Genetic Variants of lncRNA MALAT1 Exert Diverse Impacts on the Risk and Clinicopathologic Characteristics of Patients with Hepatocellular Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects, Ethics, and Consent
2.2. Genomic DNA Extraction
2.3. Selection of MALAT1 SNPs
2.4. Genotyping of MALAT1 SNPs
2.5. Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
3.1. Demographic Characteristics of Recruited Subjects
3.2. Associations between MALAT1 Candidate SNPs and HCC Susceptibility
3.3. Relationships between Clinicopathological Features of HCC and MALAT1 SNPs in Different Genders
3.4. Relationships between Clinicopathological Features and MALAT1 SNPs in HCC Patients Who Smoked Tobacco
3.5. Serum Alpha-Fetoprotein (AFP), Aspartate Aminotransferase (AST), and Alanine Aminotransferase (ALT) Levels among Different MALAT1 SNP Carriers in HCC Patients
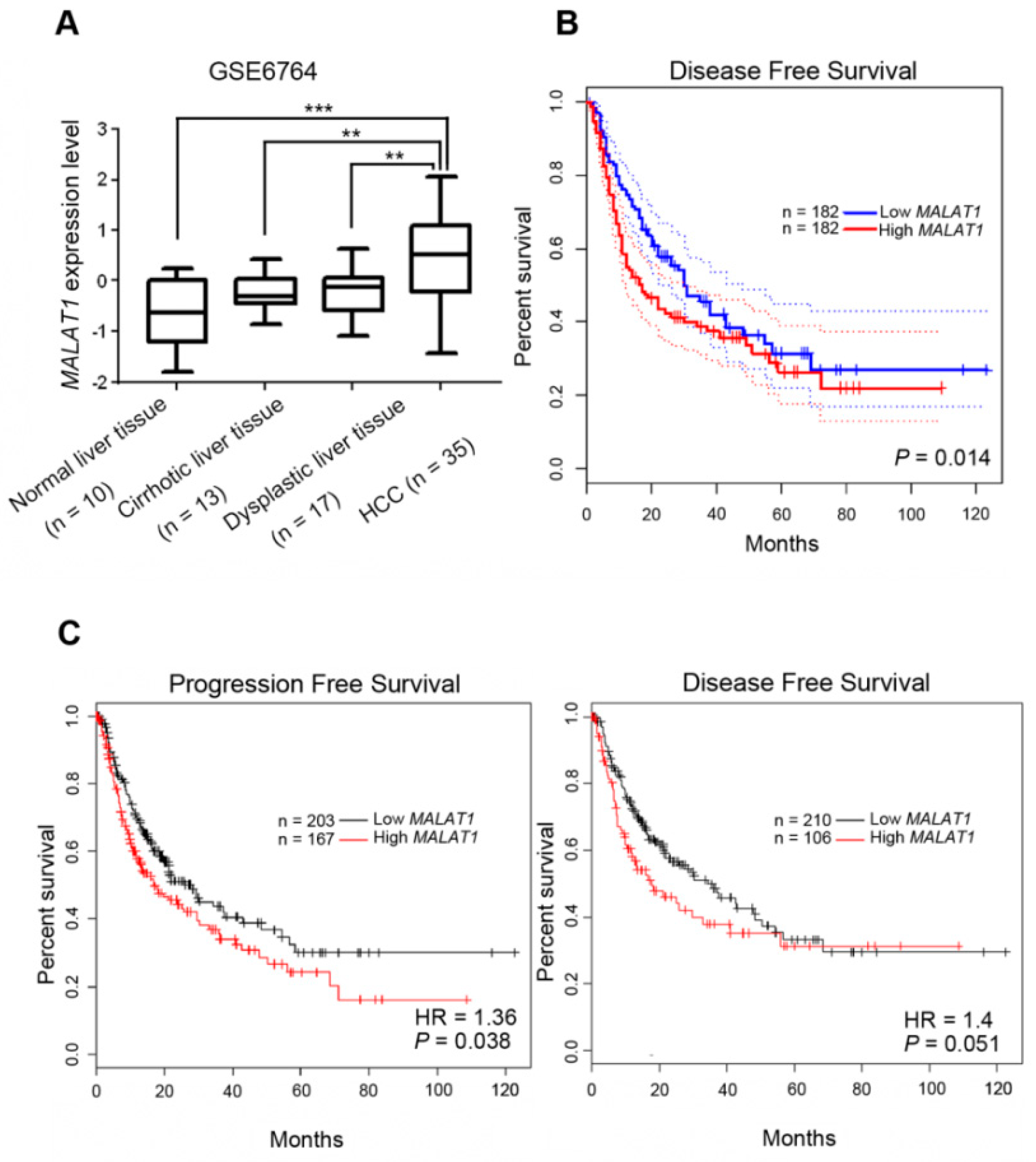
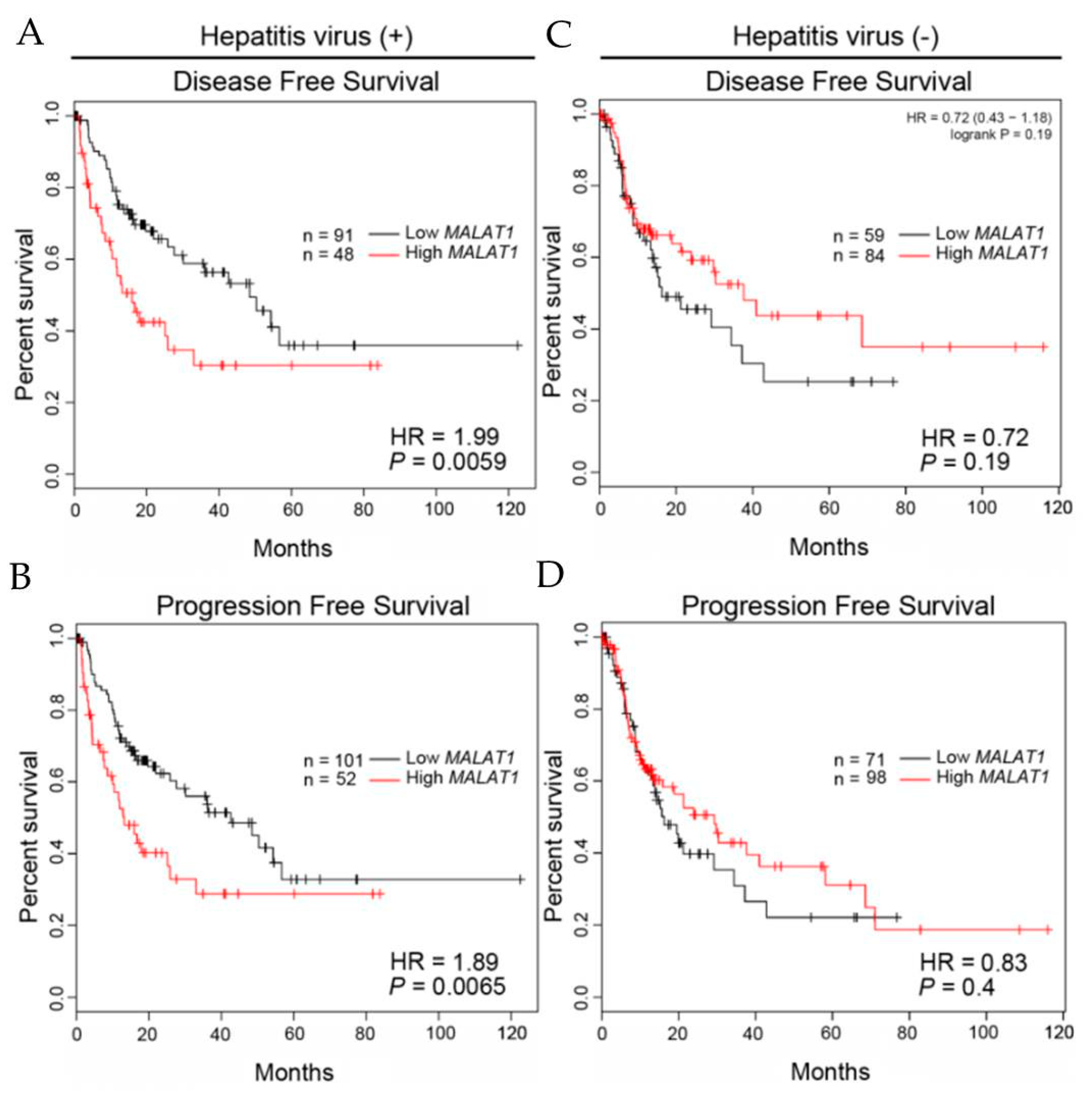
3.6. Clinical Relevance of MALAT1 Expression Levels in HCC Patients with or without Hepatitis Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Llovet, J.M. Liver cancer: Time to evolve trial design after everolimus failure. Nat. Rev. Clin. Oncol. 2014, 11, 506–507. [Google Scholar] [CrossRef]
- Roessler, S.; Budhu, A.; Wang, X.W. Deciphering cancer heterogeneity: The biological space. Front. Cell. Dev. Biol. 2014, 2, 12. [Google Scholar] [CrossRef]
- Shen, H.; Laird, P.W. Interplay between the cancer genome and epigenome. Cell 2013, 153, 38–55. [Google Scholar] [CrossRef]
- Lee, S.M.; Kim-Ha, J.; Choi, W.Y.; Lee, J.; Kim, D.; Lee, J.; Choi, E.; Kim, Y.J. Interplay of genetic and epigenetic alterations in hepatocellular carcinoma. Epigenomics 2016, 8, 993–1005. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Noncoding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef]
- Shen, L.; Chen, L.; Wang, Y.; Jiang, X.; Xia, H.; Zhuang, Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial–mesenchymal transition in lung cancer. J. Neurooncol. 2015, 121, 101–108. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, S.; Li, Q.; Ji, Q.; Guo, P.; Liu, X. MALAT1: A long noncoding RNA highly associated with human cancers. Oncol. Lett. 2018, 16, 19–26. [Google Scholar]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long noncoding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef]
