Fecal Microbiota Analysis in Patients Going through a Depressive Episode during Treatment in a Psychiatric Hospital Setting
Abstract
1. Introduction
2. Material and Methods
2.1. Patients
- Aged between 18–65 years;
- Depressive episode of at least moderate severity according to ICD-10;
- Written informed consent of participation.
- Exclusion criteria were as follows:
- Total or partial incapacitation;
- Comorbidity with severe depressive episode and psychotic symptoms (F32.3), schizophrenia, schizotypal or delusional disorders (F20–F29), bipolar disorder (F31), mental retardation (F70–F79), active addiction to alcohol or other psychoactive substances (F10–F19) excluding nicotine (F17) and caffeine (F15), organic—including symptomatic—mental disorders (F00–F09), epilepsy (G40);
- Inflammatory and infectious diseases;
- Poor general health, including cancer, gastrointestinal diseases (tumors, inflammations, celiac disease, food intolerance, colitis ulcerosa, and Crohn’s Disease);
- Diabetes and thyroid dysregulation;
- Treatment with antibiotics, non-steroidal anti-inflammatory drugs, probiotics, corticosteroids, immunosuppressive and immunomodulating drugs, or proton pump inhibitors for at least 3 months prior to baseline assessment;
- A history of treatment-resistant depression.
2.2. Processing of Raw Data and Statistical Analysis
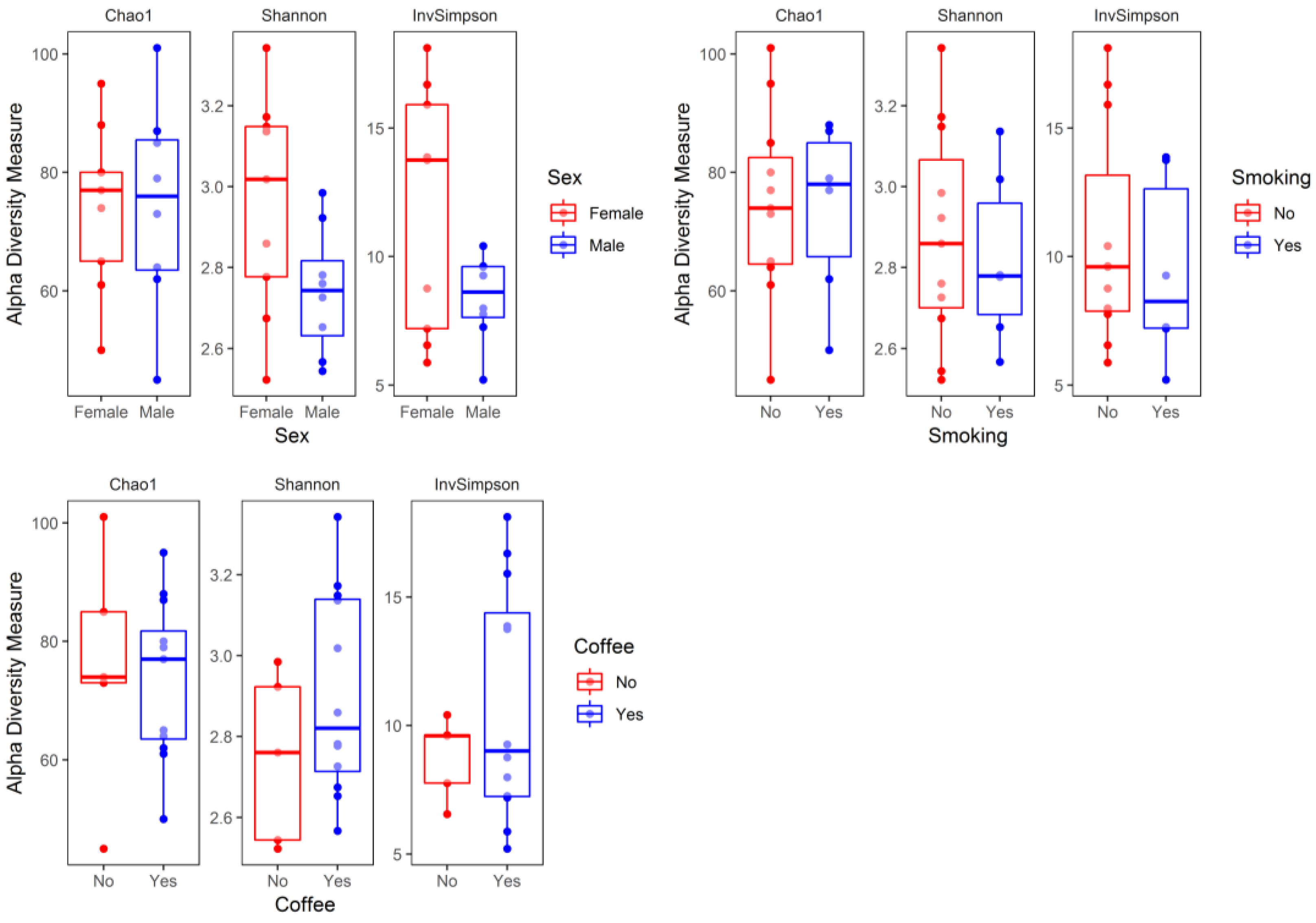
