Non-CYP2D6 Variants Selected by a GWAS Improve the Prediction of Impaired Tamoxifen Metabolism in Patients with Breast Cancer
Abstract
:1. Introduction
2. Material and Methods
2.1. Ethics Statement
2.2. Clinical Cohort
2.3. Quantifying Tamoxifen and Its Metabolites in Plasma
2.4. CYP2D6 Genotyping
2.5. Genome-Wide Microarray Analysis
2.6. Verification Genotyping
2.7. Statistical Analyses
2.7.1. GWAS and Individual Genotyping
2.7.2. Prediction Modeling
3. Results
3.1. Association Analyses
3.2. Predictive Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Desta, Z.; Ward, B.A.; Soukhova, N.V.; Flockhart, D.A. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: Prominent roles for CYP3A and CYP2D6. J. Pharmacol. Exp. Ther. 2004, 310, 1062–1075. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.D.; Zuo, H.; Lee, K.-H.; Trebley, J.P.; Rae, J.M.; Weatherman, R.V.; Desta, Z.; Flockhart, D.A.; Skaar, T.C. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res. Treat. 2004, 85, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.C.; Li, L.; Desta, Z.; Zhao, Q.; Rae, J.M.; Flockhart, D.A.; Skaar, T.C. Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J. Pharmacol. Exp. Ther. 2006, 318, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Mürdter, T.E.; Schroth, W.; Bacchus-Gerybadze, L.; Winter, S.; Heinkele, G.; Simon, W.; Fasching, P.A.; Fehm, T.; German Tamoxifen and AI Clinicians Group; Eichelbaum, M.; et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin. Pharmacol. Ther. 2011, 89, 708–717. [Google Scholar]
- Stearns, V.; Johnson, M.D.; Rae, J.M.; Morocho, A.; Novielli, A.; Bhargava, P.; Hayes, D.F.; Desta, Z.; Flockhart, D.A. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J. Natl. Cancer Inst. 2003, 95, 1758–1764. [Google Scholar] [CrossRef]
- Hawse, J.R.; Subramaniam, M.; Cicek, M.; Wu, X.; Gingery, A.; Grygo, S.B.; Sun, Z.; Pitel, K.S.; Lingle, W.L.; Goetz, M.P.; et al. Endoxifen’s molecular mechanisms of action are concentration dependent and different than that of other anti-estrogens. PLoS ONE 2013, 8, e54613. [Google Scholar] [CrossRef]
- Gong, I.Y.; Teft, W.A.; Ly, J.; Chen, Y.-H.; Alicke, B.; Kim, R.B.; Choo, E.F. Determination of clinically therapeutic endoxifen concentrations based on efficacy from human MCF7 breast cancer xenografts. Breast Cancer Res. Treat. 2013, 139, 61–69. [Google Scholar] [CrossRef]
- Wu, X.; Hawse, J.R.; Subramaniam, M.; Goetz, M.P.; Ingle, J.N.; Spelsberg, T.C. The tamoxifen metabolite, endoxifen, is a potent antiestrogen that targets estrogen receptor alpha for degradation in breast cancer cells. Cancer Res. 2009, 69, 1722–1727. [Google Scholar] [CrossRef]
- Khan, B.A.; Robinson, R.; Fohner, A.E.; Muzquiz, L.I.; Schilling, B.D.; Beans, J.A.; Olnes, M.J.; Trawicki, L.; Frydenlund, H.; Laukes, C.; et al. Cytochrome P450 Genetic Variation Associated with Tamoxifen Biotransformation in American Indian and Alaska Native People. Clin. Transl. Sci. 2018, 11, 312–321. [Google Scholar] [CrossRef] [Green Version]
- Gaedigk, A.; Ingelman-Sundberg, M.; Miller, N.A.; Leeder, J.S.; Whirl-Carrillo, M.; Klein, T.E. PharmVar Steering Committee the Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin. Pharmacol. Ther. 2018, 103, 399–401. [Google Scholar] [CrossRef]
