Common Inflammation-Related Candidate Gene Variants and Acute Kidney Injury in 2647 Critically Ill Finnish Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Definitions
2.3. Data Collection
2.4. DNA Samples and Genotyping
2.5. Variant Selection
2.6. Statistical Analyses
2.7. Power Calculations
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Nisula, S.; Kaukonen, K.-M.M.; Vaara, S.T.; Korhonen, A.-M.M.; Poukkanen, M.; Karlsson, S.; Haapio, M.; Inkinen, O.; Parviainen, I.; Suojaranta-Ylinen, R.; et al. Incidence, risk factors and 90-day mortality of patients with acute kidney injury in Finnish intensive care units: The FINNAKI study. Intensive Care Med. 2013, 39, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Hoste, E.A.J.; Bagshaw, S.M.; Bellomo, R.; Cely, C.M.; Colman, R.; Cruz, D.N.; Edipidis, K.; Forni, L.G.; Gomersall, C.D.; Govil, D.; et al. Epidemiology of acute kidney injury in critically ill patients: The multinational AKI-EPI study. Intensive Care Med. 2015, 41, 1411–1423. [Google Scholar] [CrossRef] [PubMed]
- Vilander, L.M.; Kaunisto, M.A.; Pettilä, V. Genetic predisposition to acute kidney injury—A systematic review. BMC Nephrol. 2015, 16, 197. [Google Scholar] [CrossRef] [PubMed]
- Kidney Disease: Improving Global Outcomes (KDIGO) Acute Kidney Injury Work Group. KDIGO Clinical Practice Guideline for Acute Kidney Injury. Kidney Int. Suppl. 2012, 2, 1–138. [Google Scholar]
- Bone, R.C.; Balk, R.A.; Cerra, F.B.; Dellinger, R.P.; Fein, A.M.; Knaus, W.A.; Schein, R.M.; Sibbald, W.J. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM consensus conference. Chest J. 1992, 101, 1644–1655. [Google Scholar] [CrossRef]
- Jurinke, C.; van den Boom, D.; Cantor, C.R.; Koster, H. Automated genotyping using the DNA MassArray technology. Methods Mol. Biol. 2002, 187, 179–192. [Google Scholar] [PubMed]
- Hashad, D.I.; Elsayed, E.T.; Helmy, T.A.; Elawady, S.M. Study of the role of tumor necrosis factor-α (–308 G/A) and interleukin-10 (–1082 G/A) polymorphisms as potential risk factors to acute kidney injury in patients with severe sepsis using high-resolution melting curve analysis. Ren. Fail. 2017, 39, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Susantitaphong, P.; Perianayagam, M.C.; Tighiouart, H.; Liangos, O.; Bonventre, J.V.; Jaber, B.L. Tumor necrosis factor alpha promoter polymorphism and severity of acute kidney injury. Nephron 2013, 123, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Henao-Martinez, A.; Agler, A.H.; LaFlamme, D.; Schwartz, D.A.; Yang, I. V Polymorphisms in the SUFU gene are associated with organ injury protection and sepsis severity in patients with Enterobacteriacea bacteremia. Infect. Genet. Evol. 2013, 16, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Kornek, M.; Deutsch, M.A.; Eichhorn, S.; Lahm, H.; Wagenpfeil, S.; Krane, M. COMT-Val158Met-polymorphism is not a risk factor for acute kidney injury after cardiac surgery. Dis. Mark. 2013, 35, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Haase-Fielitz, A.; Haase, M.; Bellomo, R.; Lambert, G.; Matalanis, G.; Story, D.; Doolan, L.; Buxton, B.; Gutteridge, G.; Luft, F.C.; et al. Decreased catecholamine degradation associates with shock and kidney injury after cardiac surgery. J. Am. Soc. Nephrol. 