Low-Dose and Long-Term Olaparib Treatment Sensitizes MDA-MB-231 and SUM1315 Triple-Negative Breast Cancers Spheroids to Fractioned Radiotherapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture
Spheroid Formation
2.2. Olaparib Treatment
Irradiation
2.3. Experiments in Monolayer Cell Culture
2.3.1. DNA DSBs Induction: Gamma H2AX Immunofluorescent Markings
2.3.2. SulfoRhodamine B Survival Test
2.3.3. Clonogenic Survival Test
2.4. Experiments in 3D Cell Culture
2.4.1. Spheroid Treatment
2.4.2. Spheroid Growth Monitoring
2.4.3. Spheroid Metabolic Activity Assessment With the Resazurin Test
2.4.4. Spheroid Viability and Mortality Fluorescent Profile (Live/Dead)
2.5. Transcriptomic Analysis of TNBC Cell Lines
2.6. Statistical Analysis
3. Results
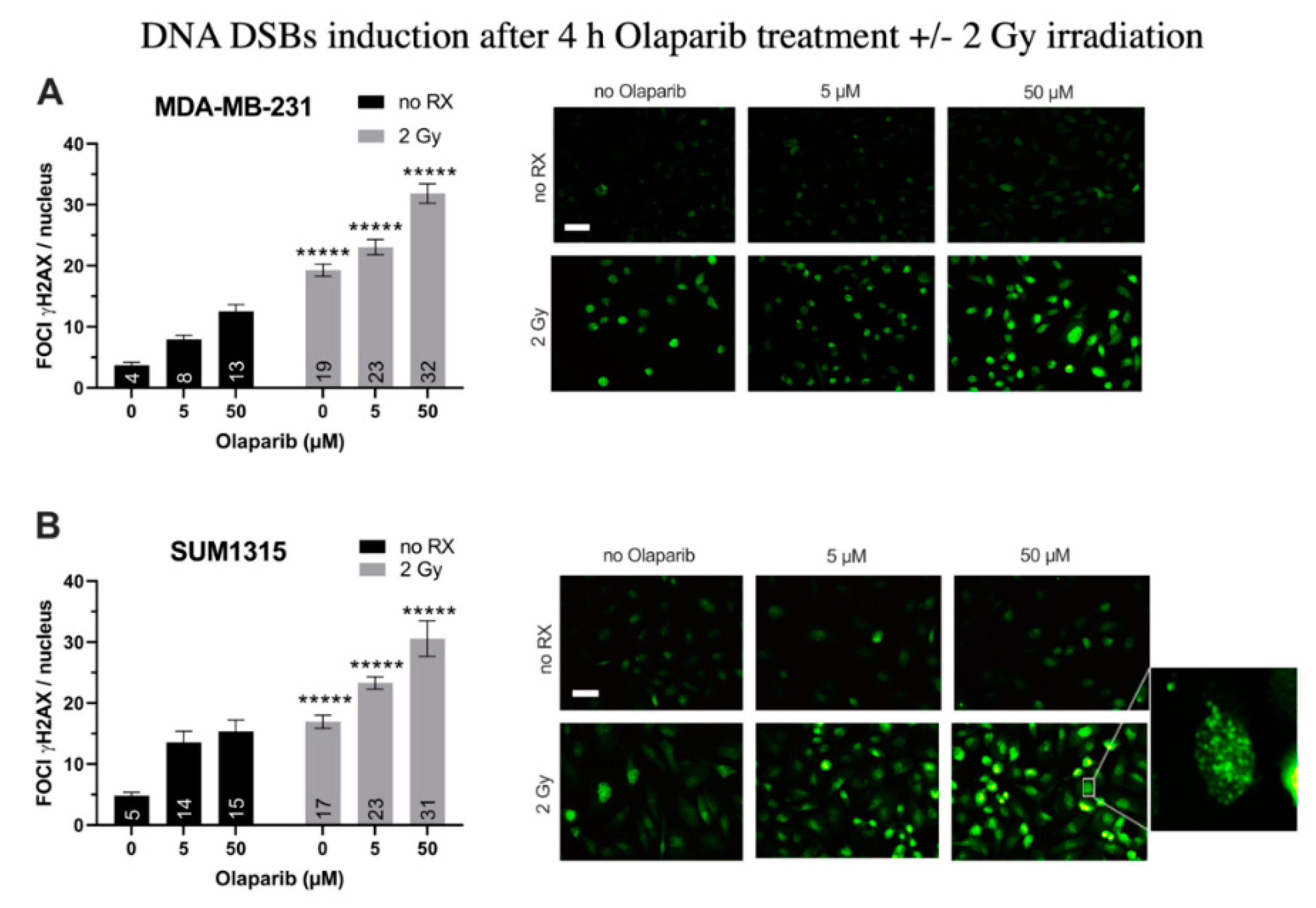
3.1. Olaparib and Radiotherapy Induce DNA Double Strand Breaks in TNBC Cell Lines
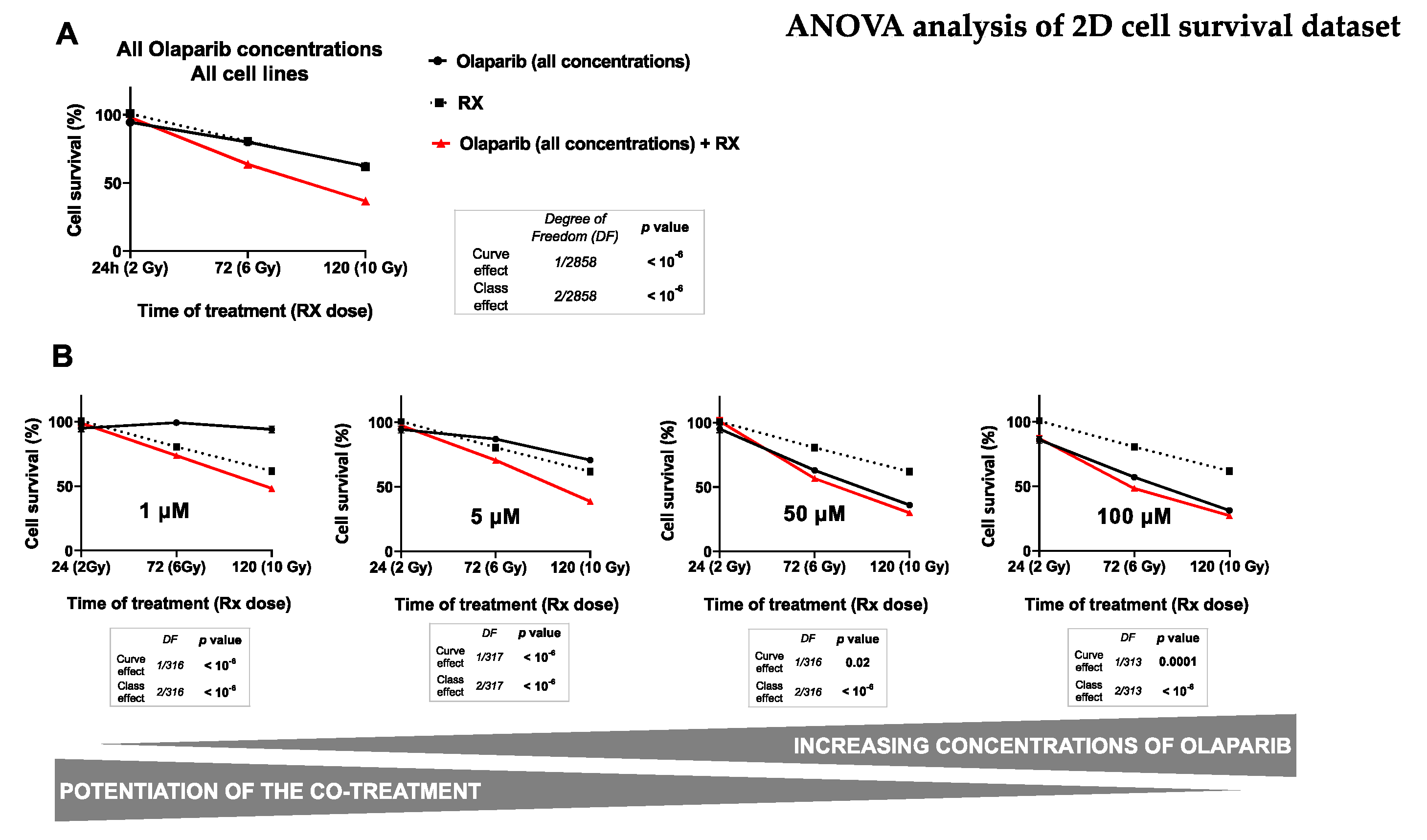
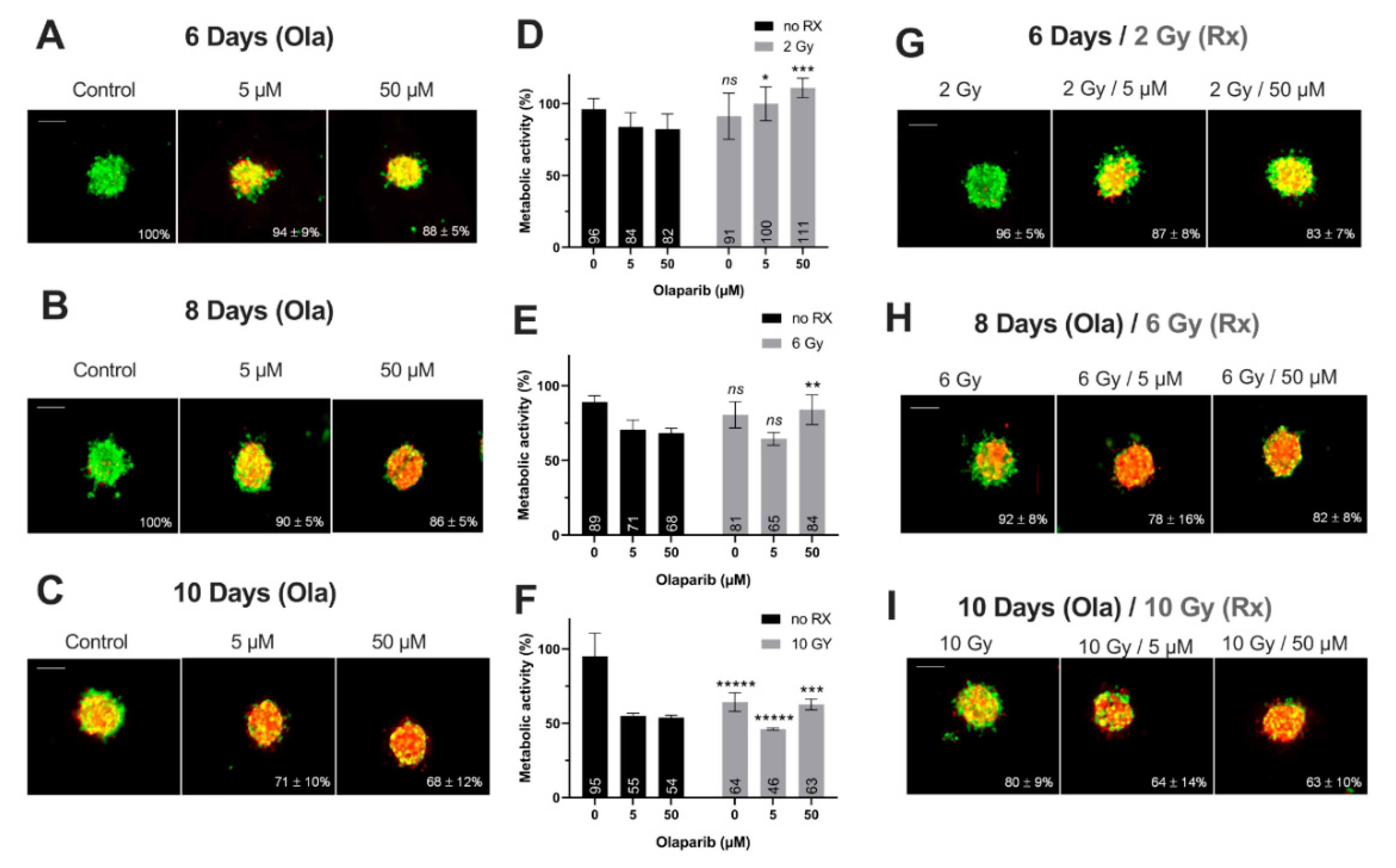
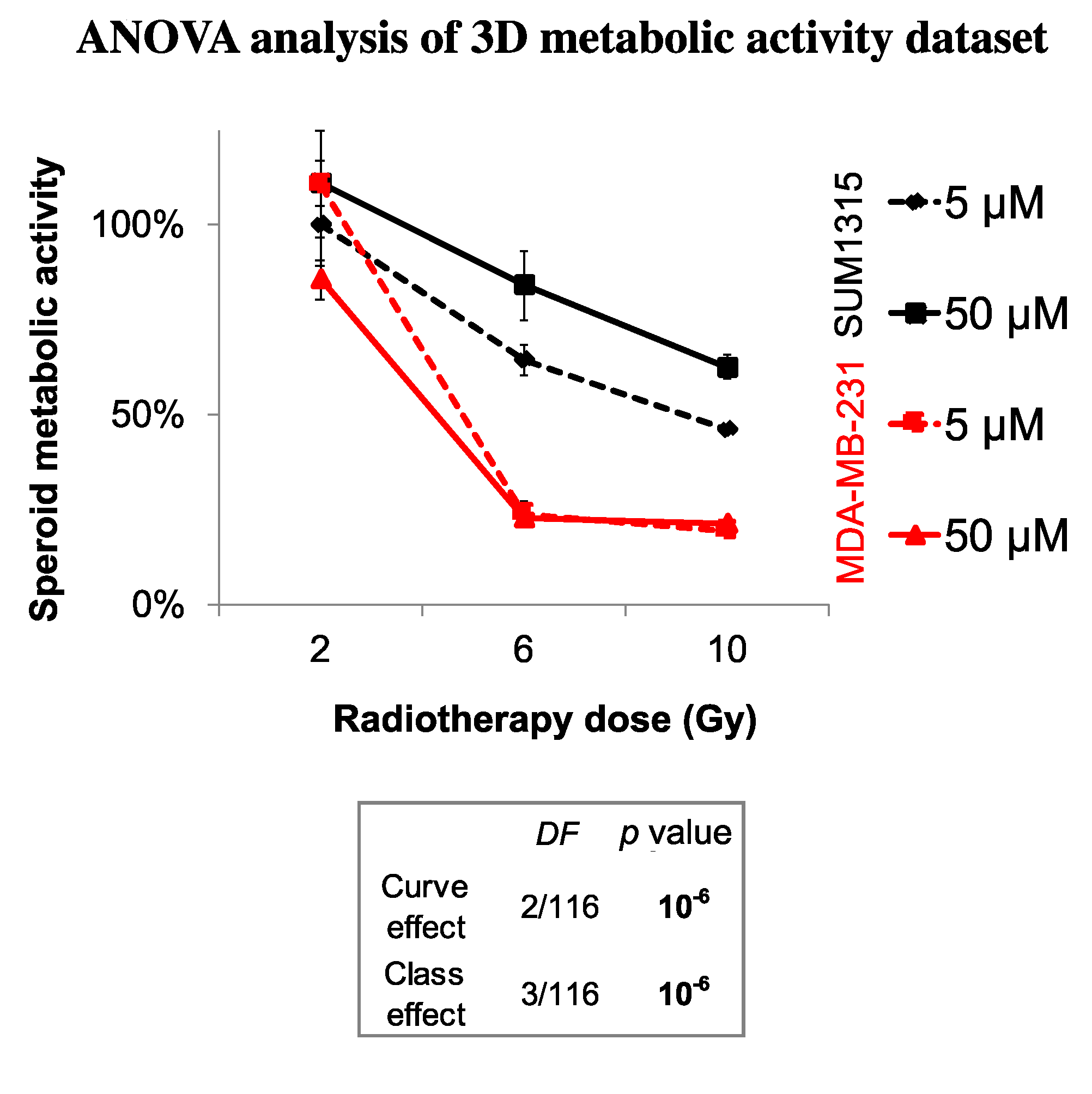
3.2. The Cytotoxicity of Radiotherapy Is Optimized by Olaparib and is Potentiated in Presence of Low Doses of Olaparib
Impact on TNBC Cell Survival in 2D Cell Culture
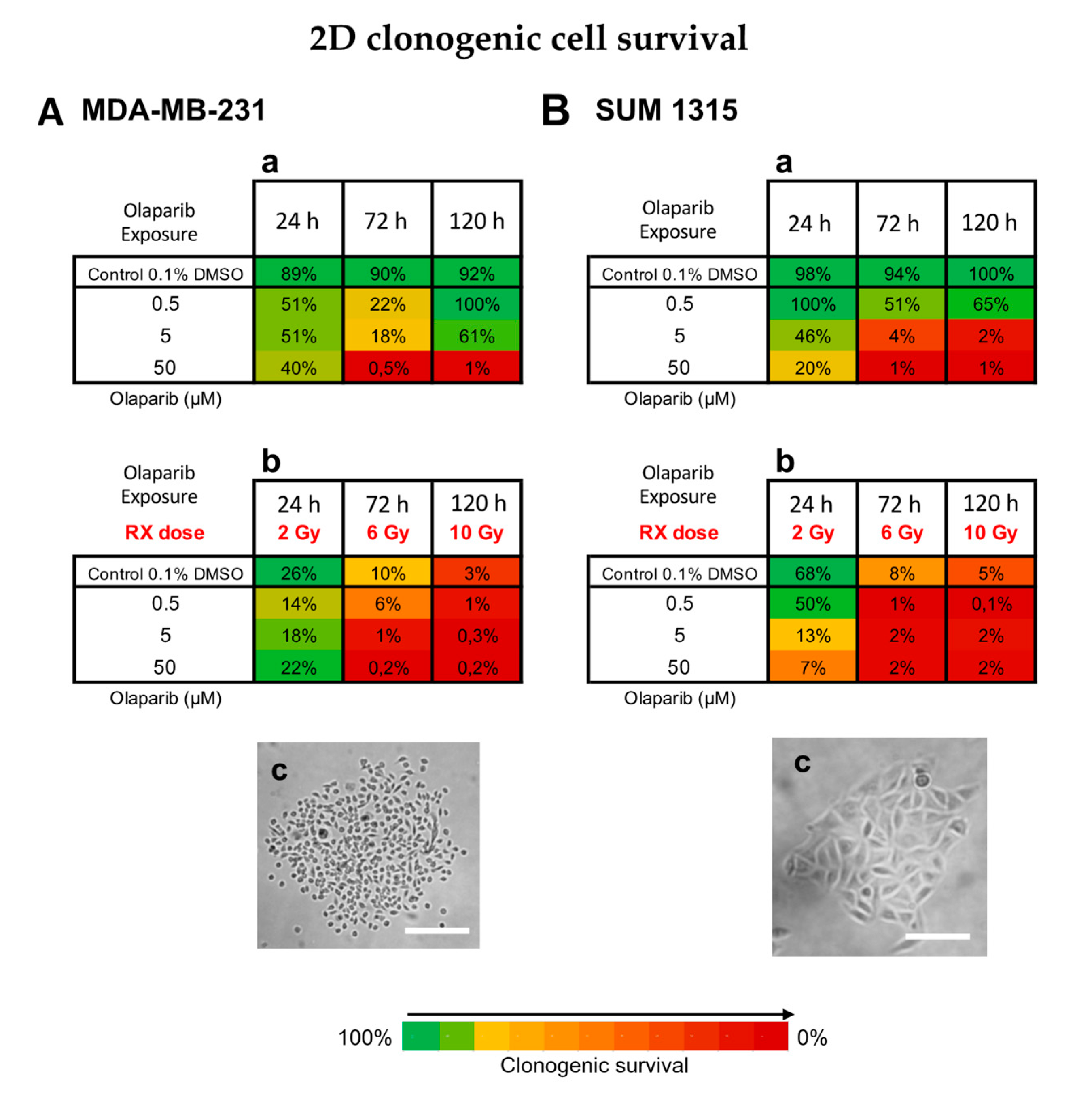
3.3. Impact on TNBC Cell Clonogenic Potential of Repopulation
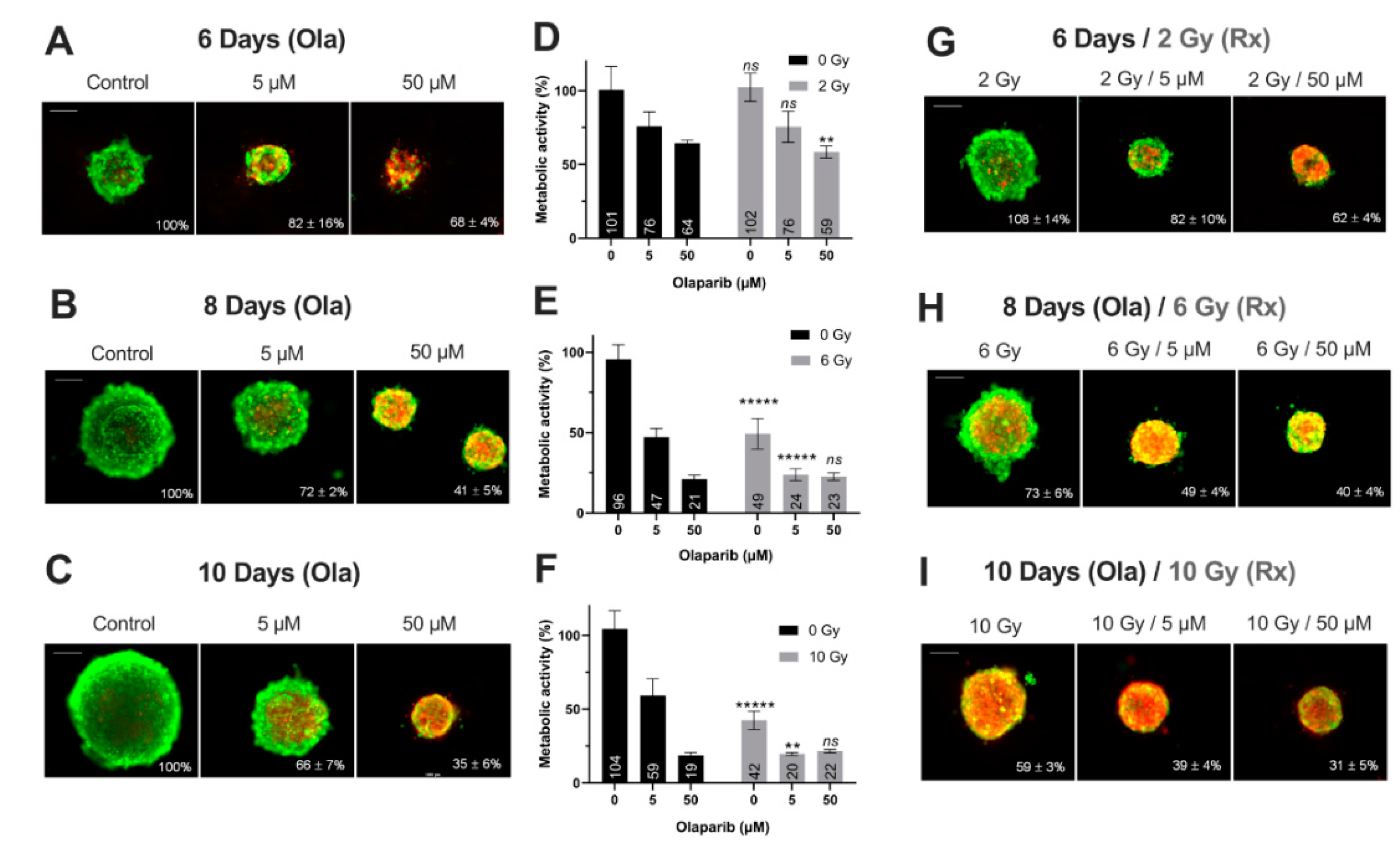
Impact on Tumor-Like Models
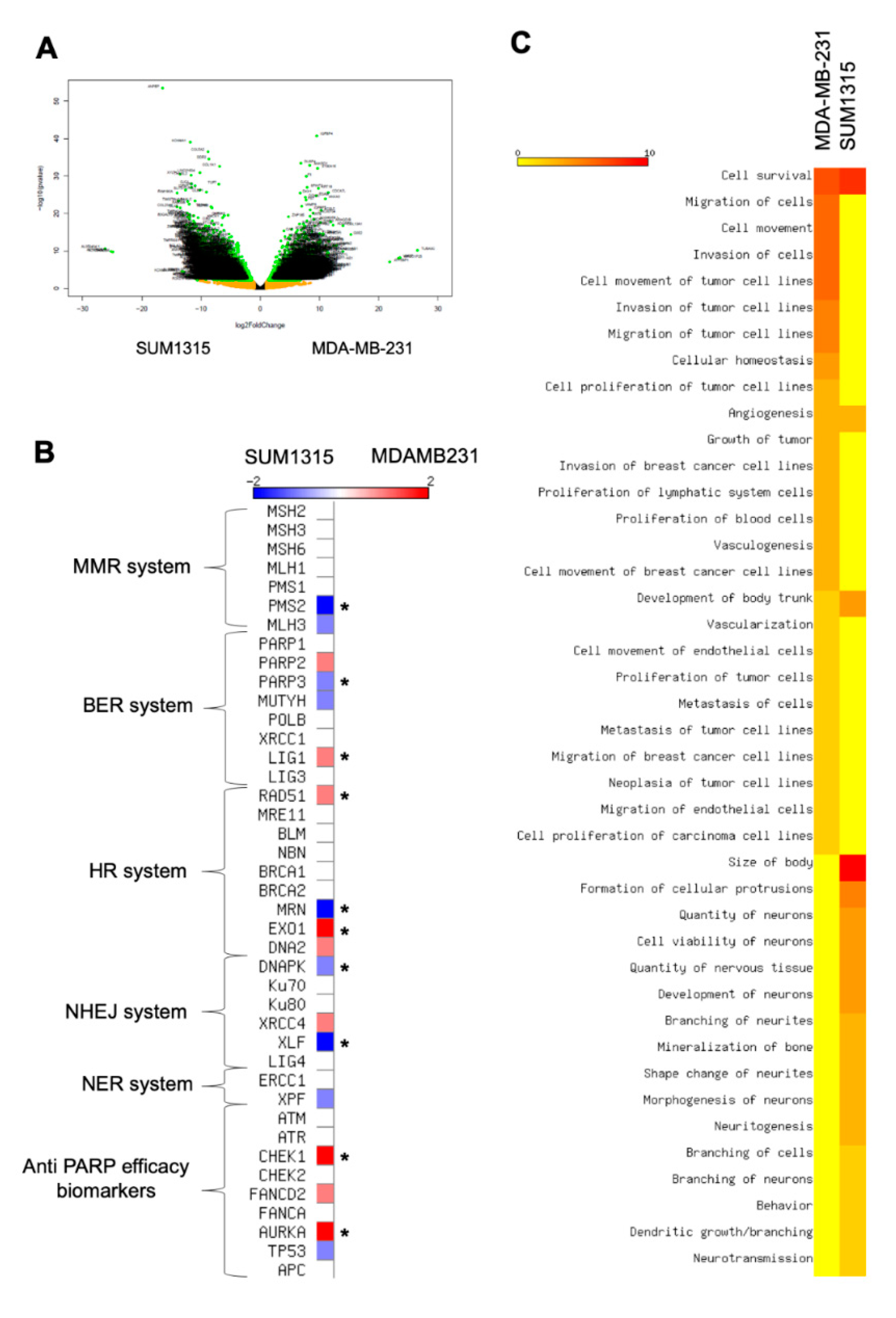
3.4. The Difference in SUM1315 and MDA-MB-231 TNBC Cell Lines Sensitivity in 2D and 3D Is Explained by a Strong Genomic Heterogeneity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Horton, J.K.; Jagsi, R.; Woodward, W.A.; Ho, A. Breast Cancer