- Rebsamen, M.C.; Desmeules, J.; Daali, Y.; Chiappe, A.; Diemand, A.; Rey, C.; Chabert, J.; Dayer, P.; Hochstrasser, D.; Rossier, M.F. The AmpliChip CYP450 test: Cytochrome P450 2D6 genotype assessment and phenotype prediction. Pharmacogenomics J. 2009, 9, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Caudle, K.E.; Dunnenberger, H.M.; Freimuth, R.R.; Peterson, J.F.; Burlison, J.D.; Whirl-Carrillo, M.; Scott, S.A.; Rehm, H.L.; Williams, M.S.; Klein, T.E.; et al. Standardizing terms for clinical pharmacogenetics test results: Consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet. Med. 2017, 19, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Suman, V.J.; Hoskin, T.L.; Gnant, M.; Filipits, M.; Safgren, S.L.; Kuffel, M.; Jakesz, R.; Rudas, M.; Greil, R.; et al. CYP2D6 metabolism and patient outcome in the Austrian Breast and Colorectal Cancer Study Group trial (ABCSG) 8. Clin. Cancer Res. 2013, 19, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Hertz, D.L.; McLeod, H.L.; Irvin, W.J. Tamoxifen and CYP2D6: A contradiction of data. Oncologist 2012, 17, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Kiyotani, K.; Mushiroda, T.; Imamura, C.K.; Hosono, N.; Tsunoda, T.; Kubo, M.; Tanigawara, Y.; Flockhart, D.A.; Desta, Z.; Skaar, T.C.; et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J. Clin. Oncol. 2010, 28, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-A.; Lim, H.-S. Association between CYP2D6 genotypes and the clinical outcomes of adjuvant tamoxifen for breast cancer: A meta-analysis. Pharmacogenomics 2014, 15, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Ro, J.; Park, S.; Lim, H.-S.; Lee, K.S.; Kang, H.S.; Jung, S.-Y.; Lee, S. Lack of any association between functionally significant CYP2D6 polymorphisms and clinical outcomes in early breast cancer patients receiving adjuvant tamoxifen treatment. Breast Cancer Res. Treat. 2012, 131, 455–461. [Google Scholar] [CrossRef]
- Rae, J.M.; Drury, S.; Hayes, D.F.; Stearns, V.; Thibert, J.N.; Haynes, B.P.; Salter, J.; Sestak, I.; Cuzick, J.; Dowsett, M.; et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J. Natl. Cancer Inst. 2012, 104, 452–460. [Google Scholar] [CrossRef]
- Regan, M.M.; Leyland-Jones, B.; Bouzyk, M.; Pagani, O.; Tang, W.; Kammler, R.; Dell’orto, P.; Biasi, M.O.; Thürlimann, B.; Lyng, M.B.; et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: The breast international group 1-98 trial. J. Natl. Cancer Inst. 2012, 104, 441–451. [Google Scholar] [CrossRef]
- Schroth, W.; Goetz, M.P.; Hamann, U.; Fasching, P.A.; Schmidt, M.; Winter, S.; Fritz, P.; Simon, W.; Suman, V.J.; Ames, M.M.; et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 2009, 302, 1429–1436. [Google Scholar] [CrossRef]
