Dissimilar Conservation Pattern in Hepatitis C Virus Mutant Spectra, Consensus Sequences, and Data Banks
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Procedures for HCV in Cell Culture
2.2. HCV Quasispecies from a Patient Cohort
2.3. Experimental Data on HCV Quasispecies
2.4. Mutation Level and Nomenclature
2.5. Sequences from the Los Alamos Database
2.6. Statistics
2.7. Sequence Accession Numbers and Data Availability
3. Results
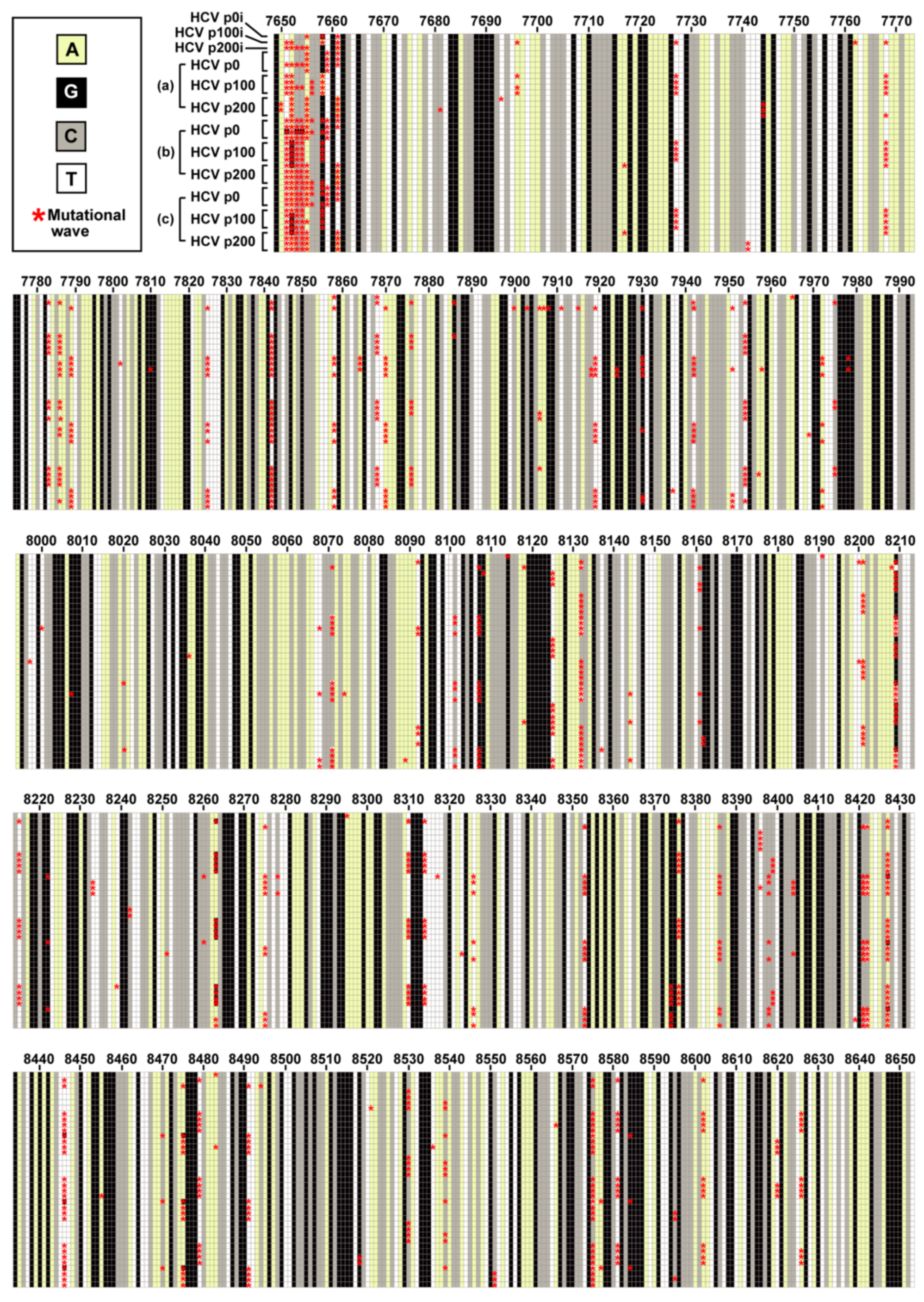
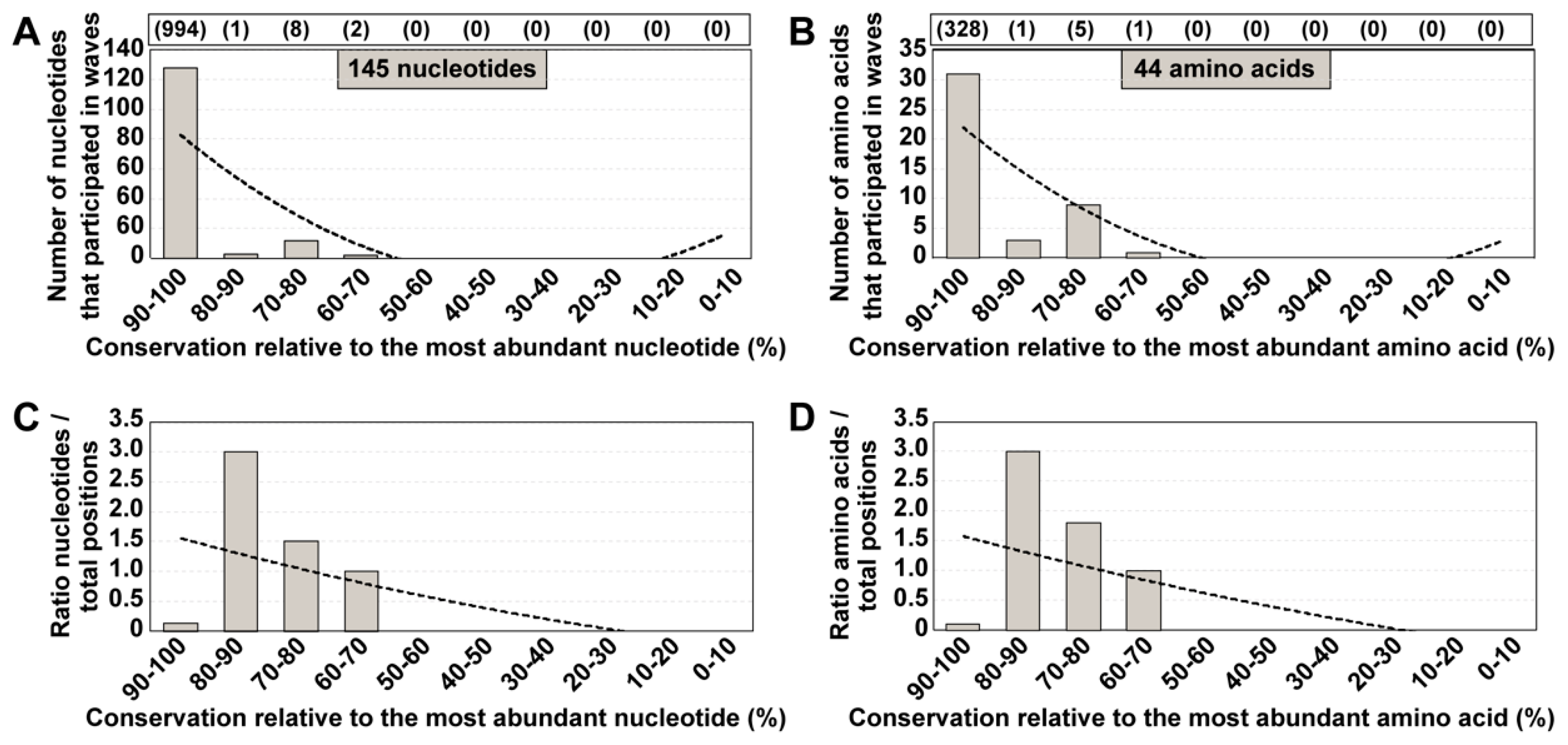
3.1. HCV Residues Involved in Mutational Waves and Their Conservation in the Consensus Sequences of the Populations
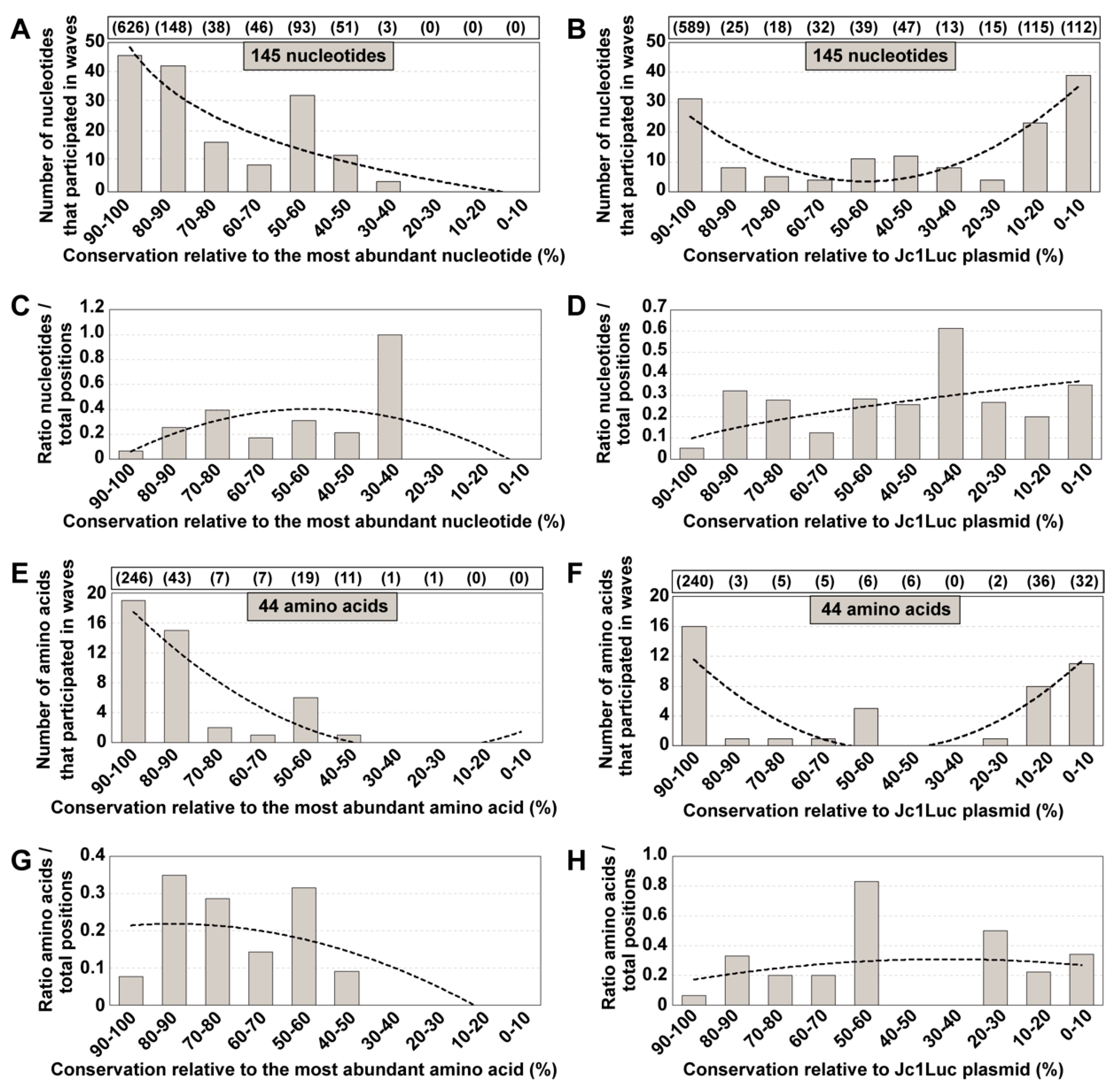
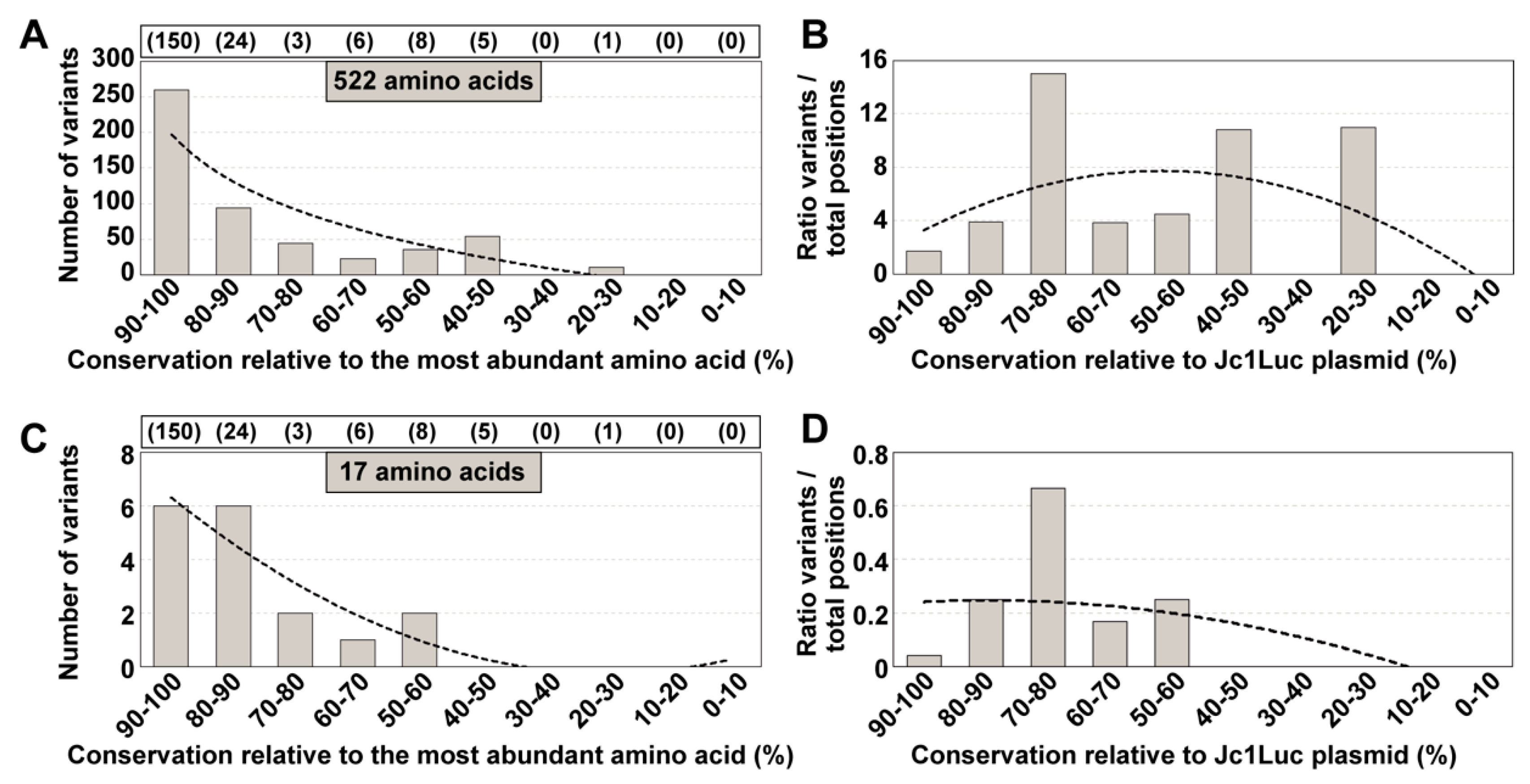
3.2. Conservation in Mutant Spectra as Compared with Conservation in the Los Alamos Data Bank
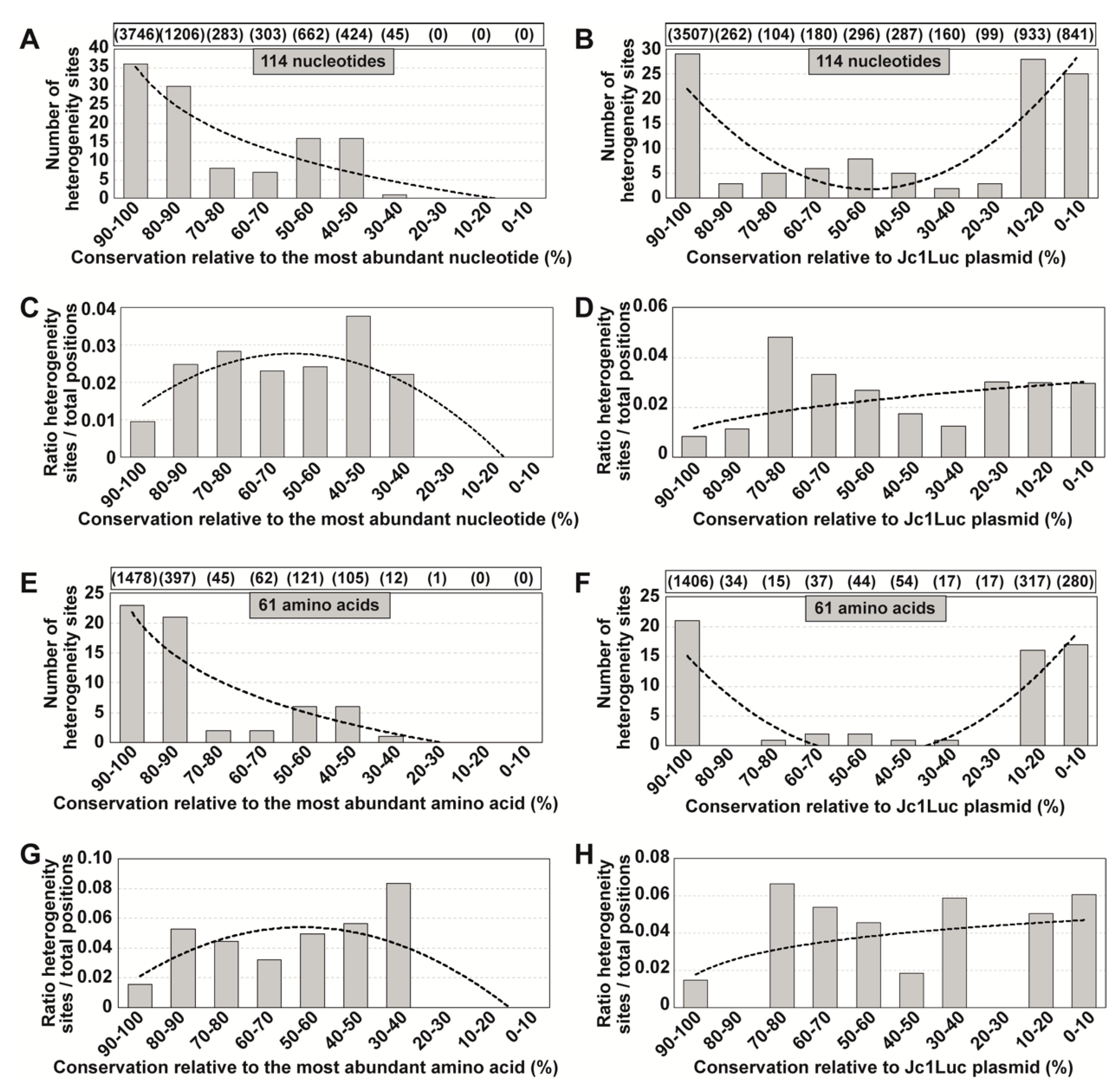
3.3. Extension of the Calculations to Mutations Associated with Sites of Heterogeneity in Consensus Sequences
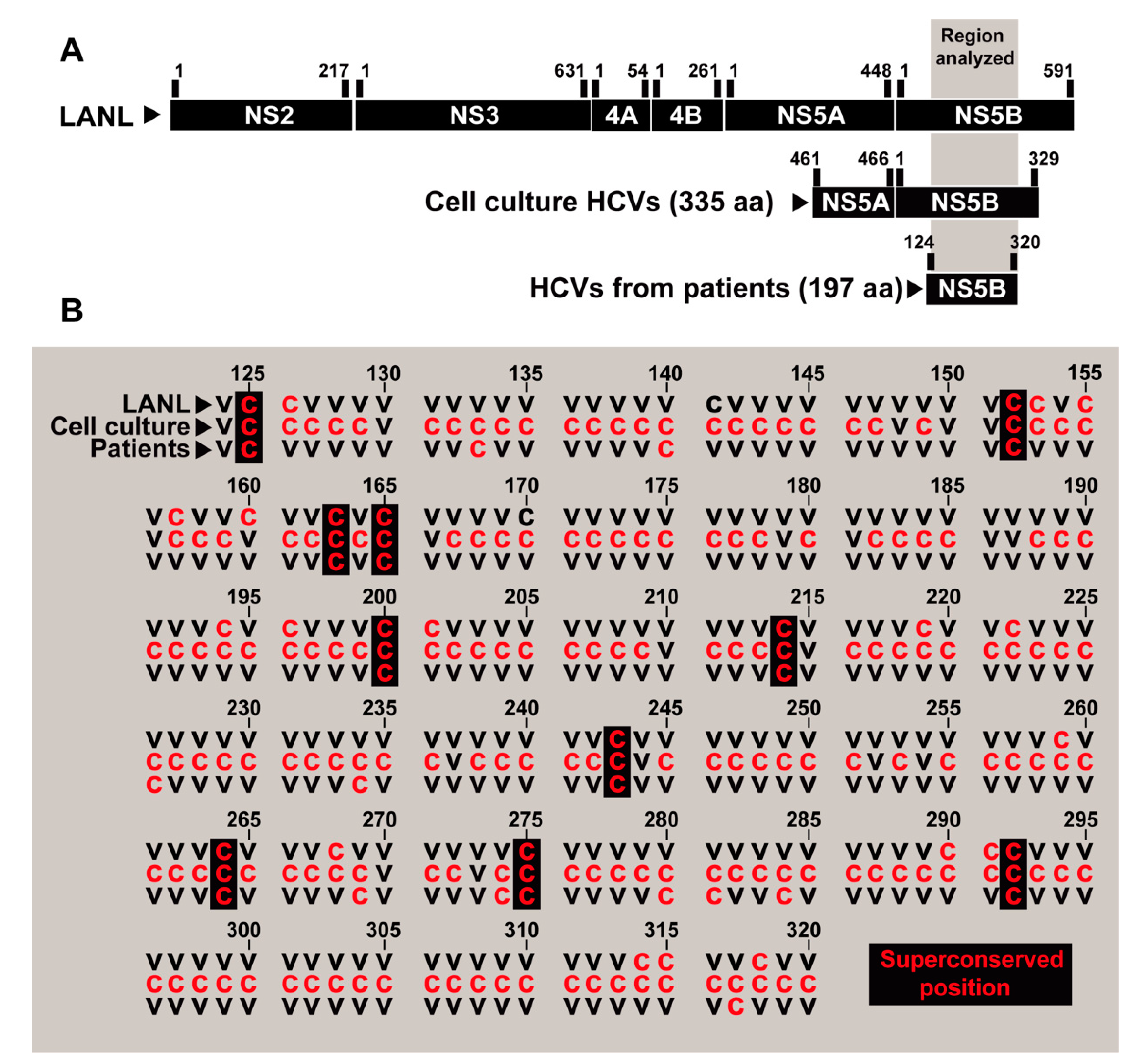
3.4. Extension to Other HCV Genomic Regions
3.5. Correlation between Quasispecies Residue Variation and Mutations Recorded in HCV from Infected Patients
3.6. Evidence of Relaxed Mutational Acceptance in HCV Quasispecies
3.7. A Possible Alternative Conservation Criterion
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartenschlager, R.; Baumert, T.F.; Bukh, J.; Houghton, M.; Lemon, S.M.; Lindenbach, B.D.; Lohmann, V.; Moradpour, D.; Pietschmann, T.; Rice, C.M.; et al. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: Considerations for scientists and funding agencies. Virus Res. 2018, 248, 53–62. [Google Scholar] [CrossRef]
