Physician-Friendly Machine Learning: A Case Study with Cardiovascular Disease Risk Prediction
Abstract
1. Introduction
- For the first time, we investigate the use of AutoML for building classifiers of cardiovascular diseases.
- We compare AutoML performances against that of a graduate student with significant experience in machine learning and computer programming.
- We provide extensive experimental results on two cardiovascular datasets.
2. Materials and Methods
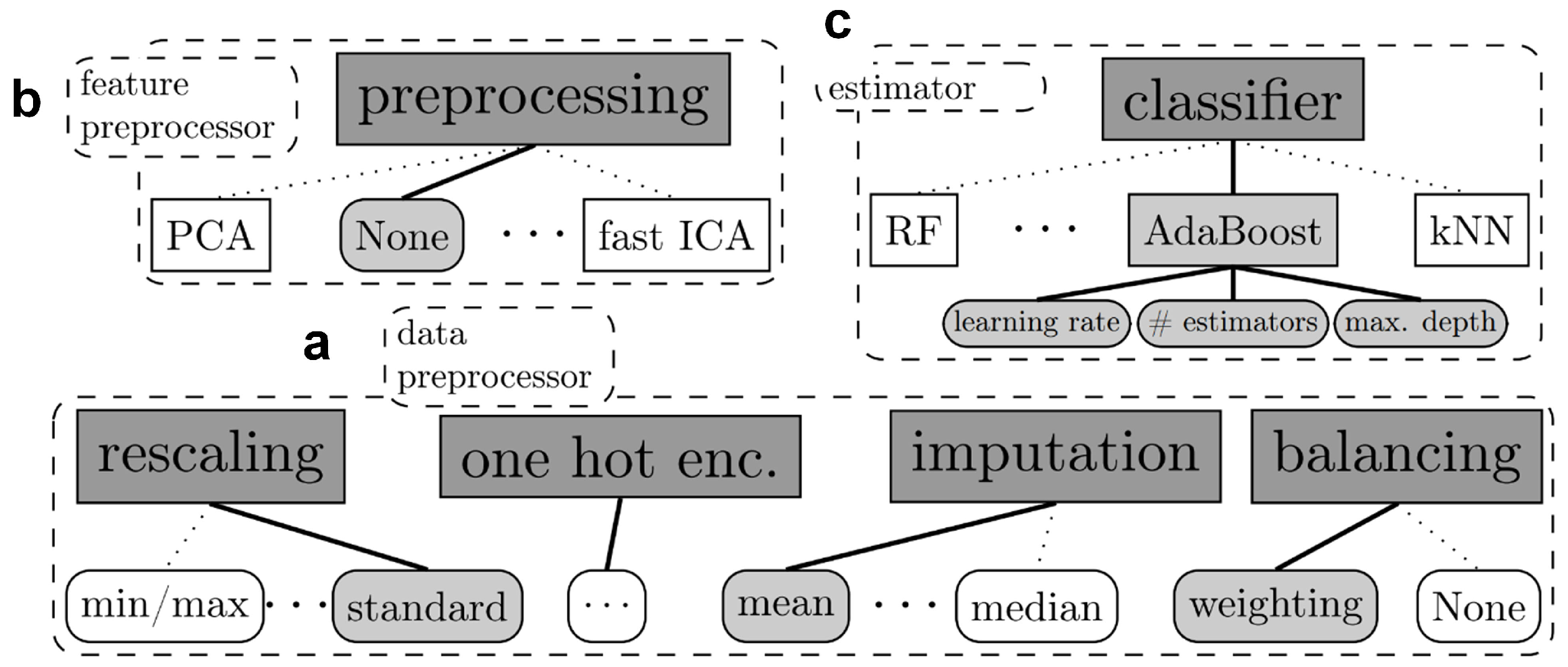
2.1. Algorithmic Description of Auto-Sklearn
2.2. Process of Building AutoML Models
2.3. Datasets and Manual Preprocessing by a Graduate Student
2.3.1. Dataset Description
2.3.2. Manual Data Preprocessing
- Looking for missing data and performing missing data imputation (both datasets did not have any missing data).
- Identifying continuous and categorical features.