- De Vries Schultink, A.H.M.; Zwart, W.; Linn, S.C.; Beijnen, J.H.; Huitema, A.D.R. Effects of Pharmacogenetics on the Pharmacokinetics and Pharmacodynamics of Tamoxifen. Clin. Pharmacokinet. 2015, 54, 797–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertz, D.L.; Kidwell, K.M.; Hilsenbeck, S.G.; Oesterreich, S.; Osborne, C.K.; Philips, S.; Chenault, C.; Hartmaier, R.J.; Skaar, T.C.; Sikora, M.J.; et al. CYP2D6 genotype is not associated with survival in breast cancer patients treated with tamoxifen: Results from a population-based study. Breast Cancer Res. Treat. 2017, 166, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Sun, J.X.; Suman, V.J.; Silva, G.O.; Perou, C.M.; Nakamura, Y.; Cox, N.J.; Stephens, P.J.; Miller, V.A.; Ross, J.S.; et al. Loss of heterozygosity at the CYP2D6 locus in breast cancer: Implications for germline pharmacogenetic studies. J. Natl. Cancer Inst. 2014, 107. [Google Scholar] [CrossRef] [PubMed]
- Province, M.A.; Goetz, M.P.; Brauch, H.; Flockhart, D.A.; Hebert, J.M.; Whaley, R.; Suman, V.J.; Schroth, W.; Winter, S.; Zembutsu, H.; et al. CYP2D6 genotype and adjuvant tamoxifen: Meta-analysis of heterogeneous study populations. Clin. Pharmacol. Ther. 2014, 95, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Schroth, W.; Hamann, U.; Fasching, P.A.; Dauser, S.; Winter, S.; Eichelbaum, M.; Schwab, M.; Brauch, H. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: Expanded polymorphism coverage improves risk stratification. Clin. Cancer Res. 2010, 16, 4468–4477. [Google Scholar] [CrossRef] [PubMed]
- Ahern, T.P.; Hertz, D.L.; Damkier, P.; Ejlertsen, B.; Hamilton-Dutoit, S.J.; Rae, J.M.; Regan, M.M.; Thompson, A.M.; Lash, T.L.; Cronin-Fenton, D.P. Cytochrome P-450 2D6 (CYP2D6) Genotype and Breast Cancer Recurrence in Tamoxifen-Treated Patients: Evaluating the Importance of Loss of Heterozygosity. Am. J. Epidemiol. 2017, 185, 75–85. [Google Scholar] [CrossRef]
- Hennig, E.E.; Piatkowska, M.; Karczmarski, J.; Goryca, K.; Brewczynska, E.; Jazwiec, R.; Kluska, A.; Omiotek, R.; Paziewska, A.; Dadlez, M.; et al. Limited predictive value of achieving beneficial plasma (Z)-endoxifen threshold level by CYP2D6 genotyping in tamoxifen-treated Polish women with breast cancer. BMC Cancer 2015, 15, 570. [Google Scholar] [CrossRef]
- Madlensky, L.; Natarajan, L.; Tchu, S.; Pu, M.; Mortimer, J.; Flatt, S.W.; Nikoloff, D.M.; Hillman, G.; Fontecha, M.R.; Lawrence, H.J.; et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clin. Pharmacol. Ther. 2011, 89, 718–725. [Google Scholar] [CrossRef]
- Borges, S.; Desta, Z.; Li, L.; Skaar, T.C.; Ward, B.A.; Nguyen, A.; Jin, Y.; Storniolo, A.M.; Nikoloff, D.M.; Wu, L.; et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clin. Pharmacol. Ther. 2006, 80, 61–74. [Google Scholar] [CrossRef]
- Saladores, P.; Mürdter, T.; Eccles, D.; Chowbay, B.; Zgheib, N.K.; Winter, S.; Ganchev, B.; Eccles, B.; Gerty, S.; Tfayli, A.; et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenomics J. 2015, 15, 84–94. [Google Scholar] [CrossRef]