2009, 20, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Albert, C.; Kube, J.; Haase-Fielitz, A.; Dittrich, A.; Schanze, D.; Zenker, M.; Kuppe, H.; Hetzer, R.; Bellomo, R.; Mertens, P.R.; et al. Pilot study of association of catechol-O-methyl transferase rs4680 genotypes with acute kidney injury and tubular stress after open heart surgery. Biomark. Med. 2014, 8, 1227–1238. [Google Scholar] [CrossRef] [PubMed]
- Popov, A.F.; Hinz, J.; Schulz, E.G.; Schmitto, J.D.; Wiese, C.H.; Quintel, M.; Seipelt, R.; Schoendube, F.A. The eNOS 786C/T polymorphism in cardiac surgical patients with cardiopulmonary bypass is associated with renal dysfunction. Eur. J. Cardio-Thoracic Surg. 2009, 36, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Popov, A.F.; Schulz, E.G.; Schmitto, J.D.; Coskun, K.O.; Tzvetkov, M.V.; Kazmaier, S.; Zimmermann, J.; Schondube, F.A.; Quintel, M.; Hinz, J. Relation between renal dysfunction requiring renal replacement therapy and promoter polymorphism of the erythropoietin gene in cardiac surgery. Artif. Organs 2010, 34, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Kolyada, A.Y.; Tighiouart, H.; Perianayagam, M.C.; Liangos, O.; Madias, N.E.; Jaber, B.L. A genetic variant of hypoxia-inducible factor-1alpha is associated with adverse outcomes in acute kidney injury. Kidney Int. 2009, 75, 1322–1329. [Google Scholar] [CrossRef] [PubMed]
- Perianayagam, M.; Tighiouart, H.; Liangos, O.; Kouznetsov, D.; Wald, R.; Rao, F.; O’Connor, D.; Jaber, B. Polymorphisms in the myeloperoxidase gene locus are associated with acute kidney injuryrelated outcomes. Kidney Int. 2012, 82, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; O’Connor, D.T.; Perianayagam, M.C.; Kolyada, A.Y.; Chen, Y.; Rao, F.; Mahata, M.; Mahata, S.; Liangos, O.; Jaber, B.L. Phenylethanolamine N-methyltransferase gene polymorphisms and adverse outcomes in acute kidney injury. Nephron 2010, 114, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.; Di Castelnuovo, A.; Zamparelli, R.; Andreotti, F.; Burzotta, F.; Iacoviello, L.; Glieca, F.; Alessandrini, F.; Nasso, G.; Donati, M.B.; et al. Genetic control of postoperative systemic inflammatory reaction and pulmonary and renal complications after coronary artery surgery. J. Thorac. Cardiovasc. Surg. 2003, 126, 1107–1112. [Google Scholar] [CrossRef]