Biology: Clinical Implications for Breast Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2018, 100, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Penault-Llorca, F.; Viale, G. Pathological and molecular diagnosis of triple-negative breast cancer: A clinical perspective. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23 (Suppl. 6), vi19–vi22. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S. Basal-like subtype of breast cancer: A review of its unique characteristics and their clinical significance. Breast Cancer Tokyo Jpn. 2008, 15, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Rakha, E.A.; Elsheikh, S.E.; Aleskandarany, M.A.; Habashi, H.O.; Green, A.R.; Powe, D.G.; El-Sayed, M.E.; Benhasouna, A.; Brunet, J.-S.; Akslen, L.A.; et al. Triple-negative breast cancer: Distinguishing between basal and nonbasal subtypes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 2302–2310. [Google Scholar] [CrossRef] [PubMed]
- De Ruijter, T.C.; Veeck, J.; de Hoon, J.P.J.; van Engeland, M.; Tjan-Heijnen, V.C. Characteristics of triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 2011, 137, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef]
- Marmé, F.; Schneeweiss, A. Targeted Therapies in Triple-Negative Breast Cancer. Breast Care Basel Switz. 2015, 10, 159–166. [Google Scholar] [CrossRef]
- Duma, N.; Gast, K.C.; Choong, G.M.; Leon-Ferre, R.A.; O’Sullivan, C.C. Where Do We Stand on the Integration of PARP Inhibitors for the Treatment of Breast Cancer? Curr. Oncol. Rep. 2018, 20, 63. [Google Scholar] [CrossRef]