- Hertz, D.L.; Snavely, A.C.; McLeod, H.L.; Walko, C.M.; Ibrahim, J.G.; Anderson, S.; Weck, K.E.; Magrinat, G.; Olajide, O.; Moore, S.; et al. In vivo assessment of the metabolic activity of CYP2D6 diplotypes and alleles. Br. J. Clin. Pharmacol. 2015, 80, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Helland, T.; Henne, N.; Bifulco, E.; Naume, B.; Borgen, E.; Kristensen, V.N.; Kvaløy, J.T.; Lash, T.L.; Alnæs, G.I.G.; van Schaik, R.H.; et al. Serum concentrations of active tamoxifen metabolites predict long-term survival in adjuvantly treated breast cancer patients. Breast Cancer Res. BCR 2017, 19, 125. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Spitman, A.; Dezentje, V.; Swen, J.; Moes, D.J.A.R.; Bohringer, S.; Batman, E.; van Druten, E.; Smorenburg, C.; van Bochove, A.; Zeillemaker, A.; et al. Tamoxifen pharmacogenetics and metabolism: Results from the prospective CYPTAM study. J. Clin. Oncol. 2019, 37, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Schroth, W.; Winter, S.; Mürdter, T.; Schaeffeler, E.; Eccles, D.; Eccles, B.; Chowbay, B.; Khor, C.C.; Tfayli, A.; Zgheib, N.K.; et al. Improved Prediction of Endoxifen Metabolism by CYP2D6 Genotype in Breast Cancer Patients Treated with Tamoxifen. Front. Pharmacol. 2017, 8, 582. [Google Scholar] [CrossRef] [PubMed]
- Teft, W.A.; Gong, I.Y.; Dingle, B.; Potvin, K.; Younus, J.; Vandenberg, T.A.; Brackstone, M.; Perera, F.E.; Choi, Y.-H.; Zou, G.; et al. CYP3A4 and seasonal variation in vitamin D status in addition to CYP2D6 contribute to therapeutic endoxifen level during tamoxifen therapy. Breast Cancer Res. Treat. 2013, 139, 95–105. [Google Scholar] [CrossRef]
- Jin, Y.; Desta, Z.; Stearns, V.; Ward, B.; Ho, H.; Lee, K.-H.; Skaar, T.; Storniolo, A.M.; Li, L.; Araba, A.; et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J. Natl. Cancer Inst. 2005, 97, 30–39. [Google Scholar] [CrossRef]
- Binkhorst, L.; Bannink, M.; de Bruijn, P.; Ruit, J.; Droogendijk, H.; van Alphen, R.J.; den Boer, T.D.; Lam, M.H.; Jager, A.; van Gelder, T.; et al. Augmentation of Endoxifen Exposure in Tamoxifen-Treated Women Following SSRI Switch. Clin. Pharmacokinet. 2016, 55, 249–255. [Google Scholar] [CrossRef]
- Zembutsu, H.; Sasa, M.; Kiyotani, K.; Mushiroda, T.; Nakamura, Y. Should CYP2D6 inhibitors be administered in conjunction with tamoxifen? Expert Rev. Anticancer Ther. 2011, 11, 185–193. [Google Scholar] [CrossRef]
- Fernández-Santander, A.; Gaibar, M.; Novillo, A.; Romero-Lorca, A.; Rubio, M.; Chicharro, L.M.; Tejerina, A.; Bandrés, F. Relationship between genotypes Sult1a2 and Cyp2d6 and tamoxifen metabolism in breast cancer patients. PLoS ONE 2013, 8, e70183. [Google Scholar] [CrossRef]
- Blevins-Primeau, A.S.; Sun, D.; Chen, G.; Sharma, A.K.; Gallagher, C.J.; Amin, S.; Lazarus, P. Functional significance of UDP-glucuronosyltransferase variants in the metabolism of active tamoxifen metabolites. Cancer Res. 2009, 69, 1892–1900. [Google Scholar] [CrossRef]
- Del Re, M.; Citi, V.; Crucitta, S.; Rofi, E.; Belcari, F.; van Schaik, R.H.; Danesi, R. Pharmacogenetics of CYP2D6 and tamoxifen therapy: Light at the end of the tunnel? Pharmacol. Res. 2016, 107, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.I.; Lee, S.K.; Kim, J.; Kim, S.W.; Yu, J.; Bae, S.Y.; Lee, J.E.; Nam, S.J.; Lee, S.