In Vitro Characterization of Dental Pulp Stem Cells Cultured in Two Microsphere-Forming Culture Plates
Abstract
:1. Introduction
2. Experimental Section
2.1. Cell Culture
2.2. Cell Proliferation Assay
2.3. In Vitro Functional Multilineage Differentiation
2.4. RNA Isolation and Sequencing
2.5. Statistical Analysis
3. Results
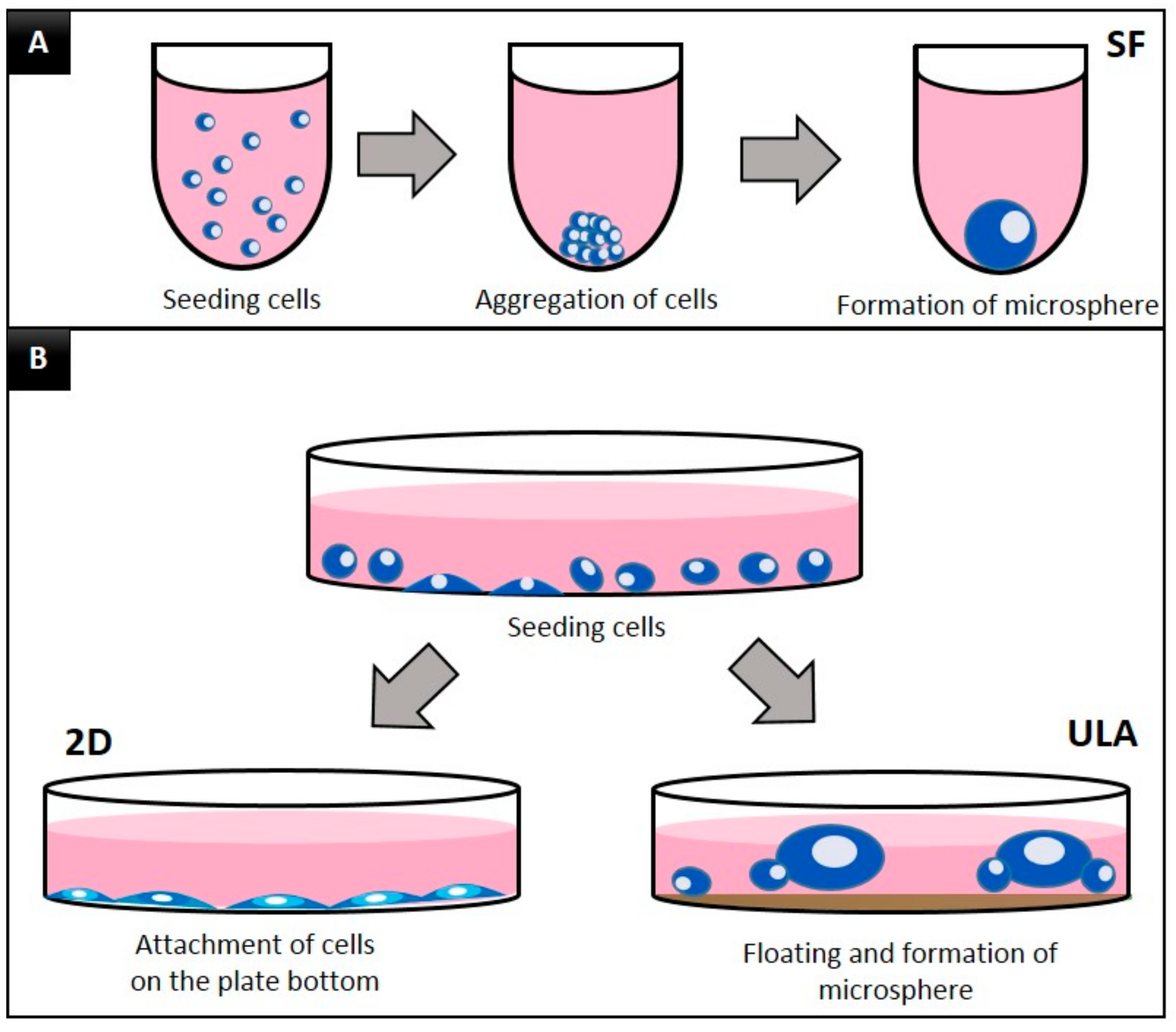
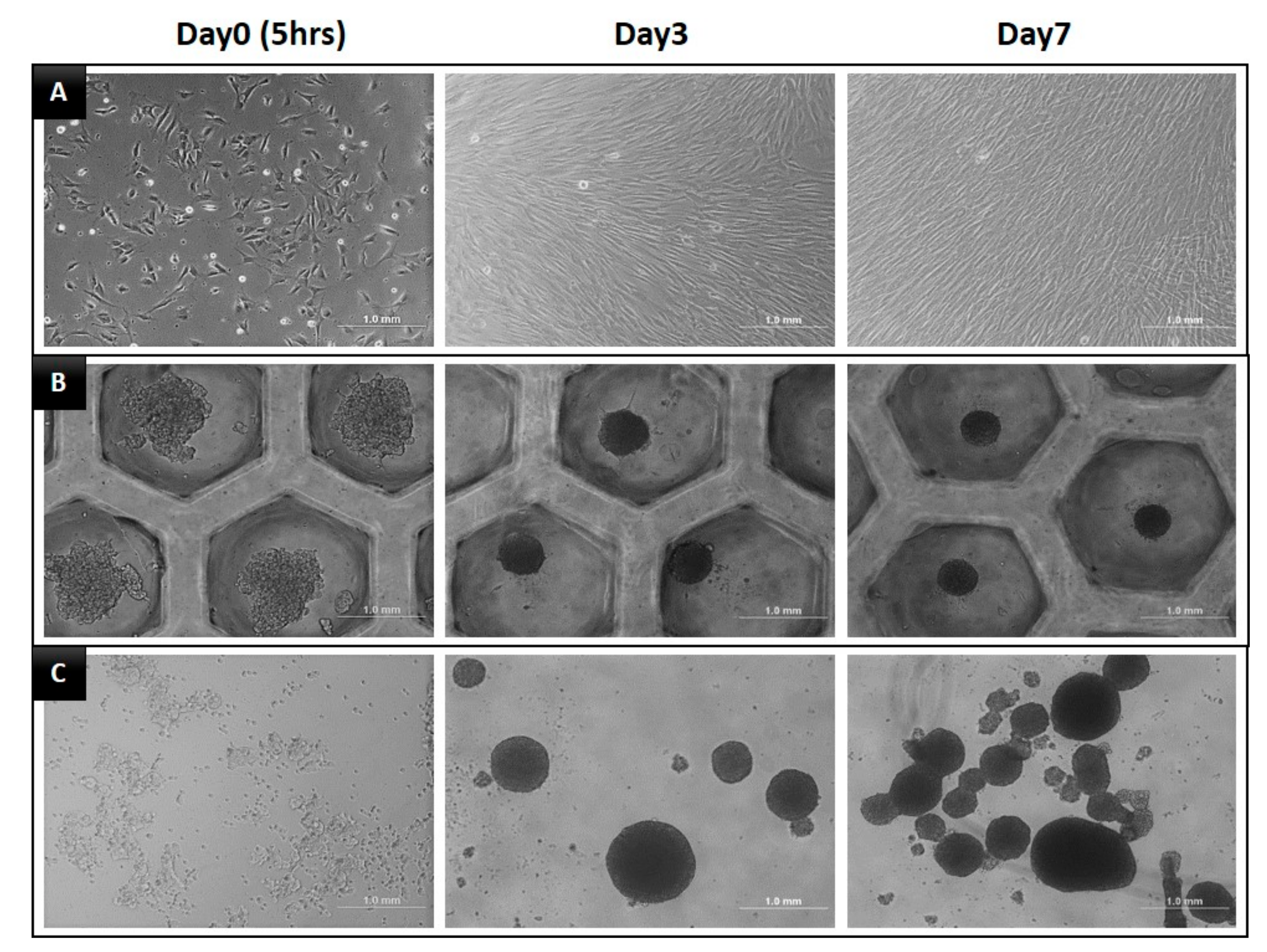
3.1. Morphology
3.2. Cell Viability
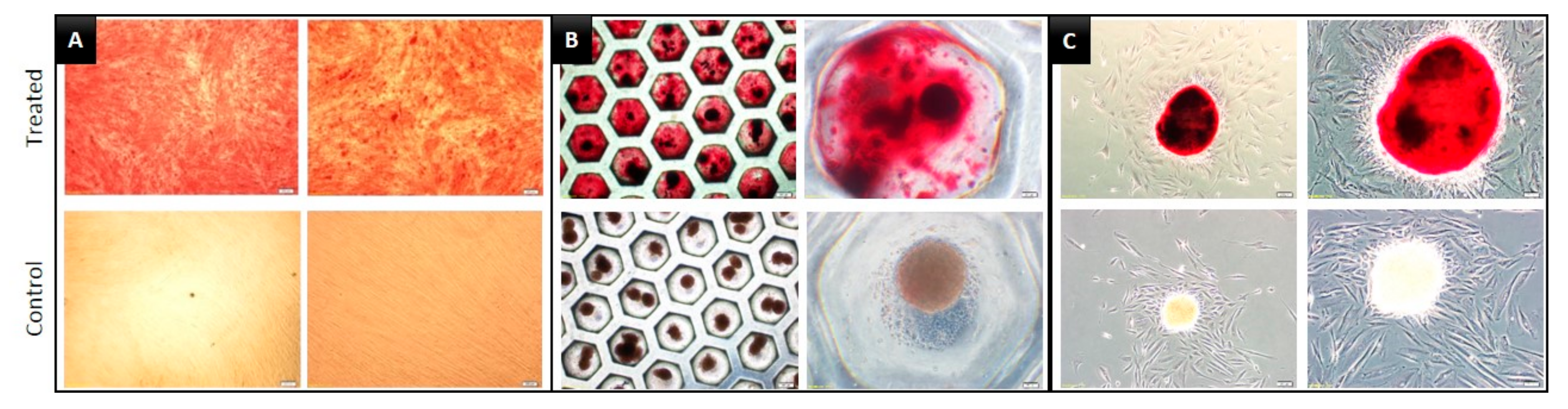
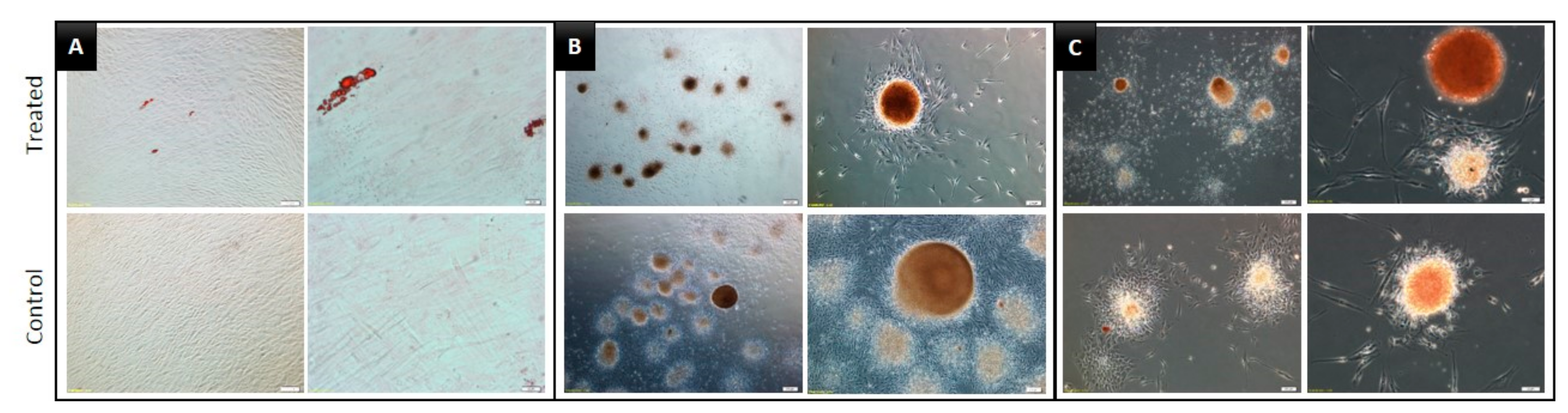
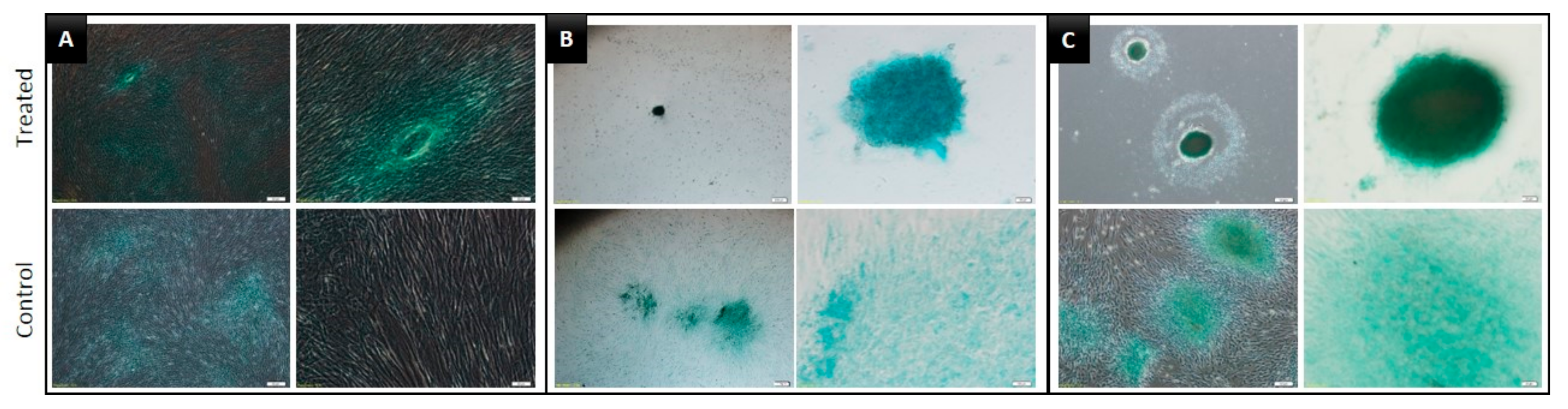
3.3. Multilineage Differentiation Capacity
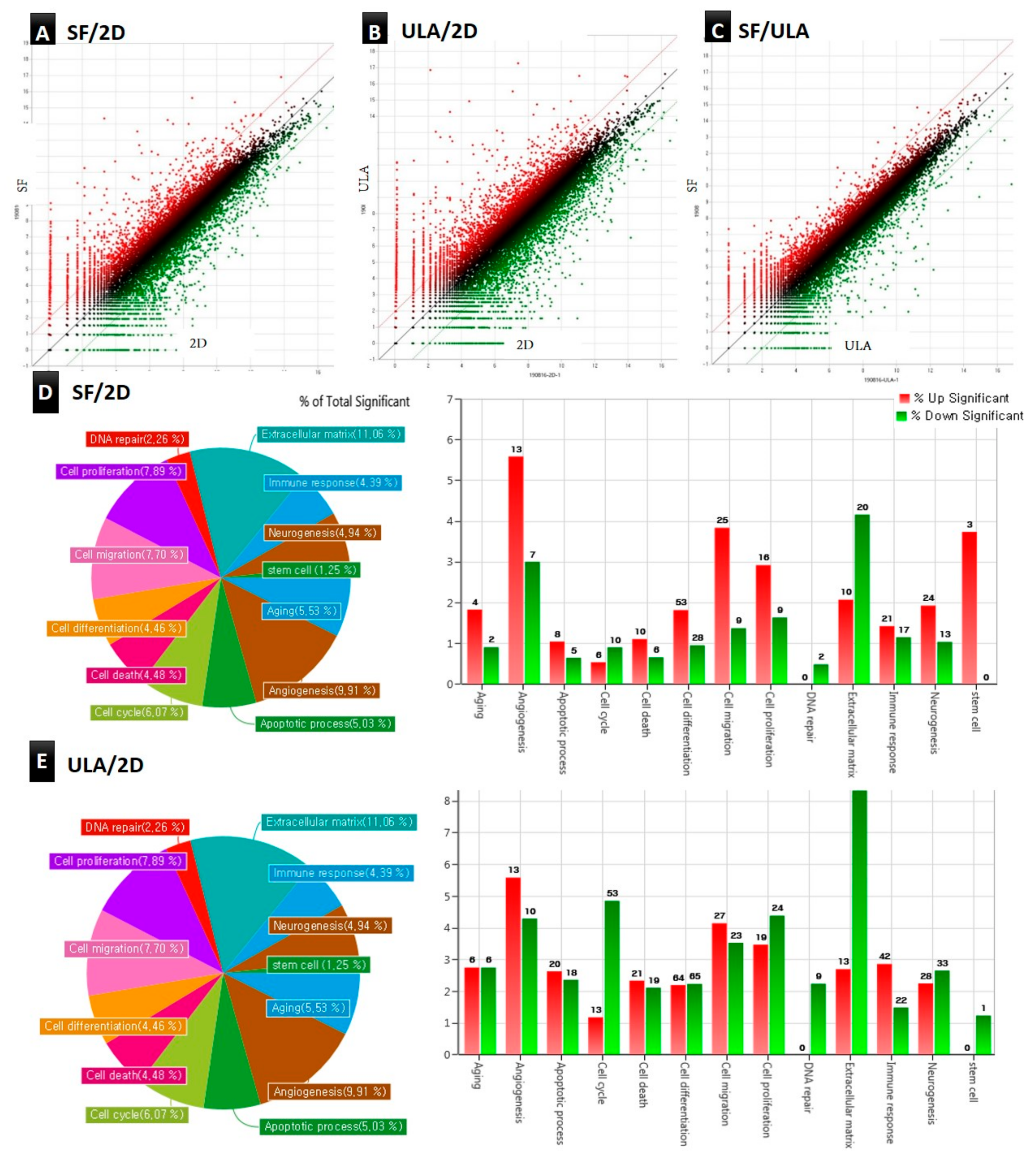
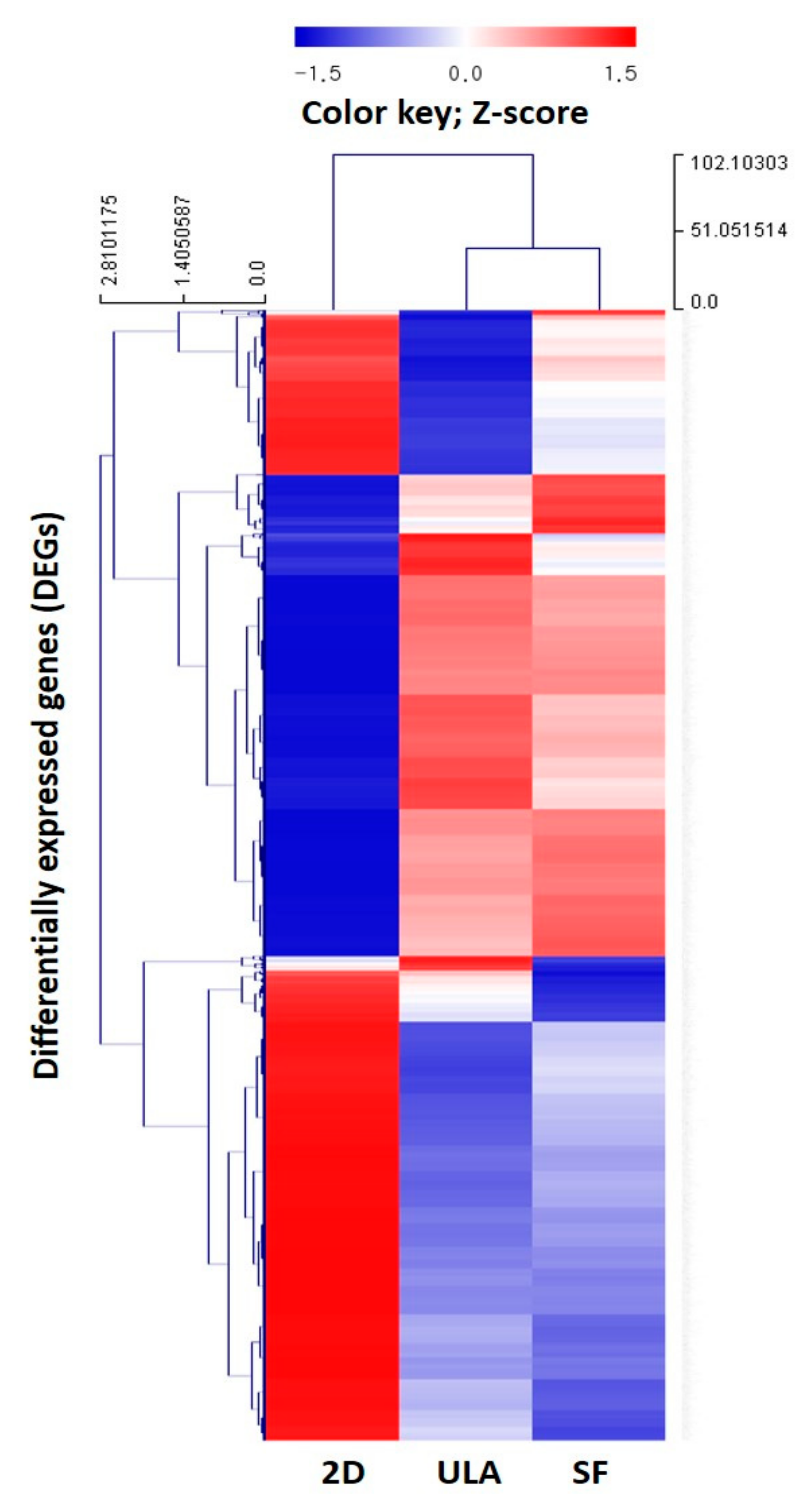
3.4. Analysis of Gene Ontology and Gene Expression Profile
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jang, J.H.; Lee, H.W.; Cho, K.M.; Shin, H.W.; Kang, M.K.; Park, S.H.; Kim, E. In vitro characterization of human dental pulp stem cells isolated by three different methods. Restor. Dent. Endod. 2016, 41, 283–295. [Google Scholar] [CrossRef] [Green Version]
