Alteration of Epigenetic Modifiers in Pancreatic Cancer and Its Clinical Implication
Abstract
1. Introduction
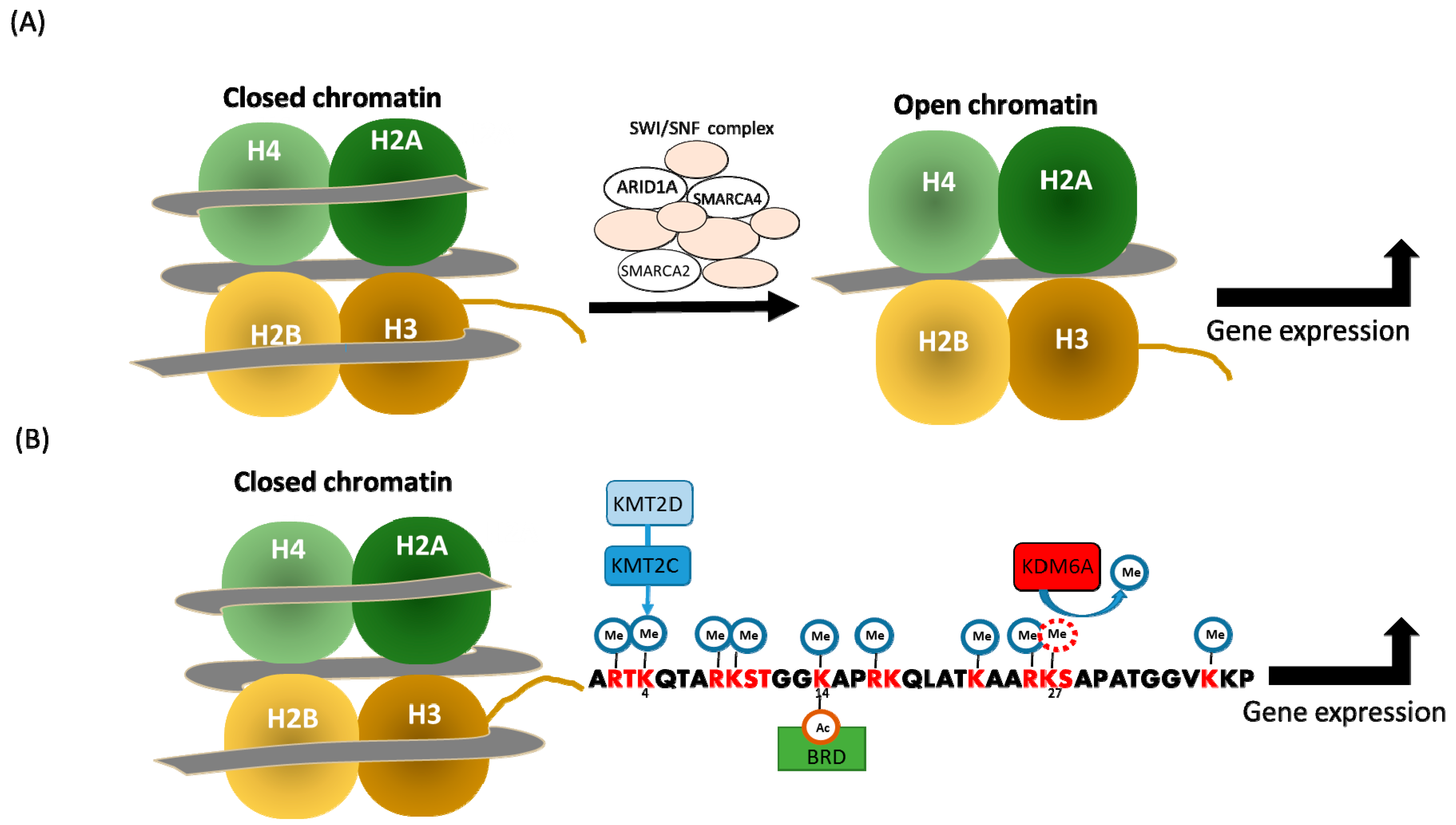
2. Expression and Mutation of SWI/SNF Genes in Pancreatic Cancer
2.1. ARID1A
2.2. SMARCA4
2.3. SMARCA2
3. Expression and Mutation of Histone Lysine Methylation Regulators in Pancreatic Cancer
3.1. KDM6A
3.2. KMT2C and KMT2D
3.3. G9a
3.4. EZH2
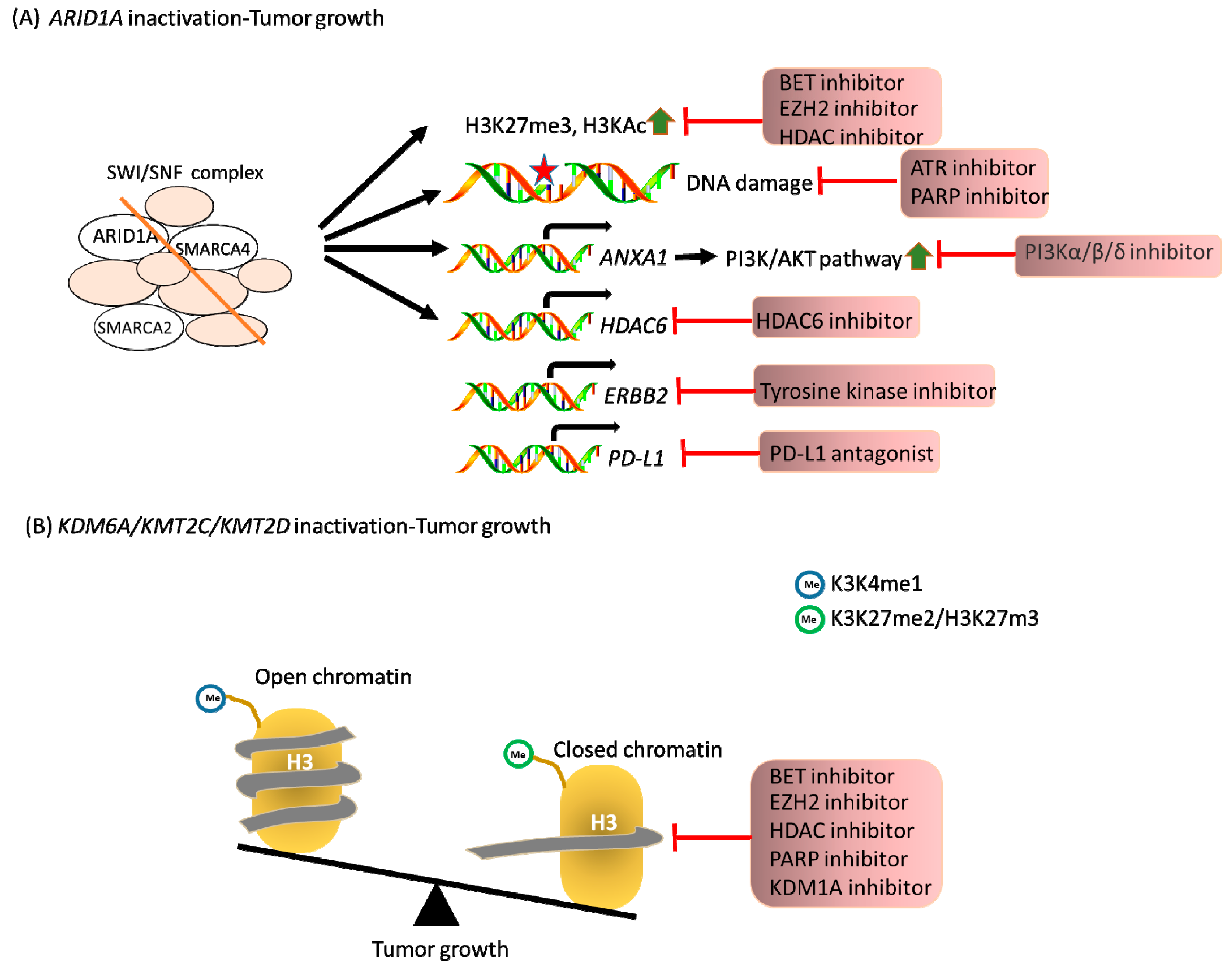
4. Therapeutics Targeting Epigenetic Regulators in Pancreatic Cancer
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.L.; Chen, Y.Y.; Lu, C.H.; Hung, C.Y.; Kuo, Y.C.; Chen, J.S.; Hsu, H.C.; Chen, P.T.; Chang, P.H.; Hung, Y.S. Effect of s-1 on survival outcomes in 838 patients with advanced pancreatic cancer: A 7-year multicenter observational cohort study in Taiwan. Cancer Med. 2019, 8, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Jansen, L.; Balavarca, Y.; van der Geest, L.; Lemmens, V.; Van Eycken, L.; De Schutter, H.; Johannesen, T.B.; Primic-Žakelj, M.; Zadnik, V.; et al. Nonsurgical therapies for resected and unresected pancreatic cancer in europe and USA in 2003–2014: A large international population-based study. Int. J. Cancer 2018, 143, 3227–3239. [Google Scholar] [CrossRef] [PubMed]
- Strobel, O.; Neoptolemos, J.; Jaeger, D.; Buechler, M.W. Optimizing the outcomes of pancreatic cancer surgery. Nat. Rev. Clin. Oncol. 2018, 16, 11–26. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Kleeff, J.; Michl, P.; Costello, E.; Greenhalf, W.; Palmer, D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Wang-Gillam, A.; Li, C.P.; Bodoky, G.; Dean, A.; Shan, Y.S.; Jameson, G.; Macarulla, T.; Lee, K.H.; Cunningham, D.; Blanc, J.F.; et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (napoli-1): A global, randomised, open-label, phase 3 trial. Lancet 2016, 387, 545–557. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, S.; Zhang, B.; Ni, Q.; Yu, X.; Xu, J. Circulating biomarkers for early diagnosis of pancreatic cancer: Facts and hopes. Am. J. Cancer Res. 2018, 8, 332–353. [Google Scholar] [PubMed]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Witkiewicz, A.K.; McMillan, E.A.; Balaji, U.; Baek, G.; Lin, W.C.; Mansour, J.; Mollaee, M.; Wagner, K.U.; Koduru, P.; Yopp, A.; et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015, 6, 6744. [Google Scholar] [CrossRef]
- Conway, J.R.; Herrmann, D.; Evans, T.J.; Morton, J.P.; Timpson, P. Combating pancreatic cancer with pi3k pathway inhibitors in the era of personalised medicine. Gut 2019, 68, 742–758. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.J.; Hahn, W.C. Synthetic lethal vulnerabilities in kras-mutant cancers. Cold Spring Harb. Perspect. Med. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Lomberk, G.; Blum, Y.; Nicolle, R.; Nair, A.; Gaonkar, K.S.; Marisa, L.; Mathison, A.; Sun, Z.; Yan, H.; Elarouci, N.; et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat. Commun. 2018, 9, 1978. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.D.; Yurgelun, M.B.; Goggins, M.G. Genetics of familial and sporadic pancreatic cancer. Gastroenterology 2019, 156, 2041–2055. [Google Scholar] [CrossRef]
- Zang, Z.J.; Cutcutache, I.; Poon, S.L.; Zhang, S.L.; McPherson, J.R.; Tao, J.; Rajasegaran, V.; Heng, H.L.; Deng, N.; Gan, A.; et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat. Genet. 2012, 44, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Chen, Y.B.; Pan, K.; Wang, W.; Chen, S.P.; Chen, J.G.; Zhao, J.J.; Lv, L.; Pan, Q.Z.; Li, Y.Q.; et al. Decreased expression of the arid1a gene is associated with poor prognosis in primary gastric cancer. PLoS ONE 2012, 7, e40364. [Google Scholar] [CrossRef]