-Y. Variations in plasma concentrations of tamoxifen metabolites and the effects of genetic polymorphisms on tamoxifen metabolism in Korean patients with breast cancer. Oncotarget 2017, 8, 100296–100311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, A.M.; Johnson, A.; Quinlan, P.; Hillman, G.; Fontecha, M.; Bray, S.E.; Purdie, C.A.; Jordan, L.B.; Ferraldeschi, R.; Latif, A.; et al. Comprehensive CYP2D6 genotype and adherence affect outcome in breast cancer patients treated with tamoxifen monotherapy. Breast Cancer Res. Treat. 2011, 125, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Lum, D.W.K.; Perel, P.; Hingorani, A.D.; Holmes, M.V. CYP2D6 genotype and tamoxifen response for breast cancer: A systematic review and meta-analysis. PLoS ONE 2013, 8, e76648. [Google Scholar] [CrossRef] [PubMed]
- Vianna-Jorge, R.; Festa-Vasconcellos, J.S.; Goulart-Citrangulo, S.M.T.; Leite, M.S. Functional polymorphisms in xenobiotic metabolizing enzymes and their impact on the therapy of breast cancer. Front. Genet. 2012, 3, 329. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Liu, Y.; Liu, Z.; You, J.; Chen, Z.; Wang, J.; Peng, Q.; Xie, L.; Li, R.; Li, S.; et al. CYP2D6 polymorphisms influence tamoxifen treatment outcomes in breast cancer patients: A meta-analysis. Cancer Chemother. Pharmacol. 2013, 72, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Binkhorst, L.; Mathijssen, R.H.J.; Jager, A.; van Gelder, T. Individualization of tamoxifen therapy: Much more than just CYP2D6 genotyping. Cancer Treat. Rev. 2015, 41, 289–299. [Google Scholar] [CrossRef]
- Del Re, M.; Michelucci, A.; Simi, P.; Danesi, R. Pharmacogenetics of anti-estrogen treatment of breast cancer. Cancer Treat. Rev. 2012, 38, 442–450. [Google Scholar] [CrossRef]
- Fleeman, N.; Martin Saborido, C.; Payne, K.; Boland, A.; Dickson, R.; Dundar, Y.; Fernández Santander, A.; Howell, S.; Newman, W.; Oyee, J.; et al. The clinical effectiveness and cost-effectiveness of genotyping for CYP2D6 for the management of women with breast cancer treated with tamoxifen: A systematic review. Health Technol. Assess. 2011, 15, 1–102. [Google Scholar] [CrossRef]
- Robertson, D.W.; Katzenellenbogen, J.A.; Long, D.J.; Rorke, E.A.; Katzenellenbogen, B.S. Tamoxifen antiestrogens. A comparison of the activity, pharmacokinetics, and metabolic activation of the cis and trans isomers of tamoxifen. J. Steroid Biochem. 1982, 16, 1–13. [Google Scholar] [CrossRef]
- Zagajewska, K.; Piątkowska, M.; Goryca, K.; Bałabas, A.; Kluska, A.; Paziewska, A.; Pośpiech, E.; Grabska-Liberek, I.; Hennig, E.E. GWAS links variants in neuronal development and actin remodeling related loci with pseudoexfoliation syndrome without glaucoma. Exp. Eye Res. 2018, 168, 138–148. [Google Scholar] [CrossRef] [PubMed]
- R: The R Project for Statistical Computing. Available online: http://www.r-project.org/ (accessed on 2 April 2015).
- CRAN—Package Epitools. Available online: https://cran.r-project.org/web/packages/epitools/index.html (accessed on 12 August 2016).
- Pośpiech, E.; Draus-Barini, J.; Kupiec, T.; Wojas-Pelc, A.; Branicki, W. Prediction of eye color from genetic data using Bayesian approach. J. Forensic Sci. 2012, 57, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T.; Trevor, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer Series of Statistics: New York, NY, USA, 2009. [Google Scholar]