- Identifying ordinal categorical features (where the categories have a natural ordered relationship between each other) and integer encoding them.
- Identifying nominal categorical features (where there is no natural ordered relationship between categories), and one-hot encoding them.
- Feature scaling, so as to bring all feature values to a similar dynamic range, which allows for faster convergence of learning algorithms optimized with the gradient descent method.
- Feature selection, which is the process of choosing relevant features from the given set and eliminating features that do not contribute much to the prediction of the target variable. This reduces training time and improves performance. Statistical tests allow ranking features according to their relation with the target. The F-statistical test (to capture linear relationships between features and target) and mutual information test (to capture linear and nonlinear relationships between features and target) are used independently on the datasets to evaluate the best set of features. In addition, recursive feature elimination, which is a greedy optimization algorithm that intends to find the best subset of features by repeatedly creating models and ranking the model’s performance with each subset, is also used. The above-mentioned statistical techniques are applied to capture complicated and nonlinear relationships of each feature with the target. Once the best set of features are extracted from training data independently using the above techniques, models are fitted on the new feature subsets and the corresponding cross validation performances are evaluated independently and compared so as to make a fool-proof decision on which feature-selection technique works well with an algorithm.
2.3.3. Train and Test Procedure
2.4. Challenges during Training Faced by a Graduate Student
2.5. Comparison of AutoML and Human
3. Results and Discussion
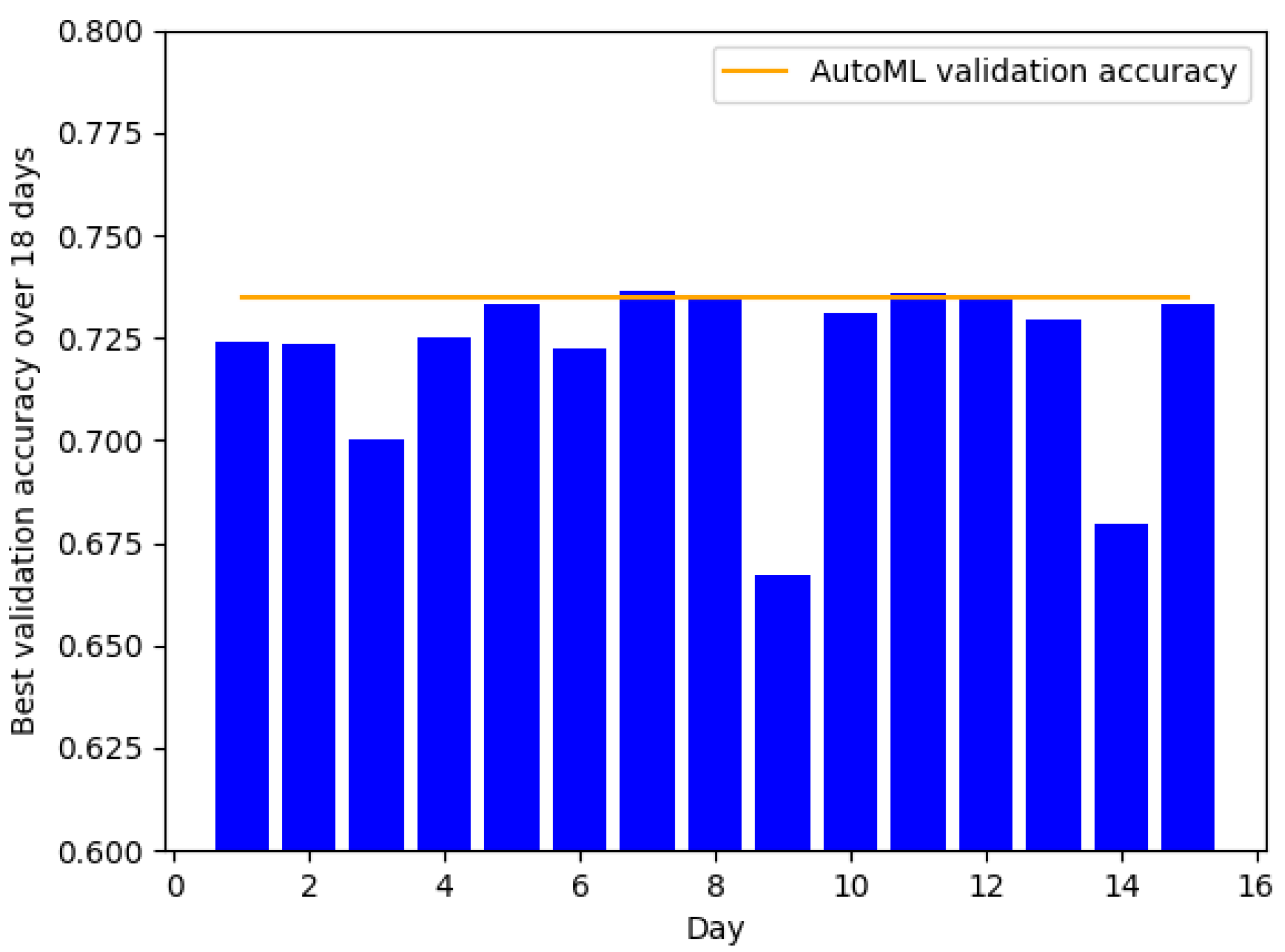
3.1. AutoML Benefits Complex Datasets More
3.2. Comparison of AutoML’s and Graduate Student’s Test-Set Performances
4. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| AutoML | Auto Machine Learning |
| UCI | University of California, Irvine |
| GPU | Graphic Processing Unit |
| TPU | Tensor Processing Unit |
| ROC | Reciever Operating Characteristic |
| PR | Precision–Recall |
| PCA | Principal Component Analysis |
References
- Bresnick, J. AI for Imaging Analytics Intrigues Healthcare Orgs, However, Starts Slow. 2018. Available online: https://healthitanalytics.com/news/ai-for-imaging-analytics-intrigues-healthcare-orgs-yet-starts-slow (accessed on 10 June 2019).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Baker, B.; Gupta, O.; Naik, N.; Raskar, R. Designing neural network architectures using reinforcement learning. arXiv Preprint 2016, arXiv:1611.02167. [Google Scholar]
