Phenotypic and Genotypic Characterization of Escherichia coli Causing Urinary Tract Infections in Kidney-Transplanted Patients
Abstract
1. Introduction
2. Experimental Section
2.1. Study Population
2.2. Patients’ Characteristics
2.3. Bacterial Strains and Culture Conditions
2.4. Phenotypic Tests
2.5. Genome Sequencing
2.6. Draft Genome Comparison and Typing
2.7. Statistical Analysis
3. Results
3.1. Patients’ Characteristics
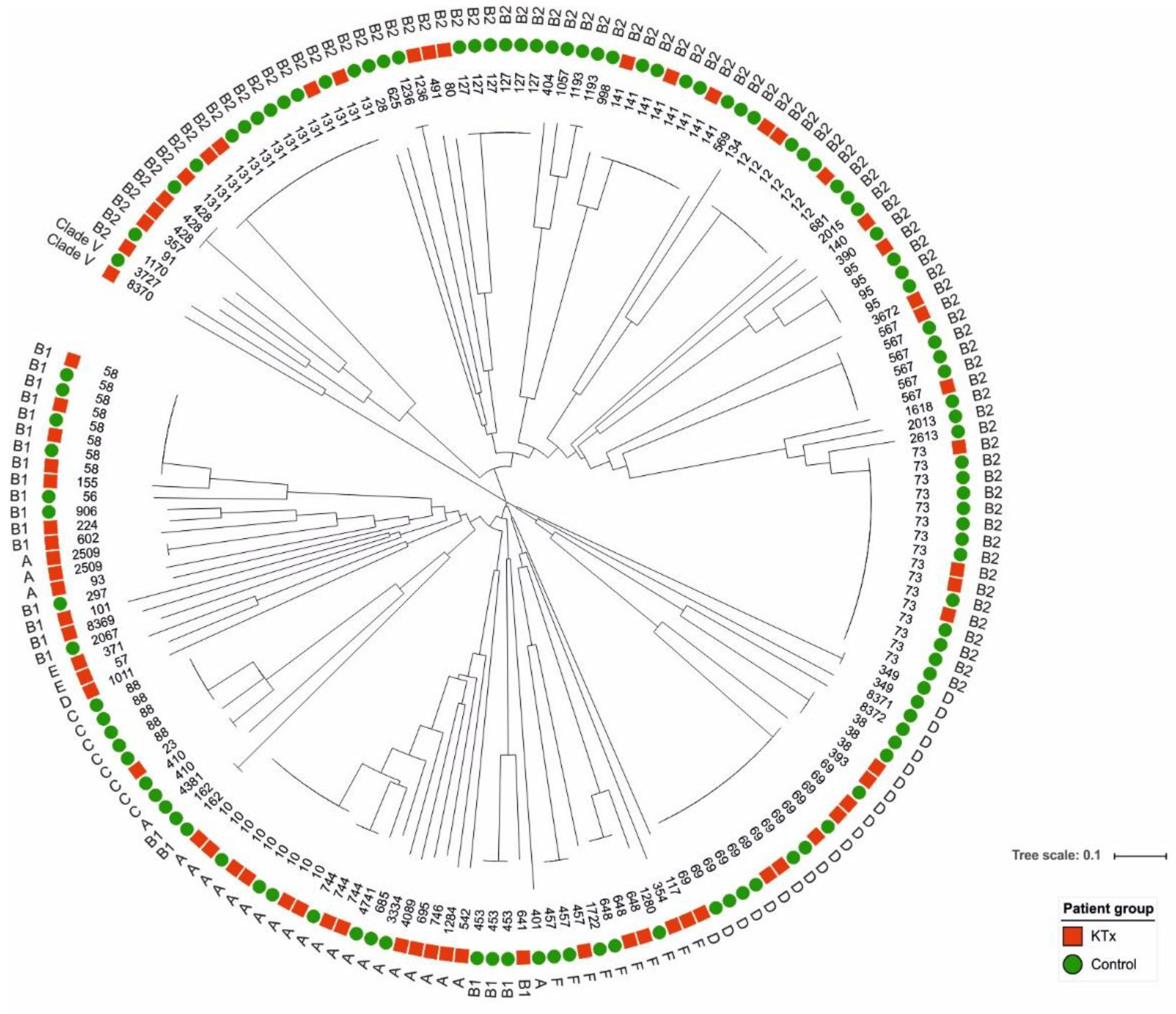
3.2. Epidemiological Classification of Urine Isolates from KTx and Control Patients
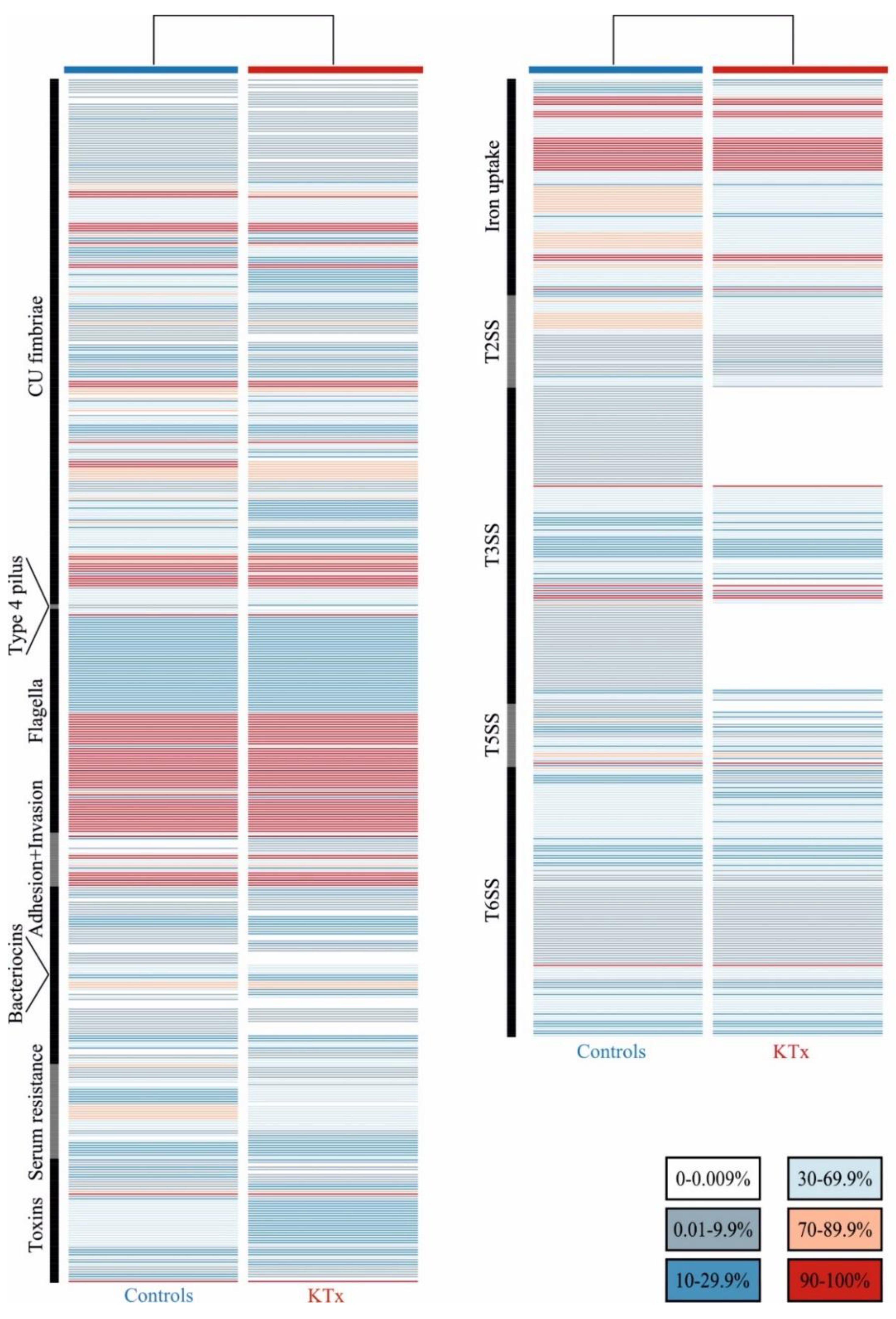
3.3. Prevalence of UPEC Virulence-ASSOCIATED genes
3.4. Antibiotic Susceptibility
3.5. Plasmid Types
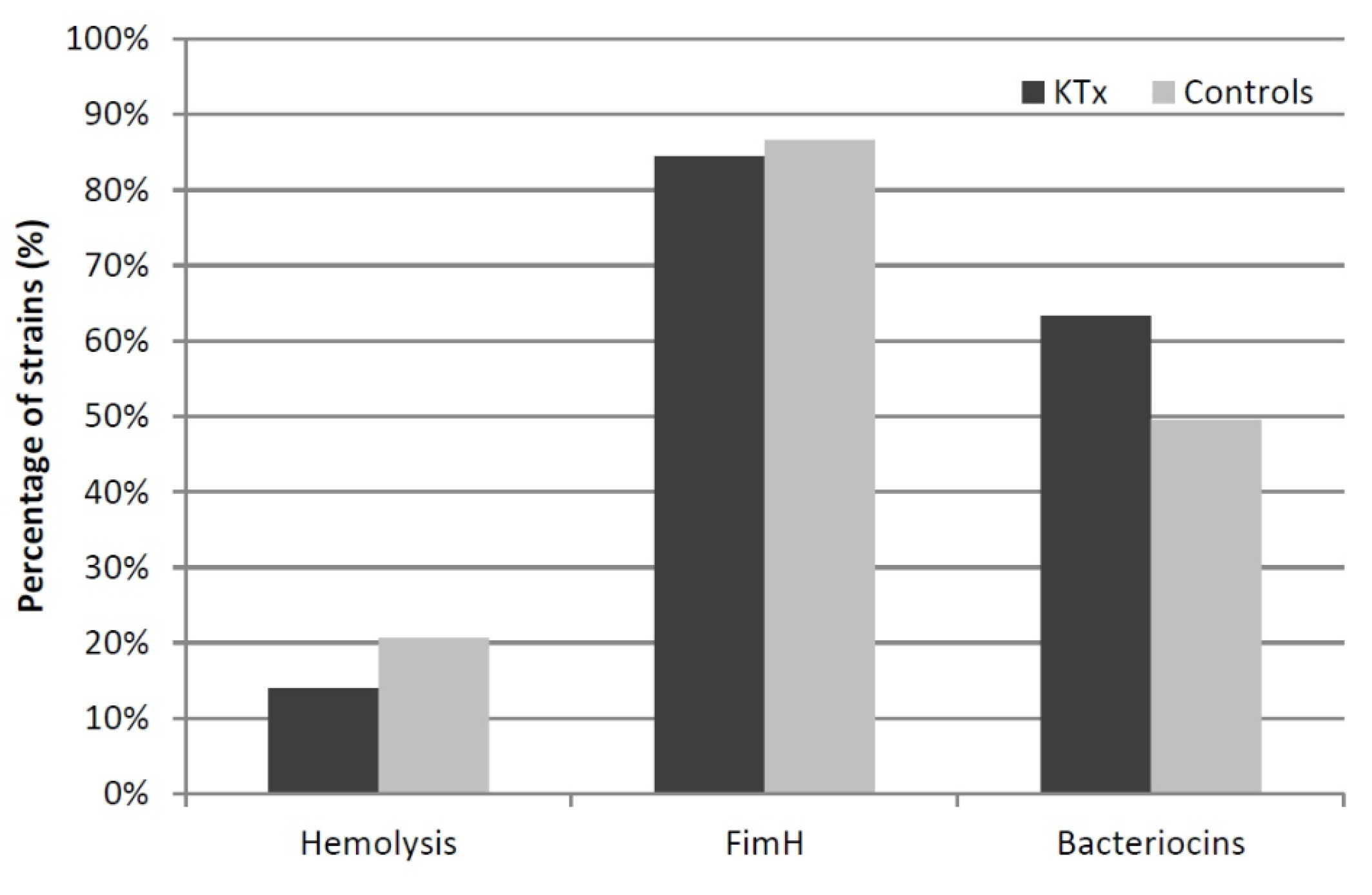
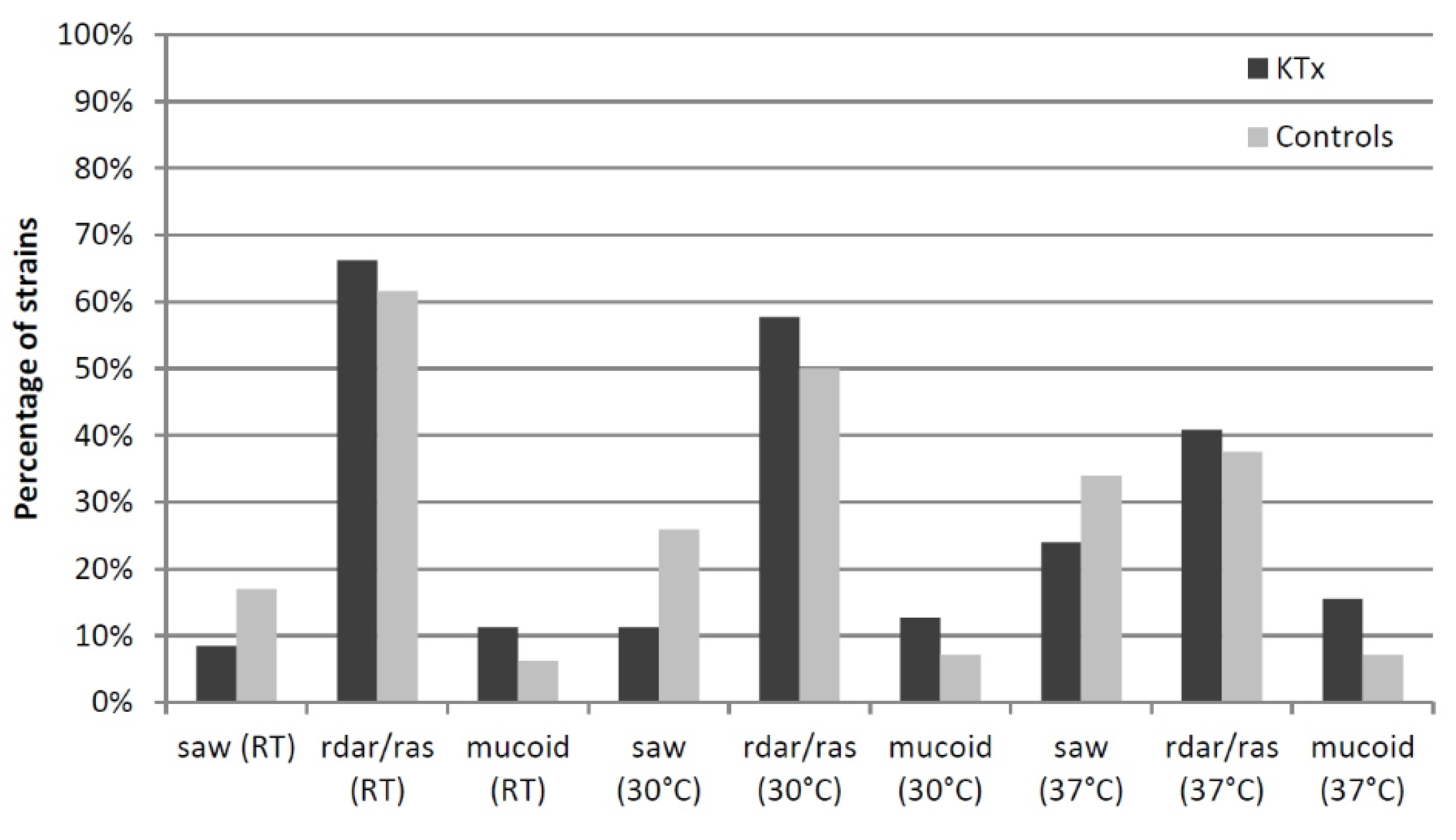