- Potdar, P.D.; Jethmalani, Y.D. Human dental pulp stem cells: Applications in future regenerative medicine. World J. Stem Cells 2015, 7, 839–851. [Google Scholar] [CrossRef]
- Riccio, M.; Resca, E.; Maraldi, T.; Pisciotta, A.; Ferrari, A.; Bruzzesi, G.; De, P.A. Human dental pulp stem cells produce mineralized matrix in 2D and 3D cultures. Eur. J. Histochem. 2010, 54, e46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volponi, A.A.; Pang, Y.; Sharpe, P.T. Stem cell-based biological tooth repair and regeneration. Trends Cell Biol. 2010, 20, 715–722. [Google Scholar] [CrossRef] [Green Version]
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension–how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Buttler-Buecher, P.; Denecke, B.; Arana-Chavez, V.E.; Apel, C.A. Comprehensive analysis of human dental pulp cell spheroids in a three-dimensional pellet culture system. Arch. Oral Biol. 2018, 91, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Kawashima, N.; Takashino, N.; Koizumi, Y.; Takimoto, K.; Suzuki, N.; Saito, M.; Suda, H. Three-dimensional spheroid culture promotes odonto/osteoblastic differentiation of dental pulp cells. Arch. Oral Biol. 2014, 59, 310–317. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, F.J. Biomaterials & scaffolds for tissue engineering. Mater. Today 2011, 14, 88–95. [Google Scholar]