- Pernin, V.; Mégnin-Chanet, F.; Pennaneach, V.; Fourquet, A.; Kirova, Y.; Hall, J. PARP inhibitors and radiotherapy: Rational and prospects for a clinical use. Cancer Radiother. J. Soc. Fr. Radiother. Oncol. 2014, 18, 790–798, quiz 799–802. [Google Scholar]
- Chalmers, A.J.; Lakshman, M.; Chan, N.; Bristow, R.G. Poly (ADP-ribose) polymerase inhibition as a model for synthetic lethality in developing radiation oncology targets. Semin. Radiat. Oncol. 2010, 20, 274–281. [Google Scholar] [CrossRef]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. Adv. Life Sci. RD 2017, 22, 456–472. [Google Scholar]
- Dubois, C.; Dufour, R.; Daumar, P.; Aubel, C.; Szczepaniak, C.; Blavignac, C.; Mounetou, E.; Penault-Llorca, F.; Bamdad, M. Development and cytotoxic response of two proliferative MDA-MB-231 and non-proliferative SUM1315 three-dimensional cell culture models of triple-negative basal-like breast cancer cell lines. Oncotarget 2017, 8, 95316–95331. [Google Scholar] [PubMed]
- Daumar, P.; Dufour, R.; Dubois, C.; Penault-Llorca, F.; Bamdad, M.; Mounetou, E. Development and validation of a high-performance liquid chromatography method for the quantitation of intracellular PARP inhibitor Olaparib in cancer cells. J. Pharm. Biomed. Anal. 2018, 152, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Dufour, R.; Daumar, P.; Mounetou, E.; Aubel, C.; Kwiatkowski, F.; Abrial, C.; Vatoux, C.; Penault-Llorca, F.; Bamdad, M. BCRP and P-gp relay overexpression in triple negative basal-like breast cancer cell line: A prospective role in resistance to Olaparib. Sci. Rep. 2015, 5, 12670. [Google Scholar] [CrossRef] [PubMed]