- Zheng, H.T.; Shi, D.B.; Wang, Y.W.; Li, X.X.; Xu, Y.; Tripathi, P.; Gu, W.L.; Cai, G.X.; Cai, S.J. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3174–3181. [Google Scholar]
- Pang, E.J.; Yang, R.; Fu, X.B.; Liu, Y.F. Overexpression of long noncoding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumor Biol. 2015, 36, 2403–2407. [Google Scholar] [CrossRef]
- Zhang, H.M.; Yang, F.Q.; Chen, S.J.; Che, J.; Zheng, J.H. Upregulation of long noncoding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumor Biol. 2015, 36, 2947–2955. [Google Scholar] [CrossRef]
- Liu, J.; Dong, H.; Yang, Y.; Qian, Y.; Liu, J.; Li, Z.; Guan, H.; Chen, Z.; Li, C.; Zhang, K.; et al. Upregulation of long noncoding RNA MALAT1 in papillary thyroid cancer and its diagnostic value. Future Oncol. (Lond. Engl.) 2018, 14, 3015–3022. [Google Scholar] [CrossRef]
- Lai, M.C.; Yang, Z.; Zhou, L.; Zhu, Q.Q.; Xie, H.Y.; Zhang, F.; Wu, L.M.; Chen, L.M.; Zheng, S.S. Long noncoding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. (Northwood Lond. Engl.) 2012, 29, 1810–1816. [Google Scholar] [CrossRef]
- Luo, F.; Sun, B.; Li, H.; Xu, Y.; Liu, Y.; Liu, X.; Lu, L.; Li, J.; Wang, Q.; Wei, S.; et al. A MALAT1/HIF-2alpha feedback loop contributes to arsenite carcinogenesis. Oncotarget 2016, 7, 5769–5787. [Google Scholar]
- Yuan, P.; Cao, W.; Zang, Q.; Li, G.; Guo, X.; Fan, J. The HIF-2alpha-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem. Biophys. Res. Commun. 2016, 478, 1067–1073. [Google Scholar] [CrossRef]
- Malakar, P.; Shilo, A.; Mogilevsky, A.; Stein, I.; Pikarsky, E.; Nevo, Y.; Benyamini, H.; Elgavish, S.; Zong, X.; Prasanth, K.V.; et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017, 77, 1155–1167. [Google Scholar] [CrossRef]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef]