- Liu, C.; Zoph, B.; Neumann, M.; Shlens, J.; Hua, W.; Li, L.J.; Fei-Fei, L.; Yuille, A.; Huang, J.; Murphy, K. Progressive neural architecture search. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 19–34. [Google Scholar]
- Miikkulainen, R.; Liang, J.; Meyerson, E.; Rawal, A.; Fink, D.; Francon, O.; Raju, B.; Shahrzad, H.; Navruzyan, A.; Duffy, N.; et al. Evolving deep neural networks. In Artificial Intelligence in the Age of Neural Networks and Brain Computing; Elsevier: Amsterdam, The Netherlands, 2019; pp. 293–312. [Google Scholar]
- Pham, H.; Guan, M.Y.; Zoph, B.; Le, Q.V.; Dean, J. Faster discovery of neural architectures by searching for paths in a large model. In Proceedings of the ICLR 2018, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Real, E.; Moore, S.; Selle, A.; Saxena, S.; Suematsu, Y.L.; Tan, J.; Le, Q.V.; Kurakin, A. Large-scale evolution of image classifiers. In Proceedings of the 34th International Conference on Machine Learning-Volume 70, Sydney, NSW, Australia, 6–11 August 2017; pp. 2902–2911. [Google Scholar]
- Thornton, C.; Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Auto-WEKA: Combined selection and hyperparameter optimization of classification algorithms. In Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; pp. 847–855. [Google Scholar]
- Feurer, M.; Klein, A.; Eggensperger, K.; Springenberg, J.; Blum, M.; Hutter, F. Efficient and robust automated machine learning. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; pp. 2962–2970. [Google Scholar]
- Real, E.; Aggarwal, A.; Huang, Y.; Le, Q.V. Regularized evolution for image classifier architecture search. arXiv Preprint 2018, arXiv:1802.01548. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; Das, S.R.; de Ferranti, S.; Després, J.P.; Fullerton, H.J.; et al. Heart disease and stroke statistics-2016 update a report from the American Heart Association. Circulation 2016, 133, e38–e48. [Google Scholar] [PubMed]
- Chitra, R.; Seenivasagam, V. Review of heart disease prediction system using data mining and hybrid intelligent techniques. ICTACT J. Soft Comput. 2013, 3, 605–609. [Google Scholar]
- Vembandasamy, K.; Sasipriya, R.; Deepa, E. Heart diseases detection using Naive Bayes algorithm. IJISET-Int. J. Innov. Sci. Eng. Technol. 2015, 2, 441–444. [Google Scholar]
- Shouman, M.; Turner, T.; Stocker, R. Using decision tree for diagnosing heart disease patients. In Proceedings of the Ninth Australasian Data Mining Conference-Volume 121, Ballarat, Australia, 1–2 December 2011; pp. 23–30. [Google Scholar]
- Srinivas, K.; Rani, B.K.; Govrdhan, A. Applications of data mining techniques in healthcare and prediction of heart attacks. Int. J. Comput. Sci. Eng. (IJCSE) 2010, 2, 250–255. [Google Scholar]