- Dalboni, M.A.; Quinto, B.M.; Grabulosa, C.C.; Narciso, R.; Monte, J.C.; Durao, M. Tumour necrosis factor-alpha plus interleukin-10 low producer phenotype predicts acute kidney injury and death in intensive care unit patients. Clin. Exp. Immunol. 2013, 173, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Wattanathum, A.; Manocha, S.; Groshaus, H.; Russell, J.A.; Walley, K.R. Interleukin-10 Haplotype Associated with Increased Mortality in Critically Ill Patients with Sepsis From Pneumonia But Not in Patients With Extrapulmonary Sepsis. Chest 2005, 128, 1690–1698. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.F.; Lu, T.M.; Yang, W.C.; Lin, S.J.; Lin, C.C.; Chung, M.Y. Gene polymorphisms of interleukin-10 and tumor necrosis factor-alpha are associated with contrast-induced nephropathy. Am. J. Nephrol. 2013, 37, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Boehm, J.; Eichhorn, S.; Kornek, M.; Hauner, K.; Prinzing, A.; Grammer, J. Apolipoprotein E genotype, TNF-alpha 308G/A and risk for cardiac surgery associated-acute kidney injury in Caucasians. Ren. Fail. 2014, 36, 237–243. [Google Scholar] [CrossRef] [PubMed]
- McBride, W.T.; Prasad, P.S.; Armstrong, M.; Patterson, C.; Gilliland, H.; Drain, A. Cytokine phenotype, genotype, and renal outcomes at cardiac surgery. Cytokine 2013, 61, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Stafford-Smith, M.; Podgoreanu, M.; Swaminathan, M.; Phillips-Bute, B.; Mathew, J.P.; Hauser, E.H. Association of genetic polymorphisms with risk of renal injury after coronary bypass graft surgery. Am. J. Kidney Dis. 2005, 45, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Jaber, B.L.; Rao, M.; Guo, D.; Balakrishnan, V.S.; Perianayagam, M.C.; Freeman, R.B.; Pereira, B.J. Cytokine gene promoter polymorphisms and mortality in acute renal failure. Cytokine 2004, 25, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Jouan, J.; Golmard, L.; Benhamouda, N.; Durrleman, N.; Golmard, J.L.; Ceccaldi, R.; Trinquart, L.; Fabiani, J.N.; Tartour, E.; Jeunemaitre, X.; et al. Gene polymorphisms and cytokine plasma levels as predictive factors of complications after cardiopulmonary bypass. J. Thorac. Cardiovasc. Surg. 2012, 144, 467–473.e2. [Google Scholar] [CrossRef] [PubMed]
- Cardinal-Fernandez, P.; Ferruelo, A.; El-Assar, M.; Santiago, C.; Gomez-Gallego, F.; Martin-Pellicer, A. Genetic predisposition to acute kidney injury induced by severe sepsis. J. Crit. Care 2013, 28, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.J.; Sheu, C.C.; Zhao, Y.; Chen, F.; Su, L.; Gong, M.N.; Bajwa, E.; Thompson, B.T.; Christiani, D.C. BCL2 genetic variants are associated with acute kidney injury in septic shock. Crit. Care Med. 2012, 40, 2116–2123. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Lu, Q.; Cheng, Y.; Belcher, J.M.; Siew, E.D.; Leaf, D.E.; Body, S.C.; Fox, A.A.; Waikar, S.S.; Collard, C.D.; et al. A Genome-Wide Association Study to Identify Single-Nucleotide Polymorphisms for Acute Kidney Injury. Am. J. Respir. Crit. Care Med. 2017, 195, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Stafford-Smith, M.; Li, Y.