3.6. Phenotypic Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wu, X.; Dong, Y.; Liu, Y.; Li, Y.; Sun, Y.; Wang, J.; Wang, S. The prevalence and predictive factors of urinary tract infection in patients undergoing renal transplantation: A meta-analysis. Am. J. Infect. Control. 2016, 44, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Al-Hasan, M.N.; Razonable, R.R.; Kremers, W.K.; Baddour, L.M. Impact of Gram-negative bloodstream infection on long-term allograft survival after kidney transplantation. Transplantation 2011, 91, 1206–1210. [Google Scholar] [CrossRef] [PubMed]
- Bodro, M.; Sanclemente, G.; Lipperheide, I.; Allali, M.; Marco, F.; Bosch, J.; Cofan, F.; Ricart, M.J.; Esforzado, N.; Oppenheimer, F.; et al. Impact of urinary tract infections on short-term kidney graft outcome. Clin. Microbiol. Infect. 2015, 21, 1104.e1–1104.e8. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tholking, G.; Schuette-Nuetgen, K.; Vogl, T.; Dobrindt, U.; Kahl, B.C.; Brand, M.; Pavenstadt, H.; Suwelack, B.; Koch, R.; Reuter, S. Male kidney allograft recipients at risk for urinary tract infection? PLoS ONE 2017, 12, e0188262. [Google Scholar] [CrossRef] [PubMed]
- de Souza, R.M.; Olsburgh, J. Urinary tract infection in the renal transplant patient. Nat. Clin. Pract. Nephrol. 2008, 4, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.; Torre-Cisneros, J.; Blanes, M.; Montejo, M.; Cervera, C.; Aguado, J.M.; Len, O.; Carratala, J.; Cordero, E.; Bou, G.; et al. Spanish Network for Research in Infectious Diseases (REIPI) Bacterial urinary tract infection after solid organ transplantation in the RESITRA cohort. Transpl. Infect. Dis. 2012, 14, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Luthje, P.; Brauner, A. Virulence factors of uropathogenic E. coli and their interaction with the host. Adv. Microb. Physiol. 2014, 65, 337–372. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- O’Brien, V.P.; Dorsey, D.A.; Hannan, T.J.; Hultgren, S.J. Host restriction of Escherichia coli recurrent urinary tract infection occurs in a bacterial strain-specific manner. PLoS Pathog. 2018, 14, e1007457. [Google Scholar] [CrossRef]
- Guidelines on Urological Infections. Available online: https://uroweb.org/wp-content/uploads/19-Urological-infections_LR2.pdf (accessed on 30 June 2019).