- Ostrowski, J.; Paziewska, A.; Lazowska, I.; Ambrozkiewicz, F.; Goryca, K.; Kulecka, M.; Rawa, T.; Karczmarski, J.; Dabrowska, M.; Zeber-Lubecka, N.; et al. Genetic architecture differences between pediatric and adult-onset inflammatory bowel diseases in the Polish population. Sci. Rep. 2016, 6, 39831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paziewska, A.; Habior, A.; Rogowska, A.; Zych, W.; Goryca, K.; Karczmarski, J.; Dabrowska, M.; Ambrozkiewicz, F.; Walewska-Zielecka, B.; Krawczyk, M.; et al. A novel approach to genome-wide association analysis identifies genetic associations with primary biliary cholangitis and primary sclerosing cholangitis in Polish patients. BMC Med. Genom. 2017, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Jager, N.G.L.; Rosing, H.; Schellens, J.H.M.; Linn, S.C.; Beijnen, J.H. Tamoxifen dose and serum concentrations of tamoxifen and six of its metabolites in routine clinical outpatient care. Breast Cancer Res. Treat. 2014, 143, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Irvin, W.J.; Walko, C.M.; Weck, K.E.; Ibrahim, J.G.; Chiu, W.K.; Dees, E.C.; Moore, S.G.; Olajide, O.A.; Graham, M.L.; Canale, S.T.; et al. Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism: A multicenter study. J. Clin. Oncol. 2011, 29, 3232–3239. [Google Scholar] [CrossRef] [PubMed]
- Martinez de Dueñas, E.; Ochoa Aranda, E.; Blancas Lopez-Barajas, I.; Ferrer Magdalena, T.; Bandrés Moya, F.; Chicharro García, L.M.; Gómez Capilla, J.A.; Zafra Ceres, M.; de Haro, T.; Romero Llorens, R.; et al. Adjusting the dose of tamoxifen in patients with early breast cancer and CYP2D6 poor metabolizer phenotype. Breast 2014, 23, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Dezentjé, V.O.; Opdam, F.L.; Gelderblom, H.; Hartigh den, J.; Van der Straaten, T.; Vree, R.; Maartense, E.; Smorenburg, C.H.; Putter, H.; Dieudonné, A.S.; et al. CYP2D6 genotype- and endoxifen-guided tamoxifen dose escalation increases endoxifen serum concentrations without increasing side effects. Breast Cancer Res. Treat. 2015, 153, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Hertz, D.L.; Deal, A.; Ibrahim, J.G.; Walko, C.M.; Weck, K.E.; Anderson, S.; Magrinat, G.; Olajide, O.; Moore, S.; Raab, R.; et al. Tamoxifen Dose Escalation in Patients with Diminished CYP2D6 Activity Normalizes Endoxifen Concentrations Without Increasing Toxicity. Oncologist 2016, 21, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Sangkuhl, K.; Guchelaar, H.-J.; Schwab, M.; Province, M.; Whirl-Carrillo, M.; Symmans, W.F.; McLeod, H.L.; Ratain, M.J.; Zembutsu, H.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin. Pharmacol. Ther. 2018, 103, 770–777. [Google Scholar] [CrossRef]
- Hicks, J.K.; Swen, J.J.; Gaedigk, A. Challenges in CYP2D6 phenotype assignment from genotype data: A critical assessment and call for standardization. Curr. Drug Metab. 2014, 15, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Sotgia, F.; Fiorillo, M.; Lisanti, M.P. Mitochondrial markers predict recurrence, metastasis and tamoxifen-resistance in breast cancer patients: Early detection of treatment failure with companion diagnostics. Oncotarget 2017, 8, 68730–68745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Wang, Z.; Liu, W.; Lei, R.; Shan, J.; Li, L.; Wang, X. Distinct prognostic values of S100 mRNA expression in breast cancer. Sci. Rep. 2017, 7, 39786. [Google Scholar] [CrossRef] [PubMed]