-J.; Mathew, J.P.; Li, Y.-W.; Ji, Y.; Phillips-Bute, B.G.; Milano, C.A.; Newman, M.F.; Kraus, W.E.; Kertai, M.D.; et al. Genome-wide association study of acute kidney injury after coronary bypass graft surgery identifies susceptibility loci. Kidney Int. 2015, 88, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Vilander, L.M.; Kaunisto, M.A.; Vaara, S.T.; Pettilä, V.; FINNAKI Study Group. Genetic variants in SERPINA4 and SERPINA5, but not BCL2 and SIK3 are associated with acute kidney injury in critically ill patients with septic shock. Crit. Care 2017, 21, 47. [Google Scholar] [CrossRef] [PubMed]
- Bhatraju, P.; Hsu, C.; Mukherjee, P.; Glavan, B.J.; Burt, A.; Mikacenic, C.; Himmelfarb, J.; Wurfel, M. Associations between single nucleotide polymorphisms in the FAS pathway and acute kidney injury. Crit. Care 2015, 19, 368. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, J.; Li, Y.; Xing, G. Cytoprotective Effect of Heat Shock Protein 27 Against Lipopolysaccharide-Induced Apoptosis of Renal Epithelial HK-2 Cells. Cell. Physiol. Biochem. 2017, 41, 2211–2220. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, G.; Li, L.; Liu, Z.; Zhou, Q.; Wang, G.; Chen, D. Surfactant protein-D (SP-D) gene polymorphisms and serum level as predictors of susceptibility and prognosis of acute kidney injury in the Chinese population. BMC Nephrol. 2017, 18, 67. [Google Scholar] [CrossRef]
- Schaalan, M.F.; Mohamed, W.A. Determinants of hepcidin levels in sepsis-associated acute kidney injury: Impact on pAKT/PTEN pathways? J. Immunotoxicol. 2016, 13, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Cherny, S.S.; Sham, P.C. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003, 19, 149–150. [Google Scholar] [CrossRef] [PubMed]
- Larach, D.B.; Engoren, M.C.; Schmidt, E.M.; Heung, M. Genetic variants and acute kidney injury: A review of the literature. J. Crit. Care 2018, 44, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P. Why most discovered true associations are inflated. Epidemiology 2008, 19, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Lohmueller, K.E.; Pearce, C.L.; Pike, M.; Lander, E.S.; Hirschhorn, J.N. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003, 33, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Kraft, P.; Zeggini, E.; Ioannidis, J.P.A. Replication in genome-wide association studies. Stat. Sci. 2009, 24, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Teuffel, O.; Ethier, M.C.; Beyene, J.; Sung, L. Association between tumor necrosis factor-α promoter −308 A/g polymorphism and susceptibility to sepsis and sepsis mortality: A systematic review and meta-analysis. Crit. Care Med. 2010, 38, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Christaki, E.; Giamarellos-Bourboulis, E.J. The beginning of personalized medicine in sepsis: Small steps to a bright future. Clin. Genet. 