- Ragnarsdottir, B.; Lutay, N.; Gronberg-Hernandez, J.; Koves, B.; Svanborg, C. Genetics of innate immunity and UTI susceptibility. Nat. Rev. Urol. 2011, 8, 449–468. [Google Scholar] [CrossRef]
- Sanchez, G.V.; Babiker, A.; Master, R.N.; Luu, T.; Mathur, A.; Bordon, J. Antibiotic Resistance among Urinary Isolates from Female Outpatients in the United States in 2003 and 2012. Antimicrob. Agents Chemother. 2016, 60, 2680–2683. [Google Scholar] [CrossRef] [PubMed]
- Khawcharoenporn, T.; Vasoo, S.; Singh, K. Urinary Tract Infections due to Multidrug-Resistant Enterobacteriaceae: Prevalence and Risk Factors in a Chicago Emergency Department. Emerg. Med. Int. 2013, 2013, 258517. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Ranjan, R.; McGee, H.S.; Andropolis, K.E.; Panchal, D.V.; Hajjiri, Z.; Brennan, D.C.; Finn, P.W.; Perkins, D.L. Urinary microbiome of kidney transplant patients reveals dysbiosis with potential for antibiotic resistance. Transl. Res. 2017, 181, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.W.; Wrenn, K.D.; Haynes, M.L. Trimethoprim-sulfamethoxazole resistance among urinary coliform isolates. J. Gen. Intern. Med. 1999, 14, 606–609. [Google Scholar] [CrossRef]
- Killgore, K.M.; March, K.L.; Guglielmo, B.J. Risk factors for community-acquired ciprofloxacin-resistant Escherichia coli urinary tract infection. Ann. Pharmacother. 2004, 38, 1148–1152. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Nahum, K.; Saidel-Odes, L.; Riesenberg, K.; Schlaeffer, F.; Borer, A. Urinary tract infections caused by multi-drug resistant Proteus mirabilis: Risk factors and clinical outcomes. Infection 2010, 38, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Smithson, A.; Chico, C.; Ramos, J.; Netto, C.; Sanchez, M.; Ruiz, J.; Porron, R.; Bastida, M.T. Prevalence and risk factors for quinolone resistance among Escherichia coli strains isolated from males with community febrile urinary tract infection. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Spaulding, C.N.; Klein, R.D.; Ruer, S.; Kau, A.L.; Schreiber, H.L.; Cusumano, Z.T.; Dodson, K.W.; Pinkner, J.S.; Fremont, D.H.; Janetka, J.W.; et al. Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature 2017, 546, 528–532. [Google Scholar] [CrossRef]
- Biehl, L.M.; Cruz Aguilar, R.; Farowski, F.; Hahn, W.; Nowag, A.; Wisplinghoff, H.; Vehreschild, M.J.G.T. Fecal microbiota transplantation in a kidney transplant recipient with recurrent urinary tract infection. Infection 2018, 46, 871–874. [Google Scholar] [CrossRef]
- Origuen, J.; Lopez-Medrano, F.; Fernandez-Ruiz, M.; Polanco, N.; Gutierrez, E.; Gonzalez, E.; Merida, E.; Ruiz-Merlo, T.; Morales-Cartagena, A.; Perez-Jacoiste Asin, M.A.; et al. Should Asymptomatic Bacteriuria Be Systematically Treated in Kidney Transplant Recipients? Results From a Randomized Controlled Trial. Am. J. Transplant. 2016, 16, 2943–2953. [Google Scholar] [CrossRef]
- Schito, G.C.; Naber, K.G.; Botto, H.; Palou, J.; Mazzei, T.; Gualco, L.; Marchese, A. The ARESC study: An international survey on the antimicrobial resistance of pathogens involved in uncomplicated urinary tract infections. Int. J. Antimicrob. Agents 2009, 34, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Azap, O.; Togan, T.; Yesilkaya, A.; Arslan, H.; Haberal, M. Antimicrobial susceptibilities of uropathogen Escherichia coli in renal transplant recipients: Dramatic increase in ciprofloxacin resistance. Transplant. Proc. 2013, 45, 956–957. [Google Scholar] [CrossRef] [PubMed]