- Yan, H.B.; Wang, X.F.; Zhang, Q.; Tang, Z.Q.; Jiang, Y.H.; Fan, H.Z.; Sun, Y.H.; Yang, P.Y.; Liu, F. Reduced expression of the chromatin remodeling gene arid1a enhances gastric cancer cell migration and invasion via downregulation of e-cadherin transcription. Carcinogenesis 2013, 35, 867–876. [Google Scholar] [CrossRef][Green Version]
- Inada, R.; Sekine, S.; Taniguchi, H.; Tsuda, H.; Katai, H.; Fujiwara, T.; Kushima, R. Arid1a expression in gastric adenocarcinoma: Clinicopathological significance and correlation with DNA mismatch repair status. World J. Gastroenterol. 2015, 21, 2159–2168. [Google Scholar] [CrossRef]
- Yang, L.; Wei, S.; Zhao, R.; Wu, Y.; Qiu, H.; Xiong, H. Loss of arid1a expression predicts poor survival prognosis in gastric cancer: A systematic meta-analysis from 14 studies. Sci. Rep. 2016, 6, 28919. [Google Scholar] [CrossRef]
- Takeshima, H.; Niwa, T.; Takahashi, T.; Wakabayashi, M.; Yamashita, S.; Ando, T.; Inagawa, Y.; Taniguchi, H.; Katai, H.; Sugiyama, T.; et al. Frequent involvement of chromatin remodeler alterations in gastric field cancerization. Cancer Lett. 2015, 357, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Rokutan, H.; Hosoda, F.; Hama, N.; Nakamura, H.; Totoki, Y.; Furukawa, E.; Arakawa, E.; Ohashi, S.; Urushidate, T.; Satoh, H.; et al. Comprehensive mutation profiling of mucinous gastric carcinoma. J. Pathol. 2016, 240, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Ozen, C.; Yildiz, G.; Dagcan, A.T.; Cevik, D.; Ors, A.; Keles, U.; Topel, H.; Ozturk, M. Genetics and epigenetics of liver cancer. N. Biotechnol. 2013, 30, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Aburatani, H. Exploration of liver cancer genomes. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, S.C.; Wei, Y.; Luo, X.; Jia, Y.; Li, L.; Gopal, P.; Zhu, M.; Nassour, I.; Chuang, J.C.; et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell 2017, 32, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.Z.; Li, C.; Liu, X.Y.; Hu, T.T.; Fan, Z.S.; Han, Z.G. Hepatocyte-specific arid1a deficiency initiates mouse steatohepatitis and hepatocellular carcinoma. PLoS ONE 2015, 10, e0143042. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Li, W.; Tian, F.; Jiang, K.; Liu, X.; Cen, J.; He, Q.; Qiu, Z.; Kienast, Y.; Wang, Z.; et al. Arid1a regulates response to anti-angiogenic therapy in advanced hepatocellular carcinoma. J. Hepatol. 2018, 68, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Totoki, Y.; Tatsuno, K.; Covington, K.R.; Ueda, H.; Creighton, C.J.; Kato, M.; Tsuji, S.; Donehower, L.A.; Slagle, B.L.; Nakamura, H.; et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014, 46, 1267–1273. [Google Scholar] [CrossRef]
- Fujimoto, A.; Totoki, Y.; Abe, T.; Boroevich, K.A.; Hosoda, F.; Nguyen, H.H.; Aoki, M.; Hosono, N.; Kubo, M.; Miya, F.; et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012, 44, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Han, K.H.; Gores, G.; Llovet, J.M.; Mazzaferro, V. Liver cancer: Approaching a personalized care. J. Hepatol. 2015, 62, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Eun, J.W.; Choi, S.K.; Shen, Q.; Choi, W.S.; Han, J.W.; Nam, S.W.; You, J.S. Epigenetic reader brd4 inhibition as a therapeutic strategy to suppress e2f2-cell cycle regulation circuit in liver cancer. Oncotarget 2016, 7, 32628–32640. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Pawlik, T.M.; Anders, R.A.; Selaru, F.M.; Streppel, M.M.; Lucas, D.J.; Niknafs, N.; Guthrie, V.B.; Maitra, A.; Argani, P.; et al. Exome sequencing identifies frequent inactivating mutations in bap1, arid1a and pbrm1 in intrahepatic cholangiocarcinomas. Nat. Genet. 2013, 45, 1470–1473. [Google Scholar] [CrossRef] [PubMed]