2014, 86, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhang, A.Q.; Yue, C.L.; Gao, J.W.; Zeng, L.; Gu, W.; Jiang, J.X. Association between interleukin-10 polymorphisms and sepsis: A meta-analysis. Epidemiol. Infect. 2015, 143, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N.; Gerber, H.-P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.O. Vascular endothelial growth factors and vascular permeability. Cardiovasc. Res. 2010, 87, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Renner, W.; Kotschan, S.; Hoffmann, C.; Obermayer-Pietsch, B.; Pilger, E. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J. Vasc. Res. 2000, 37, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Medford, A.R.L.; Keen, L.J.; Bidwell, J.L.; Millar, A.B. Vascular endothelial growth factor gene polymorphism and acute respiratory distress syndrome. Thorax 2005, 60, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Gong, M.N.; Zhou, W.; Thompson, T.B.; Kraft, P.; Su, L.; Christiani, D.C. Genotypes and haplotypes of the VEGF gene are associated with higher mortality and lower VEGF plasma levels in patients with ARDS. Thorax 2007, 62, 718–722. [Google Scholar] [CrossRef] [PubMed]
- Nazir, N.; Siddiqui, K.; Al-Qasim, S.; Al-Naqeb, D. Meta-analysis of diabetic nephropathy associated genetic variants in inflammation and angiogenesis involved in different biochemical pathways. BMC Med. Genet. 2014, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Nechemia-Arbely, Y.; Barkan, D.; Pizov, G.; Shriki, A.; Rose-John, S.; Galun, E.; Axelrod, J.H. IL-6/IL-6R axis plays a critical role in acute kidney injury. J. Am. Soc. Nephrol. 2008, 19, 1106–1115. [Google Scholar] [CrossRef] [PubMed]
- Simmons, E.M.; Himmelfarb, J.; Sezer, M.T.; Chertow, G.M.; Mehta, R.L.; Paganini, E.P.; Soroko, S.; Freedman, S.; Becker, K.; Spratt, D.; et al. Plasma cytokine levels predict mortality in patients with acute renal failure. Kidney Int. 2004, 65, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, A.; Pan, W.; Yue, C.; Zeng, L.; Gu, W.; Jiang, J. Association between IL-6-174G/C polymorphism and the risk of sepsis and mortality: A systematic review and meta-analysis. PLoS ONE 2015, 10, e0118843. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Tang, Y.; Zhou, H.; Xie, K. A meta-analysis on correlation between interleukin-6 -174G/C polymorphism and end-stage renal disease. Ren. Fail. 2017, 39, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Panayides, A.; Ioakeimidou, A.; Karamouzos, V.; Antonakos, N.; Koutelidakis, I.; Giannikopoulos, G.; Makaritsis, K.; Voloudakis, N.; Toutouzas, K.; Rovina, N.; et al. -572 G/C single nucleotide polymorphism of interleukin-6 and sepsis predisposition in chronic renal disease. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 2439–2446. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Agarwal, S.; Naik, S. Genetic contribution and associated pathophysiology in end-stage renal disease. Appl. Clin. Genet. 2010, 3, 65–84. [Google Scholar] [CrossRef] [PubMed]