- Korayem, G.B.; Zangeneh, T.T.; Matthias, K.R. Urinary Tract Infections Recurrence and Development of Urinary-Specific Antibiogram for Kidney Transplant Recipients. J. Glob. Antimicrob. Resist. 2018, 12, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Bemelman, F.J.; Hodiamont, C.J.; Idu, M.M.; Ten Berge, I.J.; Geerlings, S.E. The impact of trimethoprim-sulfamethoxazole as Pneumocystis jiroveci pneumonia prophylaxis on the occurrence of asymptomatic bacteriuria and urinary tract infections among renal allograft recipients: A retrospective before-after study. BMC Infect. Dis. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Kerstenetzky, L.; Jorgenson, M.R.; Descourouez, J.L.; Leverson, G.; Rose, W.E.; Redfield, R.R.; Smith, J.A. Fosfomycin tromethamine for the Treatment of Cystitis in Abdominal Solid Organ Transplant Recipients With Renal Dysfunction. Ann. Pharmacother. 2017, 51, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Poolman, J.T.; Wacker, M. Extraintestinal Pathogenic Escherichia coli, a Common Human Pathogen: Challenges for Vaccine Development and Progress in the Field. J. Infect. Dis. 2016, 213, 6–13. [Google Scholar] [CrossRef]
- Rice, J.C.; Peng, T.; Kuo, Y.F.; Pendyala, S.; Simmons, L.; Boughton, J.; Ishihara, K.; Nowicki, S.; Nowicki, B.J. Renal allograft injury is associated with urinary tract infection caused by Escherichia coli bearing adherence factors. Am. J. Transplant. 2006, 6, 2375–2383. [Google Scholar] [CrossRef]
- Tourret, J.; Denamur, E. Population Phylogenomics of Extraintestinal Pathogenic Escherichia coli. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Tashk, P.; Lecronier, M.; Clermont, O.; Renvoise, A.; Aubry, A.; Barrou, B.; Hertig, A.; Lescat, M.; Tenaillon, O.; Denamur, E.; et al. Molecular epidemiology and kinetics of early Escherichia coli urinary tract infections in kidney transplant recipients. Nephrol. Ther. 2017, 13, 236–244. [Google Scholar] [CrossRef]
- Yun, K.W.; Kim, D.S.; Kim, W.; Lim, I.S. Molecular typing of uropathogenic Escherichia coli isolated from Korean children with urinary tract infection. Korean J. Pediatr. 2015, 58, 20–27. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Canica, M.M.; Park, Y.J.; Lavigne, J.P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Mercon, M.; Regua-Mangia, A.H.; Teixeira, L.M.; Irino, K.; Tuboi, S.H.; Goncalves, R.T.; Santoro-Lopes, G. Urinary tract infections in renal transplant recipients: Virulence traits of uropathogenic Escherichia coli. Transplant. Proc. 2010, 42, 483–485. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.F.; Hartl, D.L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 1998, 180, 1159–1165. [Google Scholar] [PubMed]
- Cusumano, C.K.; Hung, C.S.; Chen, S.L.; Hultgren, S.J. Virulence plasmid harbored by uropathogenic Escherichia coli functions in acute stages of pathogenesis. Infect. Immun. 2010, 78, 1457–1467. [Google Scholar] [CrossRef]
- Mao, B.H.; Chang, Y.F.; Scaria, J.; Chang, C.C.; Chou, L.W.; Tien, N.; Wu, J.J.; Tseng, C.C.; Wang, M.C.; Chang, C.C.; et al. Identification of Escherichia coli genes associated with urinary tract infections. J. Clin. Microbiol. 2012, 50, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Leimbach, A.; Hacker, J.; Dobrindt, U. E. coli as an all-rounder: The thin line between commensalism and pathogenicity. Curr. Top. Microbiol. Immunol. 2013, 358, 3–32. [Google Scholar] [CrossRef]