- Simbolo, M.; Fassan, M.; Ruzzenente, A.; Mafficini, A.; Wood, L.D.; Corbo, V.; Melisi, D.; Malleo, G.; Vicentini, C.; Malpeli, G.; et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget 2014, 5, 2839–2852. [Google Scholar] [CrossRef] [PubMed]
- Ruzzenente, A.; Fassan, M.; Conci, S.; Simbolo, M.; Lawlor, R.T.; Pedrazzani, C.; Capelli, P.; D’Onofrio, M.; Iacono, C.; Scarpa, A.; et al. Cholangiocarcinoma heterogeneity revealed by multigene mutational profiling: Clinical and prognostic relevance in surgically resected patients. Ann. Surg. Oncol. 2016, 23, 1699–1707. [Google Scholar] [CrossRef] [PubMed]
- Churi, C.R.; Shroff, R.; Wang, Y.; Rashid, A.; Kang, H.C.; Weatherly, J.; Zuo, M.; Zinner, R.; Hong, D.; Meric-Bernstam, F.; et al. Mutation profiling in cholangiocarcinoma: Prognostic and therapeutic implications. PLoS ONE 2014, 9, e115383. [Google Scholar] [CrossRef] [PubMed]
- Simbolo, M.; Vicentini, C.; Ruzzenente, A.; Brunelli, M.; Conci, S.; Fassan, M.; Mafficini, A.; Rusev, B.; Corbo, V.; Capelli, P.; et al. Genetic alterations analysis in prognostic stratified groups identified tp53 and arid1a as poor clinical performance markers in intrahepatic cholangiocarcinoma. Sci. Rep. 2018, 8, 7119. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Hong, E.K.; Kong, S.Y.; Han, S.S.; Kim, S.H.; Rhee, J.K.; Hwang, S.K.; Park, S.J.; Kim, T.M. Two classes of intrahepatic cholangiocarcinoma defined by relative abundance of mutations and copy number alterations. Oncotarget 2016, 7, 23825–23836. [Google Scholar] [CrossRef]
- Ito, T.; Sakurai-Yageta, M.; Goto, A.; Pairojkul, C.; Yongvanit, P.; Murakami, Y. Genomic and transcriptional alterations of cholangiocarcinoma. J. Hepatobiliary Pancreat. Sci. 2014, 21, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, D.J.; Adélaïde, J.; Mamessier, E.; Finetti, P.; Lagarde, A.; Monges, G.; Viret, F.; Gonçalvès, A.; Turrini, O.; Delpero, J.R.; et al. Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer 2011, 50, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Fukuda, A.; Ogawa, S.; Maruno, T.; Takada, Y.; Tsuda, M.; Hiramatsu, Y.; Araki, O.; Nagao, M.; Yoshikawa, T.; et al. Arid1a maintains differentiation of pancreatic ductal cells and inhibits development of pancreatic ductal adenocarcinoma in mice. Gastroenterology 2018, 155, 194–209. [Google Scholar] [CrossRef]
- Knudsen, E.S.; O’Reilly, E.M.; Brody, J.R.; Witkiewicz, A.K. Genetic diversity of pancreatic ductal adenocarcinoma and opportunities for precision medicine. Gastroenterology 2016, 150, 48–63. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, F.; Du, C.; Guo, H.; Ma, L.; Liu, X.; Kornmann, M.; Tian, X.; Yang, Y. Brm/smarca2 promotes the proliferation and chemoresistance of pancreatic cancer cells by targeting jak2/stat3 signaling. Cancer Lett. 2017, 402, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Dal Molin, M.; Hong, S.M.; Hebbar, S.; Sharma, R.; Scrimieri, F.; de Wilde, R.F.; Mayo, S.C.; Goggins, M.; Wolfgang, C.L.; Schulick, R.D.; et al. Loss of expression of the swi/snf chromatin remodeling subunit brg1/smarca4 is frequently observed in intraductal papillary mucinous neoplasms of the pancreas. Hum. Pathol. 2012, 43, 585–591. [Google Scholar] [CrossRef] [PubMed][Green Version]