- Wuttke, M.; Köttgen, A. Insights into kidney diseases from genome-wide association studies. Nat. Rev. Nephrol. 2016, 12, 549–562. [Google Scholar] [CrossRef] [PubMed]
- O’Seaghdha, C.M.; Fox, C.S. Genome-wide association studies of chronic kidney disease: What have we learned? Nat. Rev. Nephrol. 2011, 8, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lipska, B.K.; Halim, N.; Ma, Q.D.; Matsumoto, M.; Melhem, S.; Kolachana, B.S.; Hyde, T.M.; Herman, M.M.; Apud, J.; et al. Functional Analysis of Genetic Variation in Catechol-O-Methyltransferase (COMT): Effects on mRNA, Protein, and Enzyme Activity in Postmortem Human Brain. Am. J. Hum. Genet. 2004, 75, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, J. O-Methylation of Epinephrine and Other Catechols in vitro and in vivo. Science 1957, 126, 400–401. [Google Scholar] [CrossRef] [PubMed]
- Dellamea, B.; Leitão, C.; Friedman, R.; Canani, L. Nitric oxide system and diabetic nephropathy. Diabetol. Metab. Syndr. 2014, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Bai, Y.; Xu, P.; Hu, Z.; Liu, L.; Wang, F.; Jin, G.; Wang, F.; Deng, Q.; Tu, Y.; et al. Functional promoter -1271G/C variant of HSPB1 predicts lung cancer risk and survival. J. Clin. Oncol. 2010, 28, 1928–1935. [Google Scholar] [CrossRef] [PubMed]
- Pang, Q.; Wei, Q.; Xu, T.; Yuan, X.; Lopez Guerra, J.L.; Levy, L.B.; Liu, Z.; Gomez, D.R.; Zhuang, Y.; Wang, L.-E.; et al. Functional Promoter Variant rs2868371 of HSPB1 Is Associated with Risk of Radiation Pneumonitis After Chemoradiation for Non-Small Cell Lung Cancer. Int. J. Radiat. Oncol. 2013, 85, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Liu, H.; Yue, J.; Liu, L.; Han, M.; Luo, L.; Zhao, Y.; Xiao, H. Association of Single-Nucleotide Polymorphism in the Hepcidin Promoter Gene with Susceptibility to Extrapulmonary Tuberculosis. Genet. Test. Mol. Biomark. 2017, 21, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Parajes, S.; González-Quintela, A.; Campos, J.; Quinteiro, C.; Domínguez, F.; Loidi, L. Genetic study of the hepcidin gene (HAMP) promoter and functional analysis of the c.-582A > G variant. BMC Genet. 2010, 11, 110. [Google Scholar] [CrossRef] [PubMed]
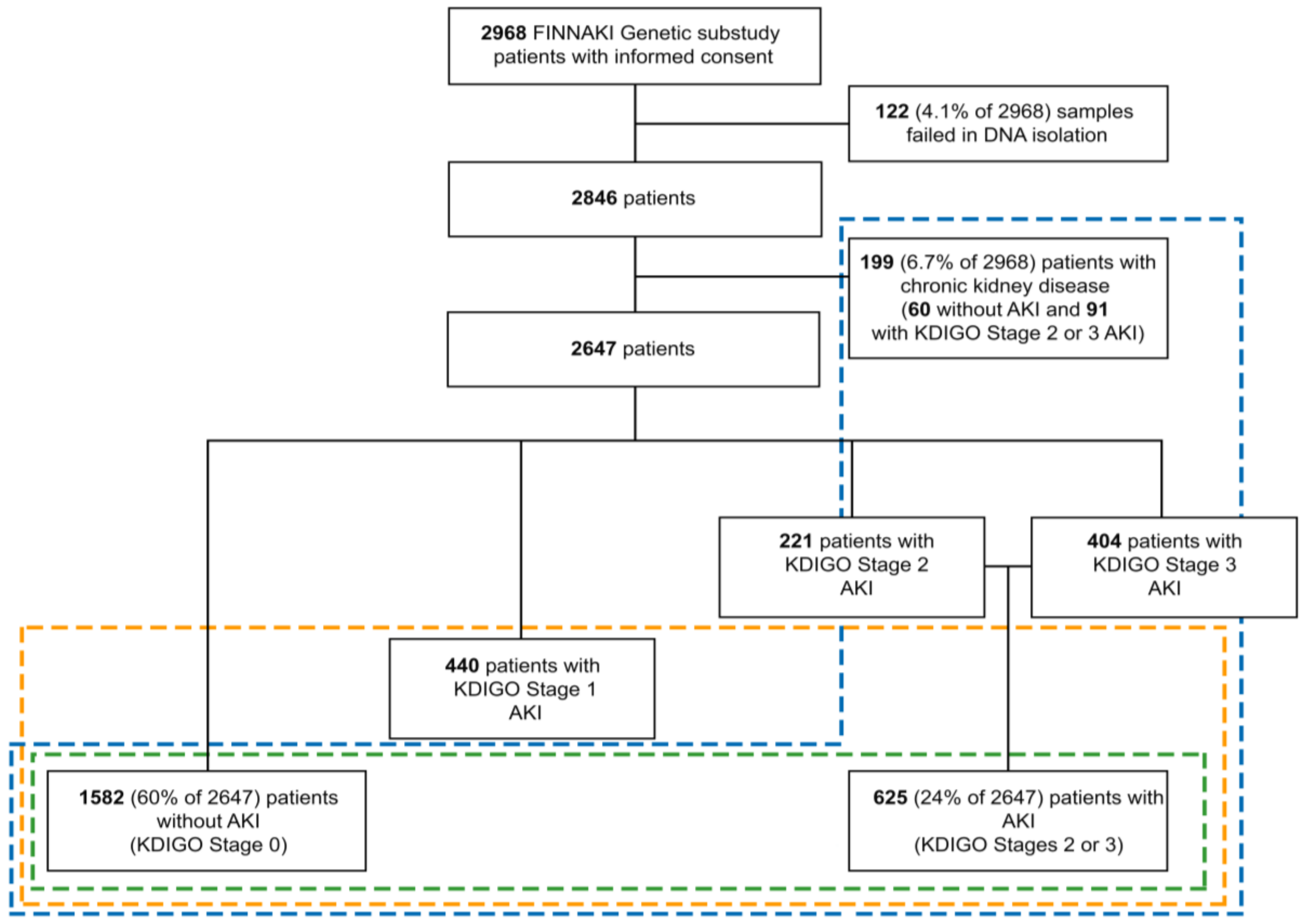
| Characteristics | Data Available | AKI | No AKI | p * | |
|---|---|---|---|---|---|
| KDIGO stage 2 or 3 | KDIGO stage 1, 2, 3 | ||||
| Age (years) | 2647 | 65 (54–74) | 65 (54–75) | 62 (48–72) | <0.001 |
| Gender (male) | 2647 | 409 (65.4) | 700 (65.7) | 980 (61.9) | 0.130 |