- Cancemi, P.; Buttacavoli, M.; Di Cara, G.; Albanese, N.N.; Bivona, S.; Pucci-Minafra, I.; Feo, S. A multiomics analysis of S100 protein family in breast cancer. Oncotarget 2018, 9, 29064–29081. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Ning, M.; Jeong, H. Transcriptional Regulation of CYP2D6 Expression. Drug Metab. Dispos. 2017, 45, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, B.; Molony, C.; Chudin, E.; Hao, K.; Zhu, J.; Gaedigk, A.; Suver, C.; Zhong, H.; Leeder, J.S.; et al. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 2010, 20, 1020–1036. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Poi, M.J.; Sun, X.; Gaedigk, A.; Leeder, J.S.; Sadee, W. Common CYP2D6 polymorphisms affecting alternative splicing and transcription: Long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum. Mol. Genet. 2014, 23, 268–278. [Google Scholar] [CrossRef]
- Wang, D.; Papp, A.C.; Sun, X. Functional characterization of CYP2D6 enhancer polymorphisms. Hum. Mol. Genet. 2015, 24, 1556–1562. [Google Scholar] [CrossRef]
| Total N = 287 | |||
|---|---|---|---|
| Genotype | Number of Patients (%) | (Z)-endoxifen (ng mL−1) Mean ± SD | Metabolic Ratio a Mean ± SD |
| NM/UM | 18 (6.3) | 6.96 ± 3.75 | 0.0201 ± 0.0076 |
| NM/NM | 90 (31.4) | 7.26 ± 3.18 | 0.0185 ± 0.0057 |
| NM/IM | 44 (15.3) | 5.60 ± 3.26 | 0.0125 ± 0.0054 |
| NM/PM | 99 (34.5) | 4.85 ± 2.46 | 0.0108 ± 0.0043 |
| IM/IM | 3 (1.0) | 3.05 ± 1.91 | 0.0063 ± 0.0022 |
| IM/PM | 12 (4.2) | 2.22 ± 0.87 | 0.0047 ± 0.0014 |
| PM/PM | 21 (7.3) | 1.79 ± 0.66 | 0.0037 ± 0.0013 |
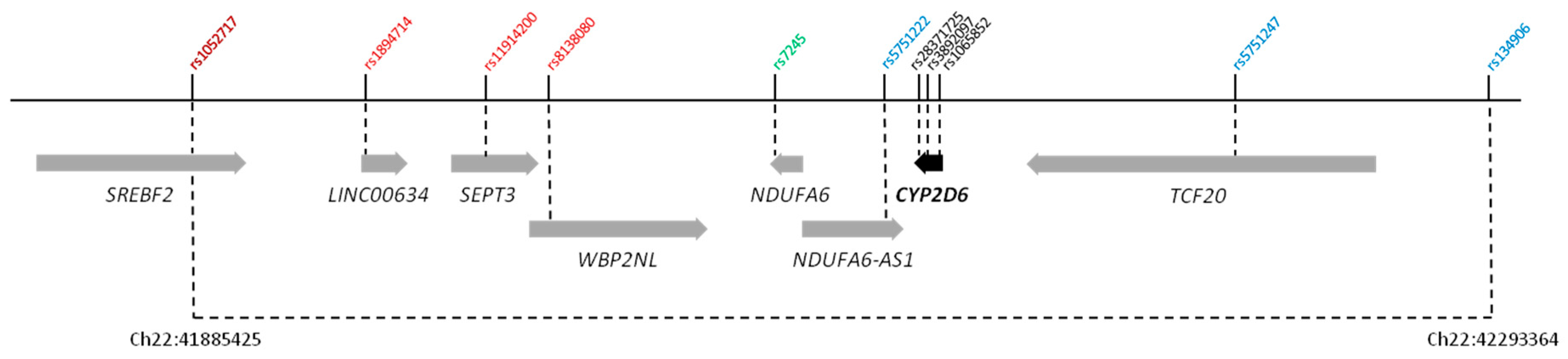
| dbSNP ID a | Region b | MA | MAF c | OR (95% CI) | p-Value |
|---|---|---|---|---|---|
| rs9844493 | Chr3:67210593 | A | 0.482 | 1.59 (1.13–2.23) | 3.74 × 10−3 |
| intergenic | |||||
| rs1320308 | Chr5:76875427 | A | 0.447 | 0.51 (0.35–0.73) | 1.59 × 10−3 |
| S100Z exon 6 (E23A) | |||||
| rs6950784 | Chr7:155902980 | G | 0.519 | 1.93 (1.37–2.72) | 4.42 × 10−4 |
| intergenic | |||||
| rs980729 | Chr7:98697127 | A | 0.361 | 0.83 (0.59–1.1) | 1.22 × 10−3 |
| intergenic | |||||
| rs11780345 | Chr8:23194453 | C | 0.196 | 0.69 (0.49–0.97) | 5.57 × 10−5 |
| TNFRSF10A intron 9 | |||||
| rs11786748 | Chr8:27924000 | G | 0.403 | 1.95 (1.35–2.82) | 6.18 × 10−4 |
| SCARA5 intron 3 | |||||
| rs7988513 | Chr13:92232033 | C | 0.300 | 0.80 (0.57–1.14) | 7.9 × 10−3 |
| GPC5 intron 7 | |||||
| rs1052717 | Chr22:41885425 | A | 0.304 | 0.36 (0.25–0.50) | 1.36 × 10−7 |
| SREBF2 intron 13 | |||||
| rs1894714 | Chr22:41953130 | T | 0.188 | 4.31 (2.65–7.00) | 7.48 × 10−9 |
| LINC00634 ncRNA | |||||
| rs11914200 | Chr22:41982066 | A | 0.239 | 4.29 (2.82–6.55) | 4.73 × 10−12 |
| SEPT3 intron 4 | |||||
| rs8138080 | Chr22:42000367 | A | 0.261 | 5.55 (3.52–8.75) | 1.78 × 10−15 |
| WBP2NL intron 2 | |||||
| rs7245 | Chr22:42085845 | G | 0.326 | 0.28 (0.2–0.4) | 7.52 × 10−13 |
| NDUFA6 3′ UTR | |||||