- Farci, P. New insights into the HCV quasispecies and compartmentalization. Semin. Liver Dis. 2011, 31, 356–374. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, L.; Thomson, E.C.; Hughes, J.; Karayiannis, P. Quasispecies Changes with Distinctive Point Mutations in the Hepatitis C Virus Internal Ribosome Entry Site (IRES) Derived from PBMCs and Plasma. Adv. Virol. 2018, 2018, 4835252. [Google Scholar] [CrossRef] [PubMed]
- Marascio, N.; Quirino, A.; Barreca, G.S.; Galati, L.; Costa, C.; Pisani, V.; Mazzitelli, M.; Matera, G.; Liberto, M.C.; Foca, A.; et al. Discussion on critical points for a tailored therapy to cure hepatitis C virus infection. Clin. Mol. Hepatol. 2019, 25, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Glebova, O.; Knyazev, S.; Melnyk, A.; Artyomenko, A.; Khudyakov, Y.; Zelikovsky, A.; Skums, P. Inference of genetic relatedness between viral quasispecies from sequencing data. BMC Genomics 2017, 18, 918. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, J.L.; Holmes, E.C. Evolutionary Virology at 40. Genetics 2018, 210, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Soria, M.E.; Gallego, I.; de Ávila, A.I.; García-Crespo, C.; Martínez-González, B.; Gómez, J.; Briones, C.; Gregori, J.; Quer, J.; et al. A new implication of quasispecies dynamics: Broad virus diversification in absence of external perturbations. Infect. Genet. Evol. 2020, 82, 104278. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Krammer, F. Universal influenza virus vaccines and therapeutic antibodies. Clin. Microbiol. Infect. 2017, 23, 222–228. [Google Scholar] [CrossRef]