- Atari, M.; Caballe-Serrano, J.; Gil-Recio, C.; Giner-Delgado, C.; Martinez-Sarra, E.; Garcia-Fernandez, D.A.; Barajas, M.; Hernández-Alfaro, F.; Ferrés-Padró, E.; Giner-Tarrida, L. The enhancement of osteogenesis through the use of dental pulp pluripotent stem cells in 3D. Bone 2012, 50, 930–941. [Google Scholar] [CrossRef]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and animal models: Are 3D cultures the ideal tool to study cancer-microenvironment interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [Green Version]
- Baharvand, H.; Hashemi, S.M.; Ashtiani, S.K.; Farrokhi, A. Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int. J. Dev. 2006, 50, 645–652. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Inaba, A.; Mohindroo, N.; Ganesh, D.; Martin, C.E.; Chugal, N.; Kim, R.H.; Kang, M.K.; Park, N.H.; Shin, K.H. Three-dimensional sphere-forming cells are unique multipotent cell population in dental pulp cells. J. Endod. 2017, 43, 1302–1308. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- Nuti, N.; Corallo, C.; Chan, B.M.; Ferrari, M.; Gerami-Naini, B. Multipotent Differentiation of Human Dental Pulp Stem Cells: A Literature Review. Stem Cell Rev. Rep. 2016, 12, 511–523. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Donor-matched comparison of dental pulp stem cells and bone marrow-derived mesenchymal stem cells in a rat model. J. Tissue Eng. Regen. Med. 2010, 4, 73–81. [Google Scholar]
- Galler, K.M.; D’souza, R.; Hartgerink, J.; Schmalz, G. Scaffolds for dental pulp tissue engineering. Adv. Dent. Res. 2011, 23, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.; Lee, S.; Hwang, Y.; Rosa, V.; Lee, K.; Min, K. Behaviour of human dental pulp cells cultured in a collagen hydrogel scaffold cross-linked with cinnamaldehyde. Int. Endod. J. 2017, 50, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Cavalcanti, B.N.; Zeitlin, B.D.; Nör, J.E. A hydrogel scaffold that maintains viability and supports differentiation of dental pulp stem cells. Dent. Mater. 