609–620. [Google Scholar] [CrossRef]



| Activity Score | Symptoms |
|---|---|
| 0 (animals to euthanize) | Immobility, bradypnea, swollen stomach, wet tail and legs, squinted eyes, bristled hair, areflexia, weight loss >20% |
| 1 | Motor retardation, trembling, swollen stomach, tachypnea, bristled hair, reaction to external stimuli, wet tail, isolation, weight loss >10% and <20% |
| 2 | Normal activity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Couturier, J.; Franconeri, L.; Janoir, C.; Ferraris, L.; Syed-Zaidi, R.; Youssouf, A.; Gateau, C.; Hoys, S.; Aires, J.; Barbut, F. Characterization of Non-Toxigenic Clostridioides difficile Strains Isolated from Preterm Neonates and In Vivo Study of Their Protective Effect. J. Clin. Med. 2020, 9, 3650. https://doi.org/10.3390/jcm9113650
Couturier J, Franconeri L, Janoir C, Ferraris L, Syed-Zaidi R, Youssouf A, Gateau C, Hoys S, Aires J, Barbut F. Characterization of Non-Toxigenic Clostridioides difficile Strains Isolated from Preterm Neonates and In Vivo Study of Their Protective Effect. Journal of Clinical Medicine. 2020; 9(11):3650. https://doi.org/10.3390/jcm9113650
Chicago/Turabian StyleCouturier, Jeanne, Léa Franconeri, Claire Janoir, Laurent Ferraris, Rabab Syed-Zaidi, Anlyata Youssouf, Cécile Gateau, Sandra Hoys, Julio Aires, and Frédéric Barbut. 2020. "Characterization of Non-Toxigenic Clostridioides difficile Strains Isolated from Preterm Neonates and In Vivo Study of Their Protective Effect" Journal of Clinical Medicine 9, no. 11: 3650. https://doi.org/10.3390/jcm9113650
APA StyleCouturier, J., Franconeri, L., Janoir, C., Ferraris, L., Syed-Zaidi, R., Youssouf, A., Gateau, C., Hoys, S., Aires, J., & Barbut, F. (2020). Characterization of Non-Toxigenic Clostridioides difficile Strains Isolated from Preterm Neonates and In Vivo Study of Their Protective Effect. Journal of Clinical Medicine, 9(11), 3650. https://doi.org/10.3390/jcm9113650