- Wijesundara, D.K.; Gummow, J.; Li, Y.; Yu, W.; Quah, B.J.; Ranasinghe, C.; Torresi, J.; Gowans, E.J.; Grubor-Bauk, B. Induction of Genotype Cross-Reactive, Hepatitis C Virus-Specific, Cell-Mediated Immunity in DNA-Vaccinated Mice. J. Virol. 2018, 92, e02133-17. [Google Scholar] [CrossRef]
- Hart, G.R.; Ferguson, A.L. Computational design of hepatitis C virus immunogens from host-pathogen dynamics over empirical viral fitness landscapes. Phys. Biol. 2018, 16, 16004. [Google Scholar] [CrossRef]
- McLean, G.R. Vaccine strategies to induce broadly protective immunity to rhinoviruses. Hum. Vaccin. Immunother. 2020, 16, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.M. Sofosbuvir-velpatasvir: A single-tablet treatment for hepatitis C infection of all genotypes. Am. J. Health Syst. Pharm. 2017, 74, 1045–1052. [Google Scholar] [CrossRef]
- Vogel, O.A.; Manicassamy, B. Broadly Protective Strategies Against Influenza Viruses: Universal Vaccines and Therapeutics. Front. Microbiol. 2020, 11, 135. [Google Scholar] [CrossRef]
- Zhong, J.; Gastaminza, P.; Cheng, G.; Kapadia, S.; Kato, T.; Burton, D.R.; Wieland, S.F.; Uprichard, S.L.; Wakita, T.; Chisari, F. V Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 9294–9299. [Google Scholar] [CrossRef]
- Wakita, T.; Pietschmann, T.; Kato, T.; Date, T.; Miyamoto, M.; Zhao, Z.; Murthy, K.; Habermann, A.; Krausslich, H.G.; Mizokami, M.; et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005, 11, 791–796. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Evans, M.J.; Syder, A.J.; Wolk, B.; Tellinghuisen, T.L.; Liu, C.C.; Maruyama, T.; Hynes, R.O.; Burton, D.R.; McKeating, J.A.; et al. Complete replication of hepatitis C virus in cell culture. Science 2005, 309, 623–626. [Google Scholar] [CrossRef]