- Elstrodt, F.; Hollestelle, A.; Nagel, J.H.A.; Gorin, M.; Wasielewski, M.; van den Ouweland, A.; Merajver, S.D.; Ethier, S.P.; Schutte, M. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006, 66, 41–45. [Google Scholar] [CrossRef]
- Marcotte, R.; Sayad, A.; Brown, K.R.; Sanchez-Garcia, F.; Reimand, J.; Haider, M.; Virtanen, C.; Bradner, J.E.; Bader, G.D.; Mills, G.B.; et al. Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance. Cell 2016, 164, 293–309. [Google Scholar] [CrossRef]
- Daemen, A.; Griffith, O.L.; Heiser, L.M.; Wang, N.J.; Enache, O.M.; Sanborn, Z.; Pepin, F.; Durinck, S.; Korkola, J.E.; Griffith, M.; et al. Modeling precision treatment of breast cancer. Genome Biol. 2013, 14, R110. [Google Scholar] [CrossRef]
- Boire, A.; Zou, Y.; Shieh, J.; Macalinao, D.G.; Pentsova, E.; Massagué, J. Complement Component 3 Adapts the Cerebrospinal Fluid for Leptomeningeal Metastasis. Cell 2017, 168, 1101–1113. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinform. Oxf. Engl. 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Ollier, M.; Radosevic-Robin, N.; Kwiatkowski, F.; Ponelle, F.; Viala, S.; Privat, M.; Uhrhammer, N.; Bernard-Gallon, D.; Penault-Llorca, F.; Bignon, Y.-J.; et al. DNA repair genes implicated in triple negative familial non-BRCA1/2 breast cancer predisposition. Am. J. Cancer Res. 2015, 5, 2113–2126. [Google Scholar] [PubMed]
- Dhillon, K.K.; Bajrami, I.; Taniguchi, T.; Lord, C.J. Synthetic lethality: The road to novel therapies for breast cancer. Endocr. Relat. Cancer 2016, 23, T39–T55. [Google Scholar] [CrossRef] [PubMed]