- Touchon, M.; Hoede, C.; Tenaillon, O.; Barbe, V.; Baeriswyl, S.; Bidet, P.; Bingen, E.; Bonacorsi, S.; Bouchier, C.; Bouvet, O.; et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009, 5, e1000344. [Google Scholar] [CrossRef]
- Bengtsson, S.; Naseer, U.; Sundsfjord, A.; Kahlmeter, G.; Sundqvist, M. Sequence types and plasmid carriage of uropathogenic Escherichia coli devoid of phenotypically detectable resistance. J. Antimicrob. Chemother. 2012, 67, 69–73. [Google Scholar] [CrossRef]
| Clinical data at UTI diagnosis | |||
| KTx Recipients | Controls | p | |
| Sex (female/male) | 49 (79%)/13 (21%) | 74 (70.5%)/31 (29.5%) | 0.1512 |
| Age (mean/median/σ in yr) | 56.67/58.75/15.09 | 53.45/ 55/21.82 | 0.2902 |
| BMI (mean/median/σ) | 26.19/26.03/4.58 | 24.62/24.21/4.41 | 0.0251 |
| Hypertension (%) | 80.6% | 35.3% | 0 |
| Diabetes mellitus (%) | 21% | 14.7% | 0.3922 |
| Immunosuppression (%) | 100% | 9.7% | 0 |
| Previous tumor diagnosis (%) | 0% | 10.7% | 0.0072 |
| Time from last KTx to UTI (mean/median/σ in yr) | 5.4/3.2/6.2 | - | - |
| Ureteral stent or urinary catheter (%) | 16.1% | 5.8% | 0.0539 |
| Lower UTI (%) | 80.3% | 80.4% | 1 |
| Upper UTI (%) | 19.7% | 19.6% | 1 |
| eGFR (mean/median/σ in mL/min) | 58.4/54.45/32.48 | 70.21/66.5/35.67 | 0.0406 |
| Acute kidney injury (%) | 28.3% | 23.1% | 0.5658 |
| Hospitalization | 24.5% | 39.3% | 0.0527 |
| Clinical data of a three-month period before the UTI diagnosis | |||
| Hospitalization | 40.3% | 20% | 0.0068 |
| Surgery | 21% | 10.5% | 0.0709 |
| Ureteral stent or urinary catheter | 29% | 12.4% | 0.0125 |
| Antibiotic therapy | |||
| Beta-lactam | 6.5% | 1.9% | 0.1961 |
| Cephalosporine | 25.8% | 4.8% | 0.0002 |
| Fluoroquinolone | 9.7% | 2.9% | 0.0787 |
| Trimethoprim/ Sulfonamide | 30.6% | 2.9% | 0 |
| Fosfomycin | 11.3% | 2.9% | 0.0401 |
| Carbapenem | 0% | 1% | 1 |
| Reason for antibiotic treatment | |||
| Prophylaxis | 29% | 4.8% | 0 |
| UTI | 25.8% | 7.6% | 0.0024 |
| Other infection | 12.9% | 7.6% | 0.2855 |
| Phylogroup | Total | KTx | Controls | p | ||
|---|---|---|---|---|---|---|
| No. of Isolates | No. of Isolates | % | No. of Isolates | % | ||
| A | 25 | 16 | 22.5 | 9 | 8.1 | 0.0061 |
| B1 | 24 | 10 | 14.1 | 14 | 12.6 | 0.4706 |
| B2 | 88 | 27 | 38 | 61 | 55 | 0.0394 |
| C | 8 | 1 | 1.4 | 7 | 6.3 | 0.1117 |
| D | 23 | 9 | 12.7 | 14 | 12.6 | 0.5805 |
| E | 2 | 2 | 2.8 | 0 | 0 | 0.1509 |
| F | 10 | 5 | 7 | 5 | 4.5 | 0.3389 |
| Clade V | 2 | 1 | 1.4 | 1 | 0.9 | 0.6293 |
| Serogroups | Total | KTx | Controls | p | ||
|---|---|---|---|---|---|---|
| No. of Isolates | No. of Isolates | % | No. of Isolates | % | ||
| O2 | 14 | 5 | 7 | 9 | 7.1 | 0.5159 |
| O4 | 8 | 3 | 4.2 | 5 | 5 | 0.6187 |
| O6 | 20 | 3 | 4.2 | 17 | 15.2 | 0.0148 |
| O8 | 18 | 7 | 9.9 | 11 | 9.8 | 0.6013 |
| O15 | 10 | 6 | 8.5 | 4 | 3.6 | 0.1435 |
| O16 | 5 | 2 | 2.8 | 3 | 2.7 | 0.6485 |
| O25 | 11 | 4 | 5.6 | 7 | 6.3 | 0.5608 |
| O83 | 5 | 1 | 1.4 | 4 | 3.6 | 0.3515 |
| O89 | 10 | 7 | 9.9 | 3 | 2.7 | 0.0432 |
| not typeable | 21 | 10 | 14.1 | 11 | 10 | nd |
| other serotypes | 60 | 23 | 32.4 | 31 | 33.9 | nd |
| Antibiotic | >1 RGs present | No RGs present | p | |||
|---|---|---|---|---|---|---|
| No. of Isolates | % | No. of Isolates | % | |||
| Beta-Lactam | KTx | 42 | 59.2 | 29 | 40.8 | 0.0033 |
| Controls | 41 | 36.9 | 70 | 63.1 | ||
| Trimethoprim | KTx | 34 | 47.9 | 37 | 52.1 | 0.0061 |
| Controls | 31 | 27.9 | 80 | 72.1 | ||
| Sulfonamides | KTx | 35 | 49.3 | 36 | 50.7 | 0.0763 |
| Controls | 40 | 36.0 | 71 | 64.0 | ||
| Fosfomycin | KTx | 0 | 0.0 | 71 | 100 | 1 |
| Controls | 1 | 0.9 | 110 | 99.1 | ||
| Fluoroquinolones | KTx | 3 | 4.2 | 68 | 95.8 | 1 |
| Controls | 6 | 5.4 | 105 | 94.6 | ||
| Aminoglycosides | KTx | 42 | 59.2 | 29 | 40.8 | 0.0101 |
| Controls | 44 | 39.6 | 67 | 60.4 | ||
| Antibiotic | Susceptible | Intermediate | Resistant | p | ||||
|---|---|---|---|---|---|---|---|---|
| No. of Isolates | % | No. of Isolates | % | No. of Isolates | % | |||
| Beta- Lactam (AMP+AMX) | KTx | 27 | 38 | 0 | 0 | 44 | 62 | 0.0015 |
| Controls | 69 | 62.2 | 0 | 0 | 42 | 37.8 | ||
| Beta- Lactam + inhibitor (SAM) | KTx | 50 | 70.4 | 0 | 0 | 21 | 29.6 | 0.1734 |
| Controls | 88 | 79.3 | 0 | 0 | 23 | 20.7 | ||
| Beta- Lactam (CFX) | KTx | 58 | 81.7 | 0 | 0 | 13 | 18.3 | 0.2147 |
| Controls | 98 | 88.3 | 0 | 0 | 13 | 11.7 | ||
| Trimethoprim | KTx | 33 | 50 | 0 | 0 | 33 | 50 | 0.0029 |
| Controls | 70 | 72.9 | 0 | 0 | 26 | 27.1 | ||
| TMP/SMX | KTx | 37 | 52.1 | 0 | 0 | 34 | 47.9 | 0.0132 |
| Controls | 78 | 70.3 | 0 | 0 | 33 | 29.7 | ||
| Fosfomycin | KTx | 66 | 98.5 | 0 | 0 | 1 | 1.5 | 1 |
| Controls | 96 | 98 | 0 | 0 | 2 | 2 | ||
| Fluoroqinolones (CIP+LVX) | KTx | 55 | 77.5 | 1 | 1.4 | 15 | 21.1 | 0.6588 |
| Controls | 92 | 82.9 | 1 | 0.9 | 18 | 16.2 | ||
| Aminoglycosides (GEN) | KTx | 66 | 93 | 0 | 0 | 5 | 7 | 0.4637 |
| Controls | 106 | 95.5 | 0 | 0 | 5 | 4.5 | ||
| Plasmid Type | Total No. of Isolates | KTx | Controls | p | ||
|---|---|---|---|---|---|---|
| No. of Isolates | % | No. of Isolates | % | |||
| IncFIA | 24 | 10 | 14.1 | 14 | 12.6 | 0.4706 |
| IncFIB | 114 | 49 | 69 | 65 | 58.6 | 0.1025 |
| IncFIC | 27 | 16 | 22.5 | 11 | 9.9 | 0.0178 |
| IncFII | 100 | 40 | 56.3 | 60 | 54.1 | 0.4411 |
| IncI1 | 33 | 17 | 23.9 | 16 | 14.4 | 0.0773 |
| IncQ1 | 20 | 13 | 18.3 | 7 | 6.3 | 0.0120 |
| Col156 | 35 | 9 | 12.7 | 26 | 23.4 | 0.0526 |
| Other replicons | 68 | 30 | 42.3 | 38 | 34.2 | 0.1751 |
| None | 33 | 7 | 9.9 | 26 | 23.4 | 0.0150 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abo Basha, J.; Kiel, M.; Görlich, D.; Schütte-Nütgen, K.; Witten, A.; Pavenstädt, H.; Kahl, B.C.; Dobrindt, U.; Reuter, S. Phenotypic and Genotypic Characterization of Escherichia coli Causing Urinary Tract Infections in Kidney-Transplanted Patients. J. Clin. Med. 2019, 8, 988. https://doi.org/10.3390/jcm8070988
Abo Basha J, Kiel M, Görlich D, Schütte-Nütgen K, Witten A, Pavenstädt H, Kahl BC, Dobrindt U, Reuter S. Phenotypic and Genotypic Characterization of Escherichia coli Causing Urinary Tract Infections in Kidney-Transplanted Patients. Journal of Clinical Medicine. 2019; 8(7):988. https://doi.org/10.3390/jcm8070988
Chicago/Turabian StyleAbo Basha, Jonas, Matthias Kiel, Dennis Görlich, Katharina Schütte-Nütgen, Anika Witten, Hermann Pavenstädt, Barbara C. Kahl, Ulrich Dobrindt, and Stefan Reuter. 2019. "Phenotypic and Genotypic Characterization of Escherichia coli Causing Urinary Tract Infections in Kidney-Transplanted Patients" Journal of Clinical Medicine 8, no. 7: 988. https://doi.org/10.3390/jcm8070988
APA StyleAbo Basha, J., Kiel, M., Görlich, D., Schütte-Nütgen, K., Witten, A., Pavenstädt, H., Kahl, B. C., Dobrindt, U., & Reuter, S. (2019). Phenotypic and Genotypic Characterization of Escherichia coli Causing Urinary Tract Infections in Kidney-Transplanted Patients. Journal of Clinical Medicine, 8(7), 988. https://doi.org/10.3390/jcm8070988