- Moreno, E.; Gallego, I.; Gregori, J.; Lucia-Sanz, A.; Soria, M.E.; Castro, V.; Beach, N.M.; Manrubia, S.; Quer, J.; Esteban, J.I.; et al. Internal Disequilibria and Phenotypic Diversification during Replication of Hepatitis C Virus in a Noncoevolving Cellular Environment. J. Virol. 2017, 91, e02505-16. [Google Scholar] [CrossRef]
- Perales, C.; Beach, N.M.; Gallego, I.; Soria, M.E.; Quer, J.; Esteban, J.I.; Rice, C.; Domingo, E.; Sheldon, J. Response of hepatitis C virus to long-term passage in the presence of alpha interferon: Multiple mutations and a common phenotype. J. Virol. 2013, 87, 7593–7607. [Google Scholar] [CrossRef]
- Gallego, I.; Soria, M.E.; Garcia-Crespo, C.; Chen, Q.; Martinez-Barragan, P.; Khalfaoui, S.; Martinez-Gonzalez, B.; Sanchez-Martin, I.; Palacios-Blanco, I.; de Avila, A.I.; et al. Broad and Dynamic Diversification of Infectious Hepatitis C Virus in a Cell Culture Environment. J. Virol. 2020, 94, e01856-19. [Google Scholar] [CrossRef]
- Gregori, J.; Salicru, M.; Domingo, E.; Sanchez, A.; Esteban, J.I.; Rodriguez-Frias, F.; Quer, J. Inference with viral quasispecies diversity indices: Clonal and NGS approaches. Bioinformatics 2014, 30, 1104–1111. [Google Scholar] [CrossRef]
- Gregori, J.; Perales, C.; Rodriguez-Frias, F.; Esteban, J.I.; Quer, J.; Domingo, E. Viral quasispecies complexity measures. Virology 2016, 493, 227–237. [Google Scholar] [CrossRef]
- Chen, Q.; Perales, C.; Soria, M.E.; Garcia-Cehic, D.; Gregori, J.; Rodriguez-Frias, F.; Buti, M.; Crespo, J.; Calleja, J.L.; Tabernero, D.; et al. Deep-sequencing reveals broad subtype-specific HCV resistance mutations associated with treatment failure. Antivir. Res. 2020, 174, 104694. [Google Scholar] [CrossRef]