- von Figura, G.; Fukuda, A.; Roy, N.; Liku, M.E.; Morris, J.P., IV; Kim, G.E.; Russ, H.A.; Firpo, M.A.; Mulvihill, S.J.; Dawson, D.W.; et al. The chromatin regulator brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat. Cell Biol. 2014, 16, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.; Malik, S.; Villanueva, K.E.; Urano, A.; Lu, X.; Von Figura, G.; Seeley, E.S.; Dawson, D.W.; Collisson, E.A.; Hebrok, M. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015, 29, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Andricovich, J.; Perkail, S.; Kai, Y.; Casasanta, N.; Peng, W.; Tzatsos, A. Loss of kdm6a activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to bet inhibitors. Cancer Cell 2018, 33, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Shimada, S.; Akiyama, Y.; Ishikawa, Y.; Ogura, T.; Ogawa, K.; Ono, H.; Mitsunori, Y.; Ban, D.; Kudo, A.; et al. Loss of kdm6a characterizes a poor prognostic subtype of human pancreatic cancer and potentiates hdac inhibitor lethality. Int. J. Cancer 2019, 145, 192–205. [Google Scholar] [CrossRef]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Kay Li, Q.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef]
- Dawkins, J.B.; Wang, J.; Maniati, E.; Heward, J.A.; Koniali, L.; Kocher, H.M.; Martin, S.A.; Chelala, C.; Balkwill, F.R.; Fitzgibbon, J.; et al. Reduced expression of histone methyltransferases kmt2c and kmt2d correlates with improved outcome in pancreatic ductal adenocarcinoma. Cancer Res. 2016, 76, 4861–4871. [Google Scholar] [CrossRef]
- Cajuso, T.; Hanninen, U.A.; Kondelin, J.; Gylfe, A.E.; Tanskanen, T.; Katainen, R.; Pitkanen, E.; Ristolainen, H.; Kaasinen, E.; Taipale, M.; et al. Exome sequencing reveals frequent inactivating mutations in arid1a, arid1b, arid2 and arid4a in microsatellite unstable colorectal cancer. Int. J. Cancer 2014, 135, 611–623. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Li, M.; Parsons, D.W.; Zhang, X.; Wesseling, J.; Kristel, P.; Schmidt, M.K.; Markowitz, S.; Yan, H.; Bigner, D.; et al. Somatic mutations in the chromatin remodeling gene arid1a occur in several tumor types. Hum. Mutat. 2012, 33, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Mathur, R.; Alver, B.H.; San Roman, A.K.; Wilson, B.G.; Wang, X.; Agoston, A.T.; Park, P.J.; Shivdasani, R.A.; Roberts, C.W. Arid1a loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 2017, 49, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Fu, L.; Han, Y.; Li, Q.; Wang, E. Decreased arid1a expression facilitates cell proliferation and inhibits 5-fluorouracil-induced apoptosis in colorectal carcinoma. Tumour Biol. 2014, 35, 7921–7927. [Google Scholar] [CrossRef] [PubMed]
- Chou, A.; Toon, C.W.; Clarkson, A.; Sioson, L.; Houang, M.; Watson, N.; DeSilva, K.; Gill, A.J. Loss of arid1a expression in colorectal carcinoma is strongly associated with mismatch repair deficiency. Hum. Pathol. 2014, 45, 1697–1703. [Google Scholar] [CrossRef]
- Ye, J.; Zhou, Y.; Weiser, M.R.; Gonen, M.; Zhang, L.; Samdani, T.; Bacares, R.; DeLair, D.; Ivelja, S.; Vakiani, E.; et al. Immunohistochemical detection of arid1a in colorectal carcinoma: Loss of staining is associated with sporadic microsatellite unstable tumors with medullary histology and high tnm stage. Hum. Pathol. 2014, 45, 2430–2436. [Google Scholar] [CrossRef]
- Lee, L.H.; Sadot, E.; Ivelja, S.; Vakiani, E.; Hechtman, J.F.; Sevinsky, C.J.; Klimstra, D.S.; Ginty, F.; Shia, J. Arid1a expression in early stage colorectal adenocarcinoma: An exploration of its prognostic significance. Hum. Pathol. 2016, 53, 97–104. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Paradise, B.; Barham, W.; Fernandez-Zapico, M. Targeting epigenetic aberrations in pancreatic cancer, a new path to improve patient outcomes? Cancers 2018, 10, 128. [Google Scholar] [CrossRef] [PubMed]