| rs5751222 | Chr22:42121918 | A | 0.229 | 5.96 (3.55–10.00) | 2.31 × 10−12 |
| NDUFA6-AS1 ncRNA | |||||
| rs5751247 | Chr22:42237048 | C | 0.290 | 4.82 (3.09–7.52) | 2.81 × 10−13 |
| TCF20 intron 3 | |||||
| rs134906 | Chr22:42293364 | T | 0.333 | 0.37 (0.26–0.52) | 2.35 × 10−8 |
| intergenic | |||||
| rs28371725 | Chr22:42127803 | T | 0.064 | 2.24 (1.11–4.50) | 6.81 ×10−2 |
| CYP2D6 intron 6 (SSV) | |||||
| rs3892097 | Chr22:42128945 | T | 0.093 | 6.93 (3.92–12.24) | 1.98 × 10−12 |
| CYP2D6 intron 3 (SSV) | |||||
| rs1065852 | Chr22:42130692 | A | 0.238 | 6.85 (3.98–11.77) | 3.5 × 10−13 |
| CYP2D6 exon 1 (P34S) |
| Variable/ dbSNP ID a | Gene | −2 Log Likelihood | Rank | R2b | OR (95% CI) | p-Value | AUC c |
|---|---|---|---|---|---|---|---|
| CYP2D6 genotype | CYP2D6 | 254.137 | 1 | 0.427 | 16.13 (7.58–34.48) | 6.28 × 10−13 | 0.758 |
| rs7245 | NDUFA6 | 238.013 | 2 | 0.482 | 0.40 (0.22–0.70) | 1.38 × 10−3 | 0.842 |
| rs6950784 | Intergenic | 223.925 | 3 | 0.527 | 2.24 (1.40–3.59) | 7.94 × 10−4 | 0.871 |
| rs1320308 | S100Z | 214.671 | 4 | 0.556 | 0.48 (0.29–0.81) | 6.26 × 10−3 | 0.879 |
| rs11786748 | SCARA5 | 210.654 | 5 | 0.568 | 1.68 (1.01–2.80) | 0.047 | 0.880 |
| dbSNP ID a | Gene | −2 Log Likelihood | Rank | R2b | OR (95% CI) | p-Value | AUC c |
|---|---|---|---|---|---|---|---|
| rs8138080 | WBP2NL | 278.432 | 1 | 0.337 | 3.73 (1.65–8.43) | 1.56 × 10−3 | 0.718 |
| rs1320308 | S100Z | 264.234 | 2 | 0.390 | 0.48 (0.29–0.79) | 3.7 × 10−3 | 0.795 |
| rs6950784 | Intergenic | 255.872 | 3 | 0.420 | 1.91 (1.25–2.92) | 2.81 × 10−3 | 0.819 |
| rs7245 | NDUFA6 | 247.300 | 4 | 0.450 | 0.54 (0.30–0.95) | 0.034 | 0.830 |
| rs1065852 | CYP2D6 | 242.012 | 5 | 0.468 | 2.78 (1.18–6.59) | 0.020 | 0.817 |
| rs11786748 | SCARA5 | 237.058 | 6 | 0.485 | 1.70 (1.06–2.74) | 0.028 | 0.813 |
| Parameter | Prediction Model Type | |
|---|---|---|
| Model 1 | Model 2 | |
| AUC | 0.879 | 0.830 |
| Sensitivity (%) | 87.8 | 80.1 |
| Specificity (%) | 70.8 | 64.2 |
| PPV (%) | 81.6 | 76.7 |
| NPV (%) | 79.8 | 68.7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hennig, E.E.; Piątkowska, M.; Goryca, K.; Pośpiech, E.; Paziewska, A.; Karczmarski, J.; Kluska, A.; Brewczyńska, E.; Ostrowski, J. Non-CYP2D6 Variants Selected by a GWAS Improve the Prediction of Impaired Tamoxifen Metabolism in Patients with Breast Cancer. J. Clin. Med. 2019, 8, 1087. https://doi.org/10.3390/jcm8081087
Hennig EE, Piątkowska M, Goryca K, Pośpiech E, Paziewska A, Karczmarski J, Kluska A, Brewczyńska E, Ostrowski J. Non-CYP2D6 Variants Selected by a GWAS Improve the Prediction of Impaired Tamoxifen Metabolism in Patients with Breast Cancer. Journal of Clinical Medicine. 2019; 8(8):1087. https://doi.org/10.3390/jcm8081087
Chicago/Turabian StyleHennig, Ewa E., Magdalena Piątkowska, Krzysztof Goryca, Ewelina Pośpiech, Agnieszka Paziewska, Jakub Karczmarski, Anna Kluska, Elżbieta Brewczyńska, and Jerzy Ostrowski. 2019. "Non-CYP2D6 Variants Selected by a GWAS Improve the Prediction of Impaired Tamoxifen Metabolism in Patients with Breast Cancer" Journal of Clinical Medicine 8, no. 8: 1087. https://doi.org/10.3390/jcm8081087