- Ali, A.; Melcher, U. Modeling of Mutational Events in the Evolution of Viruses. Viruses 2019, 11, 418. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Aiewsakun, P.; Katzourakis, A. Prisoners of war—host adaptation and its constraints on virus evolution. Nat. Rev. Microbiol. 2019, 17, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Leslie, A.J.; Pfafferott, K.J.; Chetty, P.; Draenert, R.; Addo, M.M.; Feeney, M.; Tang, Y.; Holmes, E.C.; Allen, T.; Prado, J.G.; et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 2004, 10, 282–289. [Google Scholar] [CrossRef]
- Herbeck, J.T.; Nickle, D.C.; Learn, G.H.; Gottlieb, G.S.; Curlin, M.E.; Heath, L.; Mullins, J.I. Human immunodeficiency virus type 1 env evolves toward ancestral states upon transmission to a new host. J. Virol. 2006, 80, 1637–1644. [Google Scholar] [CrossRef] [PubMed]
- Redd, A.D.; Collinson-Streng, A.N.; Chatziandreou, N.; Mullis, C.E.; Laeyendecker, O.; Martens, C.; Ricklefs, S.; Kiwanuka, N.; Nyein, P.H.; Lutalo, T.; et al. Previously transmitted HIV-1 strains are preferentially selected during subsequent sexual transmissions. J. Infect. Dis. 2012, 206, 1433–1442. [Google Scholar] [CrossRef]
- Marukian, S.; Jones, C.T.; Andrus, L.; Evans, M.J.; Ritola, K.D.; Charles, E.D.; Rice, C.M.; Dustin, L.B. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 2008, 48, 1843–1850. [Google Scholar] [CrossRef]
- Soria, M.E.; Gregori, J.; Chen, Q.; Garcia-Cehic, D.; Llorens, M.; de Avila, A.I.; Beach, N.M.; Domingo, E.; Rodriguez-Frias, F.; Buti, M.; et al. Pipeline for specific subtype amplification and drug resistance detection in hepatitis C virus. BMC Infect. Dis. 2018, 18, 446. [Google Scholar] [CrossRef]