- Peng, R.; Luo, C.; Guo, Q.; Cao, J.; Yang, Q.; Dong, K.; Wang, S.; Wang, K.; Song, C. Association analyses of genetic variants in long noncoding RNA MALAT1 with breast cancer susceptibility and mRNA expression of MALAT1 in Chinese Han population. Gene 2018, 642, 241–248. [Google Scholar] [CrossRef]
- Zhao, K.; Jin, S.; Wei, B.; Cao, S.; Xiong, Z. Association study of genetic variation of lncRNA MALAT1 with carcinogenesis of colorectal cancer. Cancer Manag. Res. 2018, 10, 6257–6261. [Google Scholar] [CrossRef]
- Zhu, H.; Lv, Z.; An, C.; Shi, M.; Pan, W.; Zhou, L.; Yang, W.; Yang, M. Onco-lncRNA HOTAIR and its functional genetic variants in papillary thyroid carcinoma. Sci. Rep. 2016, 6, 31969. [Google Scholar] [CrossRef]
- Li, Y.; Bao, C.; Gu, S.; Ye, D.; Jing, F.; Fan, C.; Jin, M.; Chen, K. Associations between novel genetic variants in the promoter region of MALAT1 and risk of colorectal cancer. Oncotarget 2017, 8, 92604–92614. [Google Scholar] [CrossRef]
- Qu, Y.; Shao, N.; Yang, W.; Wang, J.; Cheng, Y. Association of polymorphisms in MALAT1 with the risk of esophageal squamous cell carcinoma in a Chinese population. Onco. Targets Ther. 2019, 12, 2495–2503. [Google Scholar] [CrossRef]
- Zhuo, Y.; Zeng, Q.; Zhang, P.; Li, G.; Xie, Q.; Cheng, Y. Functional polymorphism of lncRNA MALAT1 contributes to pulmonary arterial hypertension susceptibility in Chinese people. Clin. Chem. Lab. Med. 2017, 55, 38–46. [Google Scholar] [CrossRef]
- Wang, J.Z.; Xiang, J.J.; Wu, L.G.; Bai, Y.S.; Chen, Z.W.; Yin, X.Q.; Wang, Q.; Guo, W.H.; Peng, Y.; Guo, H.; et al. A genetic variant in long noncoding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: A survival cohort analysis. BMC Cancer 2017, 17, 167. [Google Scholar]
- Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Menyhart, O.; Nagy, A.; Gyorffy, B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci. 2018, 5, 181006. [Google Scholar] [CrossRef]
- Gowda, S.; Desai, P.B.; Hull, V.V.; Math, A.A.K.; Vernekar, S.N.; Kulkarni, S.S. A review on laboratory liver function tests. Pan Afr. Med. J. 2009, 3, 17. [Google Scholar]
- Klingenberg, M.; Matsuda, A.; Diederichs, S.; Patel, T. Noncoding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J. Hematol. 2017, 67, 603–618. [Google Scholar]