- Längst, G.; Manelyte, L. Chromatin remodelers: From function to dysfunction. Genes 2015, 6, 299–324. [Google Scholar]
- Thomas, H. Therapy: Targeting chromatin remodelling proteins to treat pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 608. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Li, D.Q.; Müller, S.; Knapp, S. Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene 2016, 35, 4423–4436. [Google Scholar] [CrossRef] [PubMed]
- Biegel, J.A.; Busse, T.M.; Weissman, B.E. Swi/snf chromatin remodeling complexes and cancer. Am. J. Med. Genet. C Semin. Med. Genet. 2014, 166C, 350–366. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.N.; Roberts, C.W. Arid1a mutations in cancer: Another epigenetic tumor suppressor? Cancer Discov. 2013, 3, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Livshits, G.; Alonso-Curbelo, D.; Morris IV, J.P.; Koche, R.; Saborowski, M.; Wilkinson, J.E.; Lowe, S.W. Arid1a restrains kras-dependent changes in acinar cell identity. Elife 2018, 7, 35216. [Google Scholar] [CrossRef]
- McLean, M.H.; El-Omar, E.M. Genetics of gastric cancer. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 664–674. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Tian, D.A.; Li, P.Y.; He, X.X. Mouse models of liver cancer: Progress and recommendations. Oncotarget 2015, 6, 23306–23322. [Google Scholar] [CrossRef]
- Santos, N.P.; Colaco, A.A.; Oliveira, P.A. Animal models as a tool in hepatocellular carcinoma research: A review. Tumour Biol. 2017, 39, 1010428317695923. [Google Scholar] [CrossRef]
- Bultman, S.; Gebuhr, T.; Yee, D.; La Mantia, C.; Nicholson, J.; Gilliam, A.; Randazzo, F.; Metzger, D.; Chambon, P.; Crabtree, G. A brg1 null mutation in the mouse reveals functional differences among mammalian swi/snf complexes. Mol. Cell 2000, 6, 1287–1295. [Google Scholar] [CrossRef]
- Xu, Y.; Vakoc, C.R. Targeting cancer cells with bet bromodomain inhibitors. Cold Spring Harb. Perspect. Med. 2017, 7, a026674. [Google Scholar] [CrossRef] [PubMed]
- Poulard, C.; Corbo, L.; Le Romancer, M. Protein arginine methylation/demethylation and cancer. Oncotarget 2016, 7, 67532–67550. [Google Scholar] [CrossRef] [PubMed]
- Koutsioumpa, M.; Hatziapostolou, M.; Polytarchou, C.; Tolosa, E.J.; Almada, L.L.; Mahurkar-Joshi, S.; Williams, J.; Tirado-Rodriguez, A.B.; Huerta-Yepez, S.; Karavias, D. Lysine methyltransferase 2d regulates pancreatic carcinogenesis through metabolic reprogramming. Gut 2019, 68, 1271–1286. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Duns, G.; Westers, H.; Sijmons, R.; van den Berg, A.; Kok, K. Setd2: An epigenetic modifier with tumor suppressor functionality. Oncotarget 2016, 7, 50719–50734. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.; Krogan, N.J.; Dover, J.; Erdjument-Bromage, H.; Tempst, P.; Johnston, M.; Greenblatt, J.F.; Shilatifard, A. Compass: A complex of proteins associated with a trithorax-related set domain protein. Proc. Natl. Acad. Sci. USA 2001, 98, 12902–12907. [Google Scholar] [CrossRef] [PubMed]
- Van der Meulen, J.; Speleman, F.; Van Vlierberghe, P. The h3k27me3 demethylase utx in normal development and disease. Epigenetics 2014, 9, 658–668. [Google Scholar] [CrossRef]
- Pan, M.R.; Hsu, M.C.; Chen, L.T.; Hung, W.C. Orchestration of h3k27 methylation: Mechanisms and therapeutic implication. Cell. Mol. Life Sci. 2018, 75, 209–223. [Google Scholar] [CrossRef]
- Pan, M.R.; Hsu, M.C.; Luo, C.W.; Chen, L.T.; Shan, Y.S.; Hung, W.C. The histone methyltransferase g9a as a therapeutic target to override gemcitabine resistance in pancreatic cancer. Oncotarget 2016, 7, 61136–61151. [Google Scholar] [CrossRef]