- Kuiken, C.; Combet, C.; Bukh, J.; Shin, I.T.; Deleage, G.; Mizokami, M.; Richardson, R.; Sablon, E.; Yusim, K.; Pawlotsky, J.M.; et al. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology 2006, 44, 1355–1361. [Google Scholar] [CrossRef]
- Baccam, P.; Thompson, R.J.; Fedrigo, O.; Carpenter, S.; Cornette, J.L. PAQ: Partition Analysis of Quasispecies. Bioinformatics 2001, 17, 16–22. [Google Scholar] [CrossRef]
- Skums, P.; Zelikovsky, A.; Singh, R.; Gussler, W.; Dimitrova, Z.; Knyazev, S.; Mandric, I.; Ramachandran, S.; Campo, D.; Jha, D.; et al. QUENTIN: Reconstruction of disease transmissions from viral quasispecies genomic data. Bioinformatics 2018, 34, 163–170. [Google Scholar] [CrossRef]
- Ahn, S.; Ke, Z.; Vikalo, H. Viral quasispecies reconstruction via tensor factorization with successive read removal. Bioinformatics 2018, 34, i23–i31. [Google Scholar] [CrossRef] [PubMed]
- Henningsson, R.; Moratorio, G.; Borderia, A.V.; Vignuzzi, M.; Fontes, M. DISSEQT-DIStribution-based modeling of SEQuence space Time dynamics. Virus Evol. 2019, 5, vez028. [Google Scholar] [CrossRef]
- Grenfell, B.T.; Pybus, O.G.; Gog, J.R.; Wood, J.L.; Daly, J.M.; Mumford, J.A.; Holmes, E.C. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 2004, 303, 327–332. [Google Scholar] [CrossRef]
- Blackard, J.T.; Nyingi, K.; Sherman, K.E. Extrahepatic replication of HCV: Insights into clinical manifestations and biological consequences. Hepatology 2006, 44, 15–22. [Google Scholar] [CrossRef]
- Skardasi, G.; Chen, A.Y.; Michalak, T.I. Authentic Patient-Derived Hepatitis C Virus Infects and Productively Replicates in Primary CD4 + and CD8 + T Lymphocytes In Vitro. J. Virol. 2018, 92, e01790-17. [Google Scholar] [CrossRef]