| BMI (kg/m2) | 2627 | 27.5 (24.5–31.3) | 27.5 (24.2–31.2) | 25.7 (23.1–28.7) | <0.001 |
| Co-morbidities | |||||
| Arterial hypertension | 2633 | 333 (53.3) | 561 (52.9) | 641 (40.8) | <0.001 |
| Diabetes | 2643 | 169 (27.0) | 263 (24.7) | 280 (17.7) | <0.001 |
| Arteriosclerosis | 2623 | 94 (15.1) | 159 (15.0) | 160 (10.2) | 0.002 |
| Chronic obstructive pulmonary disease | 2630 | 43 (6.9) | 81 (7.7) | 136 (8.6) | 0.195 |
| Chronic liver disease | 2617 | 46 (7.4) | 59 (5.6) | 51 (3.3) | <0.001 |
| Systolic heart failure | 2628 | 79 (12.7) | 129 (12.2) | 139 (8.8) | 0.009 |
| Baseline plasma creatinine (µmol/L) | 2643 | 81.0 (68.9–94.0) | 81.0 (69.0–94.0) | 79.0 (68.0–94.0) | 0.210 |
| Pre ICU daily medication | |||||
| ACE inhibitor or ARB | 2585 | 263 (42.8) | 428 (41.1) | 475 (30.8) | <0.001 |
| NSAID | 2538 | 73 (12.1) | 112 (10.9) | 118 (7.8) | 0.002 |
| Diuretic | 2596 | 185 (29.8) | 324 (30.8) | 323 (20.9) | <0.001 |
| Metformin | 2606 | 109 (17.6) | 163 (15.5) | 164 (10.6) | <0.001 |
| Statin | 2603 | 196 (31.6) | 320 (30.5) | 397 (25.6) | 0.005 |
| Corticosteroids | 2614 | 56 (9.0) | 94 (8.9) | 105 (6.7) | 0.070 |
| Warfarin | 2608 | 107 (17.2) | 166 (15.8) | 179 (11.5) | 0.001 |
| Treatments administered 48 h before admission | |||||
| Contrast medium | 2632 | 120 (19.3) | 223 (21.1) | 417 (26.5) | <0.001 |
| ACE inhibitor or ARB | 2601 | 167 (27.3) | 287 (27.5) | 329 (21.1) | 0.002 |
| Diuretics | 2570 | 217 (35.8) | 353 (34.3) | 360 (23.4) | <0.001 |
| Colloids (gelatin or starch) | 2479 | 229 (38.3) | 395 (39.0) | 394 (26.9) | <0.001 |
| Albumin | 2584 | 14 (2.3) | 18 (1.7) | 14 (0.9) | 0.018 |
| Type of admission | |||||
| Operative | 2646 | 180 (28.8) | 343 (32.2) | 557 (35.2) | 0.004 |
| Cardiac surgery | 2647 | 35 (5.6) | 80 (7.5) | 147 (9.3) | 0.004 |
| Emergency | 2621 | 575 (92.6) | 962 (91.1) | 1386 (88.6) | 0.005 |
| SAPS II score 24 h without renal or age components | 2614 | 24.0 (16.0–34.0) | 24.0 (16.0–32.0) | 20.0 (13.0–29.3) | <0.001 |
| Mechanical ventilation | 2647 | 432 (69.1) | 776 (72.9) | 1031 (65.2) | 0.080 |
| Sepsis | 2647 | 309 (49.4) | 500 (46.9) | 362 (22.9) | <0.001 |
| White blood cell count at admission, max (109/L) | 2186 | 12.0 (8.3–17.4) | 11.7 (8.2–16.8) | 10.9 (7.8–15.3) | <0.001 |
| Platelet count at admission, min (109/L) | 2419 | 190.0 (116.5–263.5) | 194.0 (127.0–265.0) | 205.0 (153.0–268.0) | <0.001 |
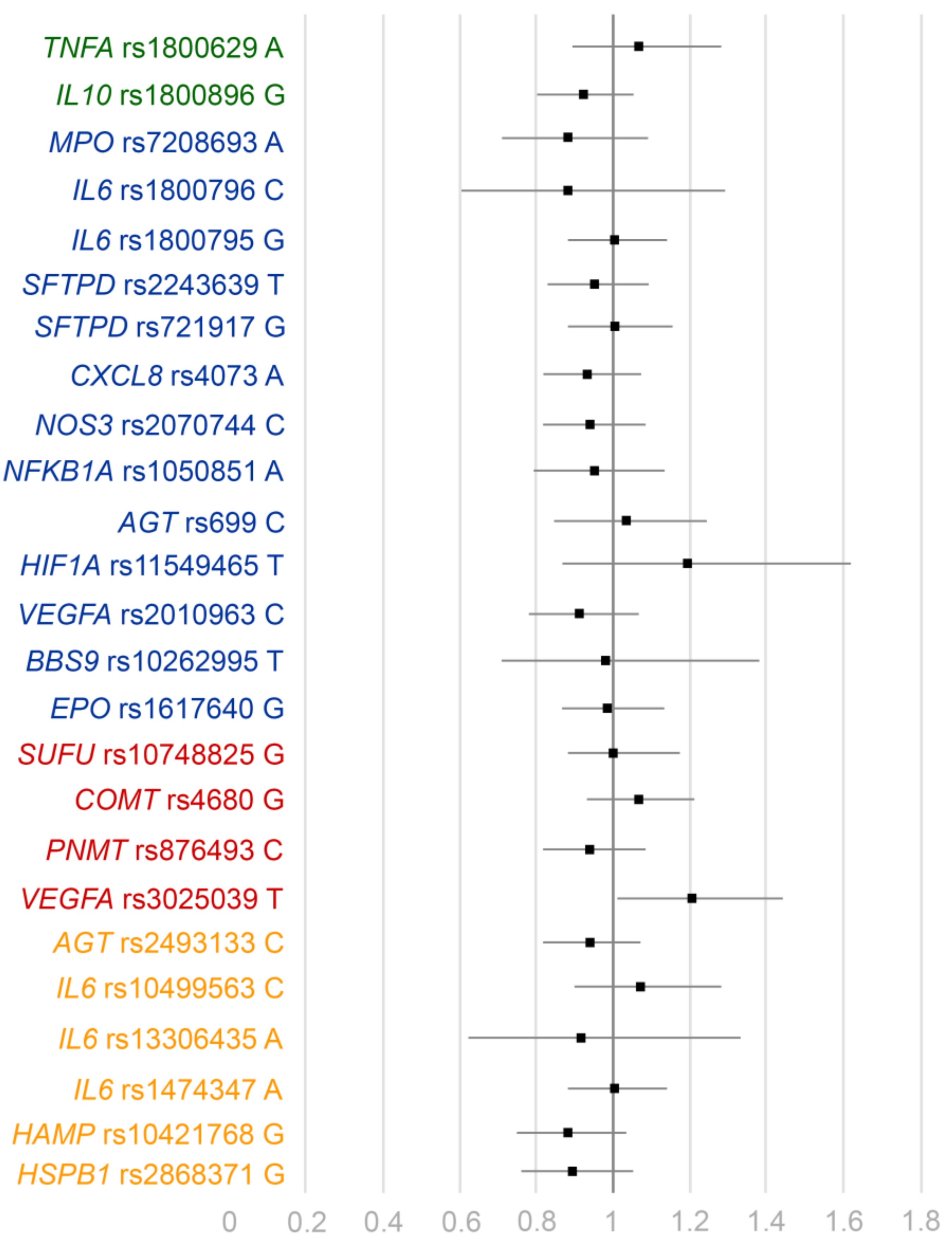
| Gene | SNP | Patients | Minor Allele | MAF (Cases/Controls) | Additive Logistic OR | 95% CI | p |
|---|---|---|---|---|---|---|---|
| TNFA | rs1800629 | 2174 | A | 0.15/0.14 | 1.06 | 0.89–1.28 | 0.51 |
| IL10 | rs1800896 | 2173 | G | 0.44/0.46 | 0.92 | 0.80–1.05 | 0.20 |
| IL6 | rs10499563 | 2192 | C | 0.15/0.14 | 1.07 | 0.90–1.28 | 0.45 |
| rs1800796 | 2197 | C | 0.03/0.03 | 0.88 | 0.60–1.29 | 0.51 | |
| rs1800795 | 2189 | G | 0.47/0.47 | 1.00 | 0.88–1.14 | 0.97 | |