- Pan, M.R.; Hsu, M.C.; Chen, L.T.; Hung, W.C. G9a orchestrates pcl3 and kdm7a to promote histone h3k27 methylation. Sci. Rep. 2015, 5, 18709. [Google Scholar] [CrossRef]
- Luo, C.W.; Wang, J.Y.; Hung, W.C.; Peng, G.; Tsai, Y.L.; Chang, T.M.; Chai, C.Y.; Lin, C.H.; Pan, M.R. G9a governs colon cancer stem cell phenotype and chemoradioresistance through pp2a-rpa axis-mediated DNA damage response. Radiother. Oncol. 2017, 124, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Toll, A.D.; Dasgupta, A.; Potoczek, M.; Yeo, C.J.; Kleer, C.G.; Brody, J.R.; Witkiewicz, A.K. Implications of enhancer of zeste homologue 2 expression in pancreatic ductal adenocarcinoma. Hum. Pathol. 2010, 41, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xie, D.; Yin Li, W.; Man Cheung, C.; Yao, H.; Chan, C.Y.; Chan, C.Y.; Xu, F.P.; Liu, Y.H.; Sung, J.J.; et al. Rnai targeting ezh2 inhibits tumor growth and liver metastasis of pancreatic cancer in vivo. Cancer Lett. 2010, 297, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Holtzinger, A.; Jagan, I.; BeGora, M.; Lohse, I.; Ngai, N.; Nostro, C.; Wang, R.; Muthuswamy, L.B.; Crawford, H.C.; et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 2015, 21, 1364–1371. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.L.; Fehling, S.C.; Garcia, P.L.; Gamblin, T.L.; Council, L.N.; van Waardenburg, R.; Yang, E.S.; Bradner, J.E.; Yoon, K.J. The bet inhibitor jq1 attenuates double-strand break repair and sensitizes models of pancreatic ductal adenocarcinoma to parp inhibitors. EBioMedicine 2019. [Google Scholar] [CrossRef]
- Bitler, B.G.; Aird, K.M.; Garipov, A.; Li, H.; Amatangelo, M.; Kossenkov, A.V.; Schultz, D.C.; Liu, Q.; Shih Ie, M.; Conejo-Garcia, J.R.; et al. Synthetic lethality by targeting ezh2 methyltransferase activity in arid1a-mutated cancers. Nat. Med. 2015, 21, 231–238. [Google Scholar] [CrossRef]
- Fukumoto, T.; Park, P.H.; Wu, S.; Fatkhutdinov, N.; Karakashev, S.; Nacarelli, T.; Kossenkov, A.V.; Speicher, D.W.; Jean, S.; Zhang, L.; et al. Repurposing pan-hdac inhibitors for arid1a-mutated ovarian cancer. Cell Rep. 2018, 22, 3393–3400. [Google Scholar] [CrossRef]
- Bitler, B.G.; Wu, S.; Park, P.H.; Hai, Y.; Aird, K.M.; Wang, Y.; Zhai, Y.; Kossenkov, A.V.; Vara-Ailor, A.; Rauscher, F.J., III; et al. ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat. Cell Biol. 2017, 19, 962–973. [Google Scholar] [CrossRef]
- Miller, R.E.; Brough, R.; Bajrami, I.; Williamson, C.T.; McDade, S.; Campbell, J.; Kigozi, A.; Rafiq, R.; Pemberton, H.; Natrajan, R.; et al. Synthetic lethal targeting of arid1a-mutant ovarian clear cell tumors with dasatinib. Mol. Cancer Ther. 2016, 15, 1472–1484. [Google Scholar] [CrossRef]
- Shen, J.; Ju, Z.; Zhao, W.; Wang, L.; Peng, Y.; Ge, Z.; Nagel, Z.D.; Zou, J.; Wang, C.; Kapoor, P.; et al. Arid1a deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 2018, 24, 556–562. [Google Scholar] [CrossRef]
- Berns, K.; Caumanns, J.J.; Hijmans, E.M.; Gennissen, A.M.C.; Severson, T.M.; Evers, B.; Wisman, G.B.A.; Jan Meersma, G.; Lieftink, C.; Beijersbergen, R.L.; et al. Arid1a mutation sensitizes most ovarian clear cell carcinomas to bet inhibitors. Oncogene 2018, 37, 4611–4625. [Google Scholar] [CrossRef]
- Williamson, C.T.; Miller, R.; Pemberton, H.N.; Jones, S.E.; Campbell, J.; Konde, A.; Badham, N.; Rafiq, R.; Brough, R.; Gulati, A.; et al. Atr inhibitors as a synthetic lethal therapy for tumours deficient in arid1a. Nat. Commun. 2016, 7, 13837. [Google Scholar] [CrossRef]