2013, 29, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Rücker, M.; Laschke, M.W.; Junker, D.; Carvalho, C.; Schramm, A.; Mülhaupt, R.; Gellrich, N.C.; Menger, M.D. Angiogenic and inflammatory response to biodegradable scaffolds in dorsal skinfold chambers of mice. Biomaterials 2006, 27, 5027–5038. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]

| Group | Day 1 | Day 3 | Day 5 | Day 7 |
|---|---|---|---|---|
| 2D | 100.00 ± 3.71 Aa | 100.66 ± 11.85 Aa | 110.15 ± 1.38 Ab | 120.56 ± 3.76 Ac |
| SF | 100.00 ± 3.23 Aa | 55.81 ± 1.27 Bb | 51.40 ± 1.66 Bc | 47.00 ± 0.95 Bd |
| ULA | 100.00 ± 3.79 Aa | 52.53 ± 1.08 Bb | 33.76 ± 0.57 Cc | 35.15 ± 0.67 Cc |
| Type III Sum of Squares | df | F | Significance | |
|---|---|---|---|---|
| Culturing Method | 127,783.6 | 2 | 3907.67 | 0.000 |
| Culturing Time | 48,252.2 | 3 | 983.71 | 0.000 |
| Method × Time | 50,905.9 | 6 | 518.91 | 0.000 |
| Gene | Description | Fold Changes | Related Biological Function |
|---|---|---|---|
| ULA/2D | |||
| PTGS2 | prostaglandin-endoperoxide synthase 2 | 930.52 | Angiogenesis, cell differentiation, inflammatory response, cellular response to hypoxia, response to oxidative stress |
| AREG | amphiregulin | 686.75 | Cell differentiation, cell proliferation, cell-cell signaling |