| rs1474347 | 2187 | A | 0.47/0.47 | 1.00 | 0.88–1.14 | 1.00 | |
| rs13306435 | 2199 | A | 0.03/0.03 | 0.91 | 0.62–1.33 | 0.62 | |
| CXCL8 | rs4073 | 2193 | A | 0.42/0.42 | 0.93 | 0.82–1.07 | 0.31 |
| NOS3 | rs2070744 | 2174 | C | 0.34/0.36 | 0.94 | 0.82–1.08 | 0.37 |
| NFKB1A | rs1050851 | 2196 | A | 0.16/0.17 | 0.95 | 0.79–1.13 | 0.54 |
| AGT | rs699 | 1047 | C | 0.43/0.42 | 1.03 | 0.85–1.24 | 0.78 |
| rs2493133 | 2196 | C | 0.41/0.42 | 0.94 | 0.82–1.07 | 0.36 | |
| VEGFA | rs2010963 | 2170 | C | 0.22/0.24 | 0.91 | 0.78–1.06 | 0.22 |
| rs3025039 | 2195 | T | 0.16/0.14 | 1.20 | 1.01–1.44 | 0.044 | |
| EPO | rs1617640 | 2173 | G | 0.44/0.45 | 0.99 | 0.87–1.13 | 0.91 |
| SUFU | rs10748825 | 2174 | G | 0.37/0.37 | 1.02 | 0.88–1.17 | 0.83 |
| HIF1A | rs11549465 | 2173 | T | 0.05/0.04 | 1.19 | 0.87–1.62 | 0.28 |
| PNMT | rs876493 | 2173 | C | 0.36/0.38 | 0.94 | 0.82–1.08 | 0.37 |
| MPO | rs7208693 | 2174 | A | 0.10/0.12 | 0.88 | 0.71–1.09 | 0.23 |
| COMT | rs4680 | 2173 | G | 0.47/0.45 | 1.06 | 0.93–1.21 | 0.39 |
| HSPB1 | rs2868371 | 2194 | G | 0.19/0.21 | 0.89 | 0.76–1.05 | 0.18 |
| SFTPD | rs2243639 | 2199 | T | 0.39/0.40 | 0.95 | 0.83–1.09 | 0.47 |
| rs721917 | 2193 | G | 0.39/0.39 | 1.00 | 0.88–1.15 | 0.97 | |
| HAMP | rs10421768 | 2199 | G | 0.23/0.25 | 0.88 | 0.75–1.03 | 0.11 |
| BBS9 | rs10262995 | 2200 | T | 0.04/0.04 | 0.98 | 0.71–1.38 | 0.93 |
| Genotype Combination | AKI (n = 615) | No AKI (n = 1558) |
|---|---|---|
| TNFA GG + IL10 AA | 138 (22%) | 340 (22%) |
| TNFA GG + IL10 GA + GG | 311 (51%) | 814 (52%) |
| TNFA GA + AA + IL10 AA | 52 (8%) | 116 (8%) |
| TNFA GA + AA + IL10 GA + GG | 114 (19%) | 288 (18%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vilander, L.M.; Vaara, S.T.; Kaunisto, M.A.; Pettilä, V.; Study Group, T.F. Common Inflammation-Related Candidate Gene Variants and Acute Kidney Injury in 2647 Critically Ill Finnish Patients. J. Clin. Med. 2019, 8, 342. https://doi.org/10.3390/jcm8030342
Vilander LM, Vaara ST, Kaunisto MA, Pettilä V, Study Group TF. Common Inflammation-Related Candidate Gene Variants and Acute Kidney Injury in 2647 Critically Ill Finnish Patients. Journal of Clinical Medicine. 2019; 8(3):342. https://doi.org/10.3390/jcm8030342
Chicago/Turabian StyleVilander, Laura M., Suvi T. Vaara, Mari A. Kaunisto, Ville Pettilä, and The FINNAKI Study Group. 2019. "Common Inflammation-Related Candidate Gene Variants and Acute Kidney Injury in 2647 Critically Ill Finnish Patients" Journal of Clinical Medicine 8, no. 3: 342. https://doi.org/10.3390/jcm8030342
APA StyleVilander, L. M., Vaara, S. T., Kaunisto, M. A., Pettilä, V., & Study Group, T. F. (2019). Common Inflammation-Related Candidate Gene Variants and Acute Kidney Injury in 2647 Critically Ill Finnish Patients. Journal of Clinical Medicine, 8(3), 342. https://doi.org/10.3390/jcm8030342