- Shen, J.; Peng, Y.; Wei, L.; Zhang, W.; Yang, L.; Lan, L.; Kapoor, P.; Ju, Z.; Mo, Q.; Shih Ie, M.; et al. Arid1a deficiency impairs the DNA damage checkpoint and sensitizes cells to parp inhibitors. Cancer Discov. 2015, 5, 752–767. [Google Scholar] [CrossRef] [PubMed]
- Chan-Penebre, E.; Armstrong, K.; Drew, A.; Grassian, A.R.; Feldman, I.; Knutson, S.K.; Kuplast-Barr, K.; Roche, M.; Campbell, J.; Ho, P.; et al. Selective killing of smarca2- and smarca4-deficient small cell carcinoma of the ovary, hypercalcemic type cells by inhibition of ezh2: In vitro and in vivo preclinical models. Mol. Cancer Ther. 2017, 16, 850–860. [Google Scholar] [CrossRef]
- Xue, Y.; Meehan, B.; Fu, Z.; Wang, X.Q.D.; Fiset, P.O.; Rieker, R.; Levins, C.; Kong, T.; Zhu, X.; Morin, G.; et al. Smarca4 loss is synthetic lethal with cdk4/6 inhibition in non-small cell lung cancer. Nat. Commun. 2019, 10, 557. [Google Scholar] [CrossRef] [PubMed]
- Tagal, V.; Wei, S.; Zhang, W.; Brekken, R.A.; Posner, B.A.; Peyton, M.; Girard, L.; Hwang, T.; Wheeler, D.A.; Minna, J.D.; et al. Smarca4-inactivating mutations increase sensitivity to aurora kinase a inhibitor vx-680 in non-small cell lung cancers. Nat. Commun. 2017, 8, 14098. [Google Scholar] [CrossRef] [PubMed]
- Ler, L.D.; Ghosh, S.; Chai, X.; Thike, A.A.; Heng, H.L.; Siew, E.Y.; Dey, S.; Koh, L.K.; Lim, J.Q.; Lim, W.K.; et al. Loss of tumor suppressor kdm6a amplifies prc2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of ezh2. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Ezponda, T.; Dupere-Richer, D.; Will, C.M.; Small, E.C.; Varghese, N.; Patel, T.; Nabet, B.; Popovic, R.; Oyer, J.; Bulic, M.; et al. UTX/KDM6A loss enhances the malignant phenotype of multiple myeloma and sensitizes cells to EZH2 inhibition. Cell Rep. 2017, 21, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Pan, X.; Chen, X.; Chen, M.; Shi, K.; Xu, J.; Zheng, J.; Niu, T.; Chen, C.; Shuai, X.; et al. Epigenetic drug library screening identified an lsd1 inhibitor to target utx-deficient cells for differentiation therapy. Signal Transduct. Target. Ther. 2019, 4, 11. [Google Scholar] [CrossRef]
- Rampias, T.; Karagiannis, D.; Avgeris, M.; Polyzos, A.; Kokkalis, A.; Kanaki, Z.; Kousidou, E.; Tzetis, M.; Kanavakis, E.; Stravodimos, K.; et al. The lysine-specific methyltransferase kmt2c/mll3 regulates DNA repair components in cancer. EMBO Rep. 2019, 20, e46821. [Google Scholar] [CrossRef]
| Gene Symbol | Gene Name | Alias |
|---|---|---|
| ARID1A | AT-rich interaction domain 1A | B120, BAF250, BAF250a, BM029, C1orf4, CSS2, ELD, MRD14, OSA1, P270, SMARCF1, hELD, hOSA1 |
| SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | BAF190, BRM, NCBRS, SNF2, SNF2L2, SNF2LA, SWI2, Sth1p, hBRM, hSNF2a |
| SMARCA4 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | BAF190, BAF190A, BRG1, CSS4, MRD16, RTPS2, SNF2, SNF2L4, SNF2LB, SWI2, hSNF2b |
| KDM6A | lysine demethylase 6A | KABUK2, UTX, bA386N14.2 |
| KMT2C | lysine methyltransferase 2C | HALR, KLEFS2, MLL3 |
| KMT2D | lysine methyltransferase 2D | AAD10, ALR, CAGL114, KABUK1, KMS, MLL2, MLL4, TNRC21 |
| BRD2 | bromodomain containing 2 | BRD2-IT1, D6S113E, FSH, FSRG1, NAT, O27.1.1, RING3, RNF3 |
| BRD3 | bromodomain containing 3 | ORFX, RING3L |
| BRD4 | bromodomain containing 4 | CAP, HUNK1, HUNKI, MCAP |
| BRDT | bromodomain testis associated | BRD6, CT9, SPGF21 |
| Cancer | Gene | Mutation | Expression | Effect | Reference |
|---|---|---|---|---|---|
| Stomach | ARID1A | Nonsense, missense, splice site | [14,15,16] | ||
| Loss | Increased proliferation | [14,17] | |||
| Loss | Increased migration and invasion | [18] | |||
| Loss | Association with tumor stage and grade | [17] | |||