| EREG | epiregulin | 271.40 | Cell differentiation, cell proliferation, angiogenesis, cell cycle, cell-cell signaling, mRNA transcription |
| GFAP | glial fibrillary acidic protein | 54.93 | Extracellular matrix organization, regulation of protein complex assembly, response to wound healing |
| NEFM | neurofilament, medium poly peptide | 54.31 | Neurofilament bundle assembly, axon development |
| NR4A3 | nuclear receptor subfamily 4 group A member 3 | 46.59 | Apoptotic process, cell proliferation, regulation of transcription, |
| MYCN | N-myc proto-oncogene protein | 35.81 | Regulation of transcription, cell proliferation, cell differentiation, cell death regulation |
| SOD2 | superoxide dismutase 2, mitochondrial | 29.84 | Apoptotic process, regulation of transcription, oxidation-reduction process |
| ZC3H12A | zinc finger CCCH-type containing 12A | 20.11 | Cell differentiation, cell death, autophagy, p38MAPK cascade |
| LIF | leukemia inhibitory factor | 18.79 | Cell proliferation, cell differentiation |
| SF/2D | |||
| AREG | amphiregulin | 2066.98 | Cell differentiation, cell proliferation |
| RANBP3L | RAN binding protein 3 like | 500.44 | Cell cycle, cell differentiation |
| MYCN | N-myc proto-onco gene protein | 116.69 | Regulation of transcription, cell proliferation, cell differentiation, cell death regulation |
| EREG | epiregulin | 111.00 | Angiogenesis, cell cycle, cell differentiation, cell proliferation |
| NKD1 | naked cuticle homolog 1 | 107.20 | DNA repair, Wnt signaling pathway, |
| PTGS2 | prostaglandin-endoperoxide synthase 2 | 97.83 | Angiogenesis, cell differentiation, inflammatory response |
| CNTN4 | contactin 4 | 36.70 | Cell adhesion, cell differentiation, axonogenesis, |
| GFAP | glial fibrillary acidic protein | 21.71 | Extracellular matrix organization, regulation of protein complex assembly, response to wound healing |
| FGFR3 | fibroblast growth factor receptor 3 | 20.27 | Bone mineralization, cell-cell signaling, apoptotic process, cell proliferation, cell differentiation, |
| NR4A3 | nuclear receptor subfamily 4 group A member 3 | 18.71 | Apoptotic process, regulation of transcription, cell proliferation |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bu, N.-U.; Lee, H.-S.; Lee, B.-N.; Hwang, Y.-C.; Kim, S.-Y.; Chang, S.W.; Choi, K.