- Mehra, M.; Chauhan, R. Long Noncoding RNAs as a Key Player in Hepatocellular Carcinoma. Biomark. Cancer 2017, 9, 1179299X17737301. [Google Scholar] [CrossRef]
- Wu, W.; Wagner, E.K.; Hao, Y.; Rao, X.; Dai, H.; Han, J.; Chen, J.; Storniolo, A.M.V.; Liu, Y.; He, C. Tissue-specific Co-expression of Long Noncoding and Coding RNAs Associated with Breast Cancer. Sci. Rep. 2016, 6, 32731. [Google Scholar] [CrossRef]
- Foroughi, K.; Amini, M.; Atashi, A.; Mahmoodzadeh, H.; Hamann, U.; Manoochehri, M. Tissue-Specific Down-Regulation of the Long Noncoding RNAs PCAT18 and LINC01133 in Gastric Cancer Development. Int. J. Mol. Sci. 2018, 19, 3881. [Google Scholar] [CrossRef]
- Fok, E.T.; Scholefield, J.; Fanucchi, S.; Mhlanga, M.M. The emerging molecular biology toolbox for the study of long noncoding RNA biology. Epigenomics 2017, 9, 1317–1327. [Google Scholar] [CrossRef]
- Abbastabar, M.; Sarfi, M.; Golestani, A.; Khalili, E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. 2018, 17, 900–913. [Google Scholar]
- Hou, Z.; Xu, X.; Fu, X.; Tao, S.; Zhou, J.; Liu, S.; Tan, D. HBx-related long noncoding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am. J. Cancer Res. 2017, 7, 845–856. [Google Scholar]
- Li, C.; Miao, R.; Liu, S.; Wan, Y.; Zhang, S.; Deng, Y.; Bi, J.; Qu, K.; Zhang, J.; Liu, C. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget 2017, 8, 28683–28695. [Google Scholar] [CrossRef]
- Liu, D.; Zhu, Y.; Pang, J.; Weng, X.; Feng, X.; Guo, Y. Knockdown of long noncoding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J. Cell. Biochem. 2018, 119, 1368–1380. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Lin, L.; Li, H.; Zhu, Y.; He, S.; Ge, H. Expression of metastasis-associated lung adenocarcinoma transcript 1 long noncoding RNA in vitro and in patients with non-small cell lung cancer. Oncol. Lett. 2018, 15, 9443–9449. [Google Scholar]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget 2017, 8, 110635–110649. [Google Scholar] [CrossRef]
- Xu, H.; Wei, Y.; Zhang, Y.; Xu, Y.; Li, F.; Liu, J.; Zhang, W.; Han, X.; Tan, R.; Shen, P. Oestrogen attenuates tumour progression in hepatocellular carcinoma. J. Pathol. 2012, 228, 216–229. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, C.; Liu, Y.; Wu, C. 17beta-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem. Biophys. Res. Commun. 2014, 445, 388–393. [Google Scholar] [CrossRef]
- Zhang, X.F.; Wei, T.; Liu, X.M.; Liu, C.; Lv, Y. Impact of cigarette smoking on outcome of hepatocellular carcinoma after surgery in patients with hepatitis B. PLoS ONE 2014, 9, e85077. [Google Scholar] [CrossRef]
- Lu, L.; Luo, F.; Liu, Y.; Liu, X.; Shi, L.; Lu, X.; Liu, Q. Posttranscriptional silencing of the lncRNA MALAT1 by miR-217 inhibits the epithelial–mesenchymal transition via enhancer of zeste homolog 2 in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol. Appl. Pharmacol. 2015, 289, 276–285. [Google Scholar] [CrossRef]
- Hrdlickova, B.; de Almeida, R.C.; Borek, Z.; Withoff, S. Genetic variation in the noncoding genome: Involvement of micro-RNAs and long noncoding RNAs in disease. Biochim. Biophys. Acta 2014, 1842, 1910–1922. [Google Scholar] [CrossRef]
- Ji, X.; Zhang, J.; Liu, L.; Lin, Z.; Pi, L.; Lin, Z.; Tian, N.; Lin, X.; Chen, S.; Yu, X.; et al. Association of tagSNPs at lncRNA MALAT-1 with HCC Susceptibility in a Southern Chinese Population. Sci. Rep. 2019, 9, 10895. [Google Scholar] [CrossRef]
| Variable | Controls (N = 1199) n (%) | Patients (N = 394) n (%) | p Value |
|---|---|---|---|
| Age (years) | |||
| ≥55 | 912 (76.1%) | 307 (77.9%) | 0.451 |
| Gender | |||
| Male | 839 (70%) | 273 (69.3%) | 0.797 |
| Cigarette smoking | |||
| Yes | 471 (39.3%) | 156 (39.6%) | 0.913 |
| Alcohol consumption | |||
| Yes | 169 (14.1%) | 139 (35.3%) | <0.001 * |
| HBsAg | |||
| Positive | 146 (12.2%) | 167 (42.4%) | <0.001 * |
| Anti-HCV | |||
| Positive | 53 (4.4%) | 178 (45.2%) | <0.001 * |
| AFP (ng/mL) a | 20.83 | ||
| AST (IU/L) a | 48 | ||
| ALT (IU/L) a | 43 | ||
| Stage | |||
| III + IV | 117 (29.7%) | ||
| Tumor T status | |||
| T3 + T4 | 112 (28.4%) | ||
| Lymph node status | |||
| N1 + N2 + N3 | 12 (3%) | ||
| Metastasis | |||
| M1 | 19 (4.8%) | ||
| Child–Pugh grade | |||
| B or C | 74 (18.8%) | ||
| Liver cirrhosis | |||
| Positive | 326 (82.7%) |
| Variable | Controls (N = 1199) n (%) | Patients (N = 394) n (%) | OR (95% CI) | AOR (95% CI) a |
|---|---|---|---|---|
| rs3200401 | ||||
| CC | 802 (66.9%) | 263 (66.8%) | 1.000 (reference) | 1.000 (reference) |
| CT | 347 (28.9%) | 117 (29.7%) | 1.028 (0.799–1.322) | 1.051 (0.805–1.372) |
| TT | 50 (4.2%) | 14 (3.6%) | 0.854 (0.465–1.570) | 0.881 (0.459–1.689) |
| CT + TT | 397 (33.1%) | 131 (33.2%) | 1.006 (0.790–1.281) | 1.030 (0.797–1.331) |
| rs619586 | ||||
| AA | 1014 (84.6%) | 330 (83.8%) | 1.000 (reference) | 1.000 (reference) |
| AG | 175 (14.6%) | 61 (15.5%) | 1.071 (0.780–1.470) | 0.982 (0.701–1.376) |
| GG | 10 (0.8%) | 3 (0.8%) | 0.922 (0.252–3.370) | 0.563 (0.141–2.250) |
| AG + GG | 185 (15.4%) | 64 (16.2%) | 1.063 (0.780–1.450) | 0.954 (0.686–1.327) |
| rs1194338 | ||||
| CC | 510 (42.5%) | 172 (43.7%) | 1.000 (reference) | 1.000 (reference) |
| CA | 537 (44.8%) | 175 (44.4%) | 0.966 (0.758–1.232) | 0.980 (0.759–1.266) |
| AA | 152 (12.7%) | 47 (11.9%) | 0.917 (0.633–1.327) | 0.995 (0.672–1.474) |
| CA + AA | 689 (57.5%) | 222 (56.3%) | 0.955 (0.759–1.202) | 0.983 (0.772–1.253) |
| Variable | Controls (N = 287) n (%) | Patients (N = 87) n (%) | OR (95% CI) | AOR (95% CI) a |
|---|---|---|---|---|
| rs3200401 | ||||
| CC | 193 (67.2%) | 55 (63.2%) | 1.000 (reference) | 1.000 (reference) |
| CT | 82 (28.6%) | 30 (34.5%) | 1.284 (0.768–2.147) | 1.452 (0.837–2.516) |
| TT | 12 (4.2%) | 2 (2.3%) | 0.585 (0.127–2.692) | 0.725 (0.148–3.548) |
| CT + TT | 94 (32.8%) | 32 (36.8%) | 1.195 (0.724–1.971) | 1.364 (0.799–2.330) |
| rs619586 | ||||
| AA | 239 (83.3%) | 82 (94.3%) | 1.000 (reference) | 1.000 (reference) |
| AG | 47 (16.4%) | 5 (5.7%) | 0.310 (0.119–0.806) p = 0.016 * | 0.289 (0.108–0.773) p = 0.013 * |
| GG | 1 (0.3%) | 0 (0%) | NA | NA |
| AG + GG | 48 (16.7%) | 5 (5.7%) | 0.304 (0.117–0.789) p = 0.014 * | 0.286 (0.107–0.765) p = 0.013 * |
| rs1194338 | ||||
| CC | 124 (43.2%) | 36 (41.4%) | 1.000 (reference) | 1.000 (reference) |
| CA | 126 (43.9%) | 42 (48.3%) | 1.148 (0.690–1.911) | 1.313 (0.760–2.266) |
| AA | 37 (12.9%) | 9 (10.3%) | 0.838 (0.370–1.898) | 1.330 (0.548–3.232) |
| CA + AA | 163 (56.8%) | 51 (58.6%) | 1.078 (0.663–1.753) | 1.316 (0.778–2.224) |
| Variable | Genotypic Frequencies | |||
|---|---|---|---|---|
| CC (%) (n = 51) | CA + AA (%) (n = 70) | OR (95% CI) | p Value | |
| Clinical Stage | ||||
| Stage III/IV | 12 (23.5%) | 21 (30%) | 1.393 (0.611–3.177) | 0.431 |
| Tumor size | ||||
| >T2 | 9 (17.6%) | 20 (28.6%) | 1.867 (0.769–4.533) | 0.168 |
| Lymph node metastasis | ||||
| Yes | 2 (3.9%) | 0 (0%) | NA | NA |
| Distant metastasis | ||||
| Yes | 3 (5.9%) | 2 (2.9%) | 0.471 (0.076–2.925) | 0.419 |
| Vascular invasion | ||||
| Yes | 13 (25.5%) | 8 (11.4%) | 0.377 (0.143–0.994) | 0.049 * |
| Child–Pugh grade | ||||
| B or C | 12 (23.5%) | 13 (18.6%) | 0.741 (0.306–1.794) | 0.507 |
| HBsAg | ||||
| Positive | 16 (31.4%) | 22 (31.4%) | 1.003 (0.461–2.182) | 0.995 |
| Anti-HCV | ||||
| Positive | 28 (54.9%) | 39 (55.7%) | 1.033 (0.500–2.135) | 0.929 |
| Liver cirrhosis | ||||
| Positive | 45 (88.2%) | 58 (82.9%) | 0.644 (0.225–1.850) | 0.414 |
| Variable | Genotypic Frequencies | |||
|---|---|---|---|---|
| CC (%) (n = 64) | CA + AA (%) (n = 92) | OR (95% CI) | p Value | |
| Clinical stage | ||||
| Stage III/IV | 23 (35.9%) | 26 (28.3%) | 0.702 (0.355–1.39) | 0.311 |
| Tumor size | ||||
| >T2 | 23 (35.9%) | 26 (28.3%) | 0.702 (0.355–1.39) | 0.311 |
| Lymph node metastasis | ||||
| Yes | 2 (3.1%) | 3 (3.3%) | 1.045 (0.170–6.439) | 0.962 |
| Distant metastasis | ||||
| Yes | 2 (3.1%) | 5 (5.4%) | 1.782 (0.335–9.482) | 0.498 |
| Vascular invasion | ||||
| Yes | 14 (21.9%) | 11 (12%) | 0.485 (0.204–1.152) | 0.101 |
| Child–Pugh grade | ||||
| B or C | 17 (26.6%) | 12 (13%) | 0.415 (0.182–0.944) | 0.036 * |
| HBsAg | ||||
| Positive | 28 (43.8%) | 38 (41.3%) | 0.905 (0.475–1.725) | 0.761 |
| Anti-HCV | ||||
| Positive | 31 (48.4%) | 40 (43.5%) | 0.819 (0.432–1.554) | 0.541 |
| Liver cirrhosis | ||||
| Positive | 53 (82.8%) | 77 (83.7%) | 1.065 (0.454–2.500) | 0.884 |
| Variable | Genotypic Frequencies | |||
|---|---|---|---|---|
| CC (%) (n = 100) | CT + TT (%) (n = 56) | OR (95% CI) | p Value | |
| Clinical stage | ||||
| Stage III/IV | 34 (34%) | 15 (26.8%) | 0.710 (0.345–1.462) | 0.353 |
| Tumor size | ||||
| >T2 | 34 (34%) | 15 (26.8%) | 0.710 (0.345–1.462) | 0.353 |
| Lymph node metastasis | ||||
| Yes | 5 (5%) | 0 (0%) | NA | NA |
| Distant metastasis | ||||
| Yes | 6 (6%) | 1 (1.8%) | 0.285 (0.033–2.428) | 0.251 |
| Vascular invasion | ||||
| Yes | 18 (18%) | 7 (12.5%) | 0.651 (0.254–1.669) | 0.371 |
| Child–Pugh grade | ||||
| B or C | 19 (19%) | 10 (17.9%) | 0.927 (0.397–2.162) | 0.860 |
| HBsAg | ||||
| Positive | 36 (36%) | 30 (53.6%) | 2.051 (1.055–3.990) | 0.034 * |
| Anti-HCV | ||||
| Positive | 48 (48%) | 23 (41.1%) | 0.755 (0.390–1.463) | 0.405 |
| Liver cirrhosis | ||||
| Positive | 80 (80%) | 50 (89.3%) | 2.083 (0.783–5.542) | 0.141 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, L.-T.; Chang, J.-H.; Lee, H.-L.; Yang, Y.-C.; Su, S.-C.; Lin, C.-L.; Yang, S.-F.; Chien, M.-H. Genetic Variants of lncRNA MALAT1 Exert Diverse Impacts on the Risk and Clinicopathologic Characteristics of Patients with Hepatocellular Carcinoma. J. Clin. Med. 2019, 8, 1406. https://doi.org/10.3390/jcm8091406
Yuan L-T, Chang J-H, Lee H-L, Yang Y-C, Su S-C, Lin C-L, Yang S-F, Chien M-H. Genetic Variants of lncRNA MALAT1 Exert Diverse Impacts on the Risk and Clinicopathologic Characteristics of Patients with Hepatocellular Carcinoma. Journal of Clinical Medicine. 2019; 8(9):1406. https://doi.org/10.3390/jcm8091406
Chicago/Turabian StyleYuan, Lan-Ting, Jer-Hwa Chang, Hsiang-Lin Lee, Yi-Chieh Yang, Shih-Chi Su, Chien-Liang Lin, Shun-Fa Yang, and Ming-Hsien Chien. 2019. "Genetic Variants of lncRNA MALAT1 Exert Diverse Impacts on the Risk and Clinicopathologic Characteristics of Patients with Hepatocellular Carcinoma" Journal of Clinical Medicine 8, no. 9: 1406. https://doi.org/10.3390/jcm8091406
APA StyleYuan, L.-T., Chang, J.-H., Lee, H.-L., Yang, Y.-C., Su, S.-C., Lin, C.-L., Yang, S.-F., & Chien, M.-H. (2019). Genetic Variants of lncRNA MALAT1 Exert Diverse Impacts on the Risk and Clinicopathologic Characteristics of Patients with Hepatocellular Carcinoma. Journal of Clinical Medicine, 8(9), 1406. https://doi.org/10.3390/jcm8091406