- Tomar, D.; Agarwal, S. Feature selection based least square twin support vector machine for diagnosis of heart disease. Int. J. Bio-Sci. Bio-Technol. 2014, 6, 69–82. [Google Scholar] [CrossRef]
- Fung, G.; Mangasarian, O.L. Incremental support vector machine classification. In Proceedings of the 2002 SIAM International Conference on Data Mining, Arlington, VA, USA, 11–13 April 2002; pp. 247–260. [Google Scholar]
- Pouriyeh, S.; Vahid, S.; Sannino, G.; De Pietro, G.; Arabnia, H.; Gutierrez, J. A comprehensive investigation and comparison of Machine Learning Techniques in the domain of heart disease. In Proceedings of the 2017 IEEE Symposium on Computers and Communications (ISCC), Heraklion, Greece, 3–6 July 2017; pp. 204–207. [Google Scholar]
- Bashir, S.; Qamar, U.; Khan, F.H. BagMOOV: A novel ensemble for heart disease prediction bootstrap aggregation with multi-objective optimized voting. Aust. Phys. Eng. Sci. Med. 2015, 38, 305–323. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Jolliffe, I. Principal Component Analysis; Springer: New York, NY, USA, 2011. [Google Scholar]
- Hyvärinen, A.; Karhunen, J.; Oja, E. Independent Component Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2004; Volume 46. [Google Scholar]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- CSV File Reading and Writing Tutorial. Available online: https://docs.python.org/3/library/csv.html (accessed on 10 July 2019).
- Zhou, Z.H.; Jiang, Y. NeC4. 5: Neural ensemble based C4. 5. IEEE Trans. Knowl. Data Eng. 2004, 16, 770–773. [Google Scholar] [CrossRef]
- Bouckaert, R.R.; Frank, E. Evaluating the replicability of significance tests for comparing learning algorithms. In Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining, Sydney, NSW, Australia, 26–28 May 2004; pp. 3–12. [Google Scholar]
- To, G.B.; Brown, G.; To, G.; Brown, G. Diversity in Neural Network Ensembles; University of Birmingham: Birmingham, UK, 2004. [Google Scholar]
- Chai, X.; Deng, L.; Yang, Q.; Ling, C.X. Test-cost sensitive naive bayes classification. In Proceedings of the Fourth IEEE International Conference on Data Mining (ICDM’04), Brighton, UK, 1–4 November 2004; pp. 51–58. [Google Scholar]
- Mason, L.; Bartlett, P.L.; Baxter, J. Improved generalization through explicit optimization of margins. Mach. Learn. 2000, 38, 243–255. [Google Scholar] [CrossRef]
- Duch, W.; Adamczak, R.; Grabczewski, K. A new methodology of extraction, optimization and application of crisp and fuzzy logical rules. IEEE Trans. Neural Netw. 2001, 12, 277–306. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.J.; Mathew, A.; Chen, Y.; Xi, L.F.; Ma, L.; Lee, J. Empirical analysis of support vector machine ensemble classifiers. Expert Syst. Appl. 2009, 36, 6466–6476. [Google Scholar] [CrossRef]
- Ulianova, S. Cardiovascular Disease Dataset. Available online: https://www.kaggle.com/sulianova/cardiovascular-disease-dataset (accessed on 10 July 2019).
- Janosi, A.; Steinbrunn, W.; Pfisterer, M.; Detrano, R. Heart Disease Data Set. Available online: https://archive.ics.uci.edu/ml/datasets/Heart+Disease (accessed on 10 July 2019).


| Attribute | Type | Description |
|---|---|---|
| Age | Continuous | Age in years |
| Cp | Discrete | Chest pain type (4 values) |
| Trestbps | Continuous | Resting blood pressure (in mm Hg on admission to the hospital) |
| Chol | Continuous | Serum cholestoral in mg/dL |
| Fbs | Discrete | Fasting blood sugar > 120 mg/dL 1 = true; 0 = false |
| Restecg | Discrete | Resting electrocardiographic results (values 0,1,2) |
| Thalach | Continuous | Maximum heart rate achieved |
| Exang | Discrete | Exercise induced angina (1 = yes; 0 = no) |
| Oldpeak | Continuous | ST depression induced by exercise relative to rest |
| Slope | Discrete | The slope of the peak Exercise ST segment (values 0,1,2) |
| Ca | Discrete | Number of major vessels (0–4) colored by flourosopy |
| Thal | Discrete | Nature of defect, values (0–3) |
| Target | Discrete | Presence or absence of heart disease, values (1,0) |
| Attribute | Type | Description |
|---|---|---|
| Age | Continuous | Age of the patient in days |
| Gender | Discrete | 1: women, 2: men |
| Height (cm) | Continuous | Height of the patient in cm |
| Weight (kg) | Continuous | Weight of the patient in kg |
| Ap_hi | Continuous | Systolic blood pressure |
| Ap_lo | Continuous | Diastolic blood pressure |
| Cholesterol | Discrete | 1: normal, 2: above normal, 3: well above normal |
| Gluc | Discrete | 1: normal, 2: above normal, 3: well above normal |
| Smoke | Discrete | whether patient smokes or not |
| Alco | Discrete | Alcohol intake-Binary feature |
| Active | Discrete | Physical activity-Binary feature |
| Cardio | Discrete | Presence or absence of cardiovascular disease |
| Accuracy | AUC-ROC | AUC-PR | Total Ttime (h) | |
|---|---|---|---|---|
| Graduate student | 0.84 | 0.82 | 0.80 | 432 |
| AutoML | 0.85 | 0.93 | 0.94 | 0.5 |
| Accuracy | AUC-ROC | AUC-PR | Total Time (h) | |
|---|---|---|---|---|
| Graduate student | 0.74 | 0.73 | 0.68 | 360 |
| AutoML | 0.74 | 0.8 | 0.79 | 0.5 |
| Author | Reported Accuracy |
|---|---|
| Shouman et al. [18] | 0.841 |
| Duch et al. [35] | 0.856 |
| Wang et al. [36] | 0.8337 |
| Srinivas et al. [19] | 0.837 |
| Tomar and Agarwal [20] | 0.8559 |
| Graduate student (this paper) | 0.84 |
| AutoML (this paper) | 0.85 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padmanabhan, M.; Yuan, P.; Chada, G.; Nguyen, H.V. Physician-Friendly Machine Learning: A Case Study with Cardiovascular Disease Risk Prediction. J. Clin. Med. 2019, 8, 1050. https://doi.org/10.3390/jcm8071050
Padmanabhan M, Yuan P, Chada G, Nguyen HV. Physician-Friendly Machine Learning: A Case Study with Cardiovascular Disease Risk Prediction. Journal of Clinical Medicine. 2019; 8(7):1050. https://doi.org/10.3390/jcm8071050
Chicago/Turabian StylePadmanabhan, Meghana, Pengyu Yuan, Govind Chada, and Hien Van Nguyen. 2019. "Physician-Friendly Machine Learning: A Case Study with Cardiovascular Disease Risk Prediction" Journal of Clinical Medicine 8, no. 7: 1050. https://doi.org/10.3390/jcm8071050
APA StylePadmanabhan, M., Yuan, P., Chada, G., & Nguyen, H. V. (2019). Physician-Friendly Machine Learning: A Case Study with Cardiovascular Disease Risk Prediction. Journal of Clinical Medicine, 8(7), 1050. https://doi.org/10.3390/jcm8071050