-K.; Kim, D.-S.; Jang, J.-H. In Vitro Characterization of Dental Pulp Stem Cells Cultured in Two Microsphere-Forming Culture Plates. J. Clin. Med. 2020, 9, 242. https://doi.org/10.3390/jcm9010242
Bu N-U, Lee H-S, Lee B-N, Hwang Y-C, Kim S-Y, Chang SW, Choi K-K, Kim D-S, Jang J-H. In Vitro Characterization of Dental Pulp Stem Cells Cultured in Two Microsphere-Forming Culture Plates. Journal of Clinical Medicine. 2020; 9(1):242. https://doi.org/10.3390/jcm9010242
Chicago/Turabian StyleBu, Nam-Ung, Hyo-Seol Lee, Bin-Na Lee, Yun-Chan Hwang, Sun-Young Kim, Seok Woo Chang, Kyoung-Kyu Choi, Duck-Su Kim, and Ji-Hyun Jang. 2020. "In Vitro Characterization of Dental Pulp Stem Cells Cultured in Two Microsphere-Forming Culture Plates" Journal of Clinical Medicine 9, no. 1: 242. https://doi.org/10.3390/jcm9010242
APA StyleBu, N.-U., Lee, H.-S., Lee, B.-N., Hwang, Y.-C., Kim, S.-Y., Chang, S. W., Choi, K.-K., Kim, D.-S., & Jang, J.-H. (2020). In Vitro Characterization of Dental Pulp Stem Cells Cultured in Two Microsphere-Forming Culture Plates. Journal of Clinical Medicine, 9(1), 242. https://doi.org/10.3390/jcm9010242