| Loss | Association with lymphatic invasion and lymph node metastasis | [19] | |||
| Loss | Association with poor prognosis | [17,19,20] | |||
| SMARCA2 | Mutation | [21] | |||
| SMARCA4 | Missense | [21] | |||
| KDM6A | Nonsense, missense | [22] | |||
| KMT2C | Mutation | [14] | |||
| Liver | ARID1A | Nonsense, missense | [23,24,25] | ||
| Loss | Decreased tumorigenesis; increased metastasis | [26] | |||
| Loss | Increased steatohepatitis and tumorigenesis | [27] | |||
| Loss | Increased tumorigenesis and angiogenesis | [28] | |||
| SMARCA2 | Missense | [29] | |||
| KMT2C | Mutation | [30] | |||
| KMT2D | Mutation | [31] | |||
| BRD4 | Gain | Increased tumorigenesis | [32] | ||
| Biliary duct | ARID1A | Nonsense, missense, splice site | [33,34,35,36] | ||
| Nonsense, missense | Association with poor prognosis | [37] | |||
| SMARCA4 | Missense | [38] | |||
| KMT2C | Mutation | [39] | |||
| KMT2D | Mutation | [38] | |||
| Pancreas | ARID1A | Nonsense, missense | [40,41] | ||
| Loss | Decreased differentiation | [42] | |||
| SMARCA2 | Mutation | [43] | |||
| Gain | Decreased patient survival and drug sensitivity | [44] | |||
| SMARCA4 | Mutation | [45] | |||
| Loss | Increased IPMN; decreased PanIN | [46] | |||
| Loss | Decreased late stage tumorigenicity | [47] | |||
| KDM6A | Mutation | [48] | |||
| Loss | Increased squamous-like cancer | [48] | |||
| Loss | Decreased overall/recurrence-free survival | [49] | |||
| KMT2C | Missense | [50] | |||
| KMT2D | Missense | [50] | |||
| Loss | Increased apoptosis and drug sensitivity | [51] | |||
| Colon | ARID1A | Missense | [52,53,54] | ||
| Loss | Increased aggressive adenocarcinoma | [55] | |||
| Loss | Increased proliferation and drug resistance | [56] | |||
| Loss | Association with ageing | [57,58] | |||
| Loss | Association with poor tumor differentiation | [56,58,59] | |||
| Loss | Association with tumor size | [57] | |||
| Loss | Association with tumor grade | [57,58] | |||
| Loss | Association with metastasis | [58] |
| Gene | Cancer | Therapeutic Target | Reference |
|---|---|---|---|
| ARID1A | Ovary | EZH2 | [86] |
| Pan HDAC | [87] | ||
| HDAC6 | [88] | ||
| tyrosine kinases | [89] | ||
| CD274 | [90] | ||
| BET | [91] | ||
| Colon | ATR | [92] | |
| PARP | [93] | ||
| SMARCA2 | Ovary | EZH2 | [94] |
| SMARCA4 | Lung | CDK4/6 | [95] |
| AURKA | [96] | ||
| Ovary | EZH2 | [94] | |
| KDM6A | Bladder | EZH2 | [97] |
| Multiple myeloma | EZH2 | [98] | |
| Acute myeloid leukemia | KDM1A | [99] | |
| Pancreas | BET | [48] | |
| HDAC | [49] | ||
| KMT2C | Epithelial cancer | PARP | [100] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hung, Y.-H.; Hsu, M.-C.; Chen, L.-T.; Hung, W.-C.; Pan, M.-R. Alteration of Epigenetic Modifiers in Pancreatic Cancer and Its Clinical Implication. J. Clin. Med. 2019, 8, 903. https://doi.org/10.3390/jcm8060903
Hung Y-H, Hsu M-C, Chen L-T, Hung W-C, Pan M-R. Alteration of Epigenetic Modifiers in Pancreatic Cancer and Its Clinical Implication. Journal of Clinical Medicine. 2019; 8(6):903. https://doi.org/10.3390/jcm8060903
Chicago/Turabian StyleHung, Yu-Hsuan, Ming-Chuan Hsu, Li-Tzong Chen, Wen-Chun Hung, and Mei-Ren Pan. 2019. "Alteration of Epigenetic Modifiers in Pancreatic Cancer and Its Clinical Implication" Journal of Clinical Medicine 8, no. 6: 903. https://doi.org/10.3390/jcm8060903
APA StyleHung, Y.-H., Hsu, M.-C., Chen, L.-T., Hung, W.-C., & Pan, M.-R. (2019). Alteration of Epigenetic Modifiers in Pancreatic Cancer and Its Clinical Implication. Journal of Clinical Medicine, 8(6), 903. https://doi.org/10.3390/jcm8060903