- Fang, B. Development of synthetic lethality anticancer therapeutics. J. Med. Chem. 2014, 57, 7859–7873. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Barekati, Z.; Zhang, B.; Liu, Z.; Zhong, X. Targeted therapy in breast cancer: What’s new? Swiss Med. Wkly. 2011, 141, w13231. [Google Scholar] [CrossRef] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- O’Neil, N.J.; Bailey, M.L.; Hieter, P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017, 18, 613–623. [Google Scholar] [CrossRef]
- Gani, C.; Coackley, C.; Kumareswaran, R.; Schütze, C.; Krause, M.; Zafarana, G.; Bristow, R.G. In vivo studies of the PARP inhibitor, AZD-2281, in combination with fractionated radiotherapy: An exploration of the therapeutic ratio. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2015, 116, 486–494. [Google Scholar] [CrossRef]
- Kötter, A.; Cornils, K.; Borgmann, K.; Dahm-Daphi, J.; Petersen, C.; Dikomey, E.; Mansour, W.Y. Inhibition of PARP1-dependent end-joining contributes to Olaparib-mediated radiosensitization in tumor cells. Mol. Oncol. 2014, 8, 1616–1625. [Google Scholar] [CrossRef]
- Senra, J.M.; Telfer, B.A.; Cherry, K.E.; McCrudden, C.M.; Hirst, D.G.; O’Connor, M.J.; Wedge, S.R.; Stratford, I.J. Inhibition of PARP-1 by olaparib (AZD2281) increases the radiosensitivity of a lung tumor xenograft. Mol. Cancer Ther. 2011, 10, 1949–1958. [Google Scholar] [CrossRef]
- Verhagen, C.V.M.; de Haan, R.; Hageman, F.; Oostendorp, T.P.D.; Carli, A.L.E.; O’Connor, M.J.; Jonkers, J.; Verheij, M.; van den Brekel, M.W.; Vens, C. Extent of radiosensitization by the PARP inhibitor olaparib depends on its dose, the radiation dose and the integrity of the homologous recombination pathway of tumor cells. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2015, 116, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Mah, L.-J.; El-Osta, A.; Karagiannis, T.C. gammaH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Olive, P.L.; Banáth, J.P. Phosphorylation of histone H2AX as a measure of radiosensitivity. Int. J. Radiat. Oncol. Biol. Phys. 2004, 58, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Huang, X.; Shuang, Z.; Lin, G.; Wang, J.; Duan, F.; Chen, J.; Li, S. PARP inhibitor olaparib sensitizes cholangiocarcinoma cells to radiation. Cancer Med. 2018, 7, 1285–1296. [Google Scholar] [CrossRef]
- Efimova, E.V.; Mauceri, H.J.; Golden, D.W.; Labay, E.; Bindokas, V.P.; Darga, T.E.; Chakraborty, C.; Barreto-Andrade, J.C.; Crawley, C.; Sutton, H.G.; et al. Poly (ADP-ribose) polymerase inhibitor induces accelerated senescence in irradiated breast cancer cells and tumors. Cancer Res. 2010, 70, 6277–6282. [Google Scholar] [CrossRef]
- Ihnen, M.; zu Eulenburg, C.; Kolarova, T.; Qi, J.W.; Manivong, K.; Chalukya, M.; Dering, J.; Anderson, L.; Ginther, C.; Meuter, A.; et al. Therapeutic potential of the poly (ADP-ribose) polymerase inhibitor rucaparib for the treatment of sporadic human ovarian cancer. Mol. Cancer Ther. 2013, 12, 1002–1015. [Google Scholar] [CrossRef]
- Ohmoto, A.; Yachida, S. Current status of poly (ADP-ribose) polymerase inhibitors and future directions. Onco Targets Ther. 2017, 10, 5195–5208. [Google Scholar] [CrossRef]
- Telli, M.L.; Stover, D.G.; Loi, S.; Aparicio, S.; Carey, L.A.; Domchek, S.M.; Newman, L.; Sledge, G.W.; Winer, E.P. Homologous recombination deficiency and host anti-tumor immunity in triple-negative breast cancer. Breast Cancer Res. Treat. 2018, 171, 21–31. [Google Scholar] [CrossRef]
- Wang, X.; Shi, Y.; Huang, D.; Guan, X. Emerging therapeutic modalities of PARP inhibitors in breast cancer. Cancer Treat. Rev. 2018, 68, 62–68. [Google Scholar] [CrossRef]
- Fillmore, C.M.; Kuperwasser, C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. BCR 2008, 10, R25. [Google Scholar] [CrossRef]
- Barnabas, N.; Cohen, D. Phenotypic and Molecular Characterization of MCF10DCIS and SUM Breast Cancer Cell Lines. Int. J. Breast Cancer 2013, 2013, 872743. [Google Scholar] [CrossRef] [PubMed]
| Name | Accession N | Library Type | Sequencing | End Type | Reference |
|---|---|---|---|---|---|
| MDAMB231_A | GSE73526 | TruSeq Stranded mRNA | Illumina HiSeq 2000 | paired-end | [17] |
| MDAMB231_B | GSE48213 | TruSeq RNA Sample Preparation Kit | Illumina Genome Analyzer IIx | paired-end | [18] |
| MDAMB231_C | GSE83132 | TruSeq RNA Sample Prep Kit v2 | Illumina HiSeq 2000 | paired-end | [19] |
| MDAMB231_D | GSE83132 | TruSeq RNA Sample Prep Kit v2 | Illumina HiSeq 2000 | paired-end | [19] |
| MDAMB231_E | GSE83132 | TruSeq RNA Sample Prep Kit v2 | Illumina HiSeq 2000 | paired-end | [19] |
| SUM1315_B | GSE48213 | TruSeq RNA Sample Preparation Kit | Illumina Genome Analyzer IIx | paired-end | [18] |
| SUM1315_A | GSE73526 | TruSeq Stranded mRNA | Illumina HiSeq 2000 | Illumina HiSeq 2000 | [17] |
| Experimental Parameters | Probability of Variance Influence | % of Variance Explanation |
|---|---|---|
| Type of cell line (MDA-MB-231; SUM1315) | <0.0000001 | 8% |
| Dose Olaparib (0; 0.1; 1; 5; 10; 25; 50; 75; 100 µM) | <0.0000001 | 14% |