- Domingo, E.; Perales, C. Viral quasispecies. PLoS Genet. 2019, 15, e1008271. [Google Scholar] [CrossRef]
- Soria, M.E.; García-Crespo, C.; Martínez-González, B.; Vazquez-Sirvent, L.; Lobo-Vega, R.; de Ávila, A.I.; Gallego, I.; Ferrer-Orta, C.; Verdaguer, N.; Chen, Q.; et al. Amino acid substitution associated with treatment failure of hepatitis C virus infection. J. Clin. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.A.; Verdaguer, N.; Mateu, M.G.; Domingo, E. Evolution subverting essentiality: Dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc. Natl. Acad. Sci. USA 1997, 94, 6798–6802. [Google Scholar] [CrossRef] [PubMed]
- Tami, C.; Taboga, O.; Berinstein, A.; Nuñez, J.I.; Palma, E.L.; Domingo, E.; Sobrino, F.; Carrillo, E. Evidence of the coevolution of antigenicity and host cell tropism of foot-and-mouth disease virus in vivo. J. Virol. 2003, 77, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Brun, A.; Núñez, J.I.; Cristina, J.; Briones, C.; Escarmís, C. Genomics of Viruses. In Pathogenomics: Genome Analysis of Pathogenic Microbes; Hacker, J., Dobrindt, U., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2006; pp. 369–388. [Google Scholar]
- Eigen, M. From Strange Simplicity to Complex Familiarity; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Van Slyke, G.A.; Arnold, J.J.; Lugo, A.J.; Griesemer, S.B.; Moustafa, I.M.; Kramer, L.D.; Cameron, C.E.; Ciota, A.T. Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes. PLoS Pathog. 2015, 11, e1005009. [Google Scholar] [CrossRef]
- Shatoff, E.; Bundschuh, R. Single nucleotide polymorphisms affect RNA-protein interactions at a distance through modulation of RNA secondary structures. PLoS Comput. Biol. 2020, 16, e1007852. [Google Scholar] [CrossRef]