| Olaparib exposure (24 h; 72 h; 120 h) | <0.0000001 | 40% |
| Irradiation dose (0; 2; 6; 10 Gy) | <0.0000001 | 38% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubois, C.; Martin, F.; Hassel, C.; Magnier, F.; Daumar, P.; Aubel, C.; Guerder, S.; Mounetou, E.; Penault-Lorca, F.; Bamdad, M. Low-Dose and Long-Term Olaparib Treatment Sensitizes MDA-MB-231 and SUM1315 Triple-Negative Breast Cancers Spheroids to Fractioned Radiotherapy. J. Clin. Med. 2020, 9, 64. https://doi.org/10.3390/jcm9010064
Dubois C, Martin F, Hassel C, Magnier F, Daumar P, Aubel C, Guerder S, Mounetou E, Penault-Lorca F, Bamdad M. Low-Dose and Long-Term Olaparib Treatment Sensitizes MDA-MB-231 and SUM1315 Triple-Negative Breast Cancers Spheroids to Fractioned Radiotherapy. Journal of Clinical Medicine. 2020; 9(1):64. https://doi.org/10.3390/jcm9010064
Chicago/Turabian StyleDubois, Clémence, Fanny Martin, Chervin Hassel, Florian Magnier, Pierre Daumar, Corinne Aubel, Sylvie Guerder, Emmanuelle Mounetou, Frédérique Penault-Lorca, and Mahchid Bamdad. 2020. "Low-Dose and Long-Term Olaparib Treatment Sensitizes MDA-MB-231 and SUM1315 Triple-Negative Breast Cancers Spheroids to Fractioned Radiotherapy" Journal of Clinical Medicine 9, no. 1: 64. https://doi.org/10.3390/jcm9010064
APA StyleDubois, C., Martin, F., Hassel, C., Magnier, F., Daumar, P., Aubel, C., Guerder, S., Mounetou, E., Penault-Lorca, F., & Bamdad, M. (2020). Low-Dose and Long-Term Olaparib Treatment Sensitizes MDA-MB-231 and SUM1315 Triple-Negative Breast Cancers Spheroids to Fractioned Radiotherapy. Journal of Clinical Medicine, 9(1), 64. https://doi.org/10.3390/jcm9010064