- Holland, J.J.; de La Torre, J.C.; Steinhauer, D.A. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 1992, 176, 1–20. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Crespo, C.; Soria, M.E.; Gallego, I.; Ávila, A.I.d.; Martínez-González, B.; Vázquez-Sirvent, L.; Gómez, J.; Briones, C.; Gregori, J.; Quer, J.; et al. Dissimilar Conservation Pattern in Hepatitis C Virus Mutant Spectra, Consensus Sequences, and Data Banks. J. Clin. Med. 2020, 9, 3450. https://doi.org/10.3390/jcm9113450
García-Crespo C, Soria ME, Gallego I, Ávila AId, Martínez-González B, Vázquez-Sirvent L, Gómez J, Briones C, Gregori J, Quer J, et al. Dissimilar Conservation Pattern in Hepatitis C Virus Mutant Spectra, Consensus Sequences, and Data Banks. Journal of Clinical Medicine. 2020; 9(11):3450. https://doi.org/10.3390/jcm9113450
Chicago/Turabian StyleGarcía-Crespo, Carlos, María Eugenia Soria, Isabel Gallego, Ana Isabel de Ávila, Brenda Martínez-González, Lucía Vázquez-Sirvent, Jordi Gómez, Carlos Briones, Josep Gregori, Josep Quer, and et al. 2020. "Dissimilar Conservation Pattern in Hepatitis C Virus Mutant Spectra, Consensus Sequences, and Data Banks" Journal of Clinical Medicine 9, no. 11: 3450. https://doi.org/10.3390/jcm9113450
APA StyleGarcía-Crespo, C., Soria, M. E., Gallego, I., Ávila, A. I. d., Martínez-González, B., Vázquez-Sirvent, L., Gómez, J., Briones, C., Gregori, J., Quer, J., Perales, C., & Domingo, E. (2020). Dissimilar Conservation Pattern in Hepatitis C Virus Mutant Spectra, Consensus Sequences, and Data Banks. Journal of Clinical Medicine, 9(11), 3450. https://doi.org/10.3390/jcm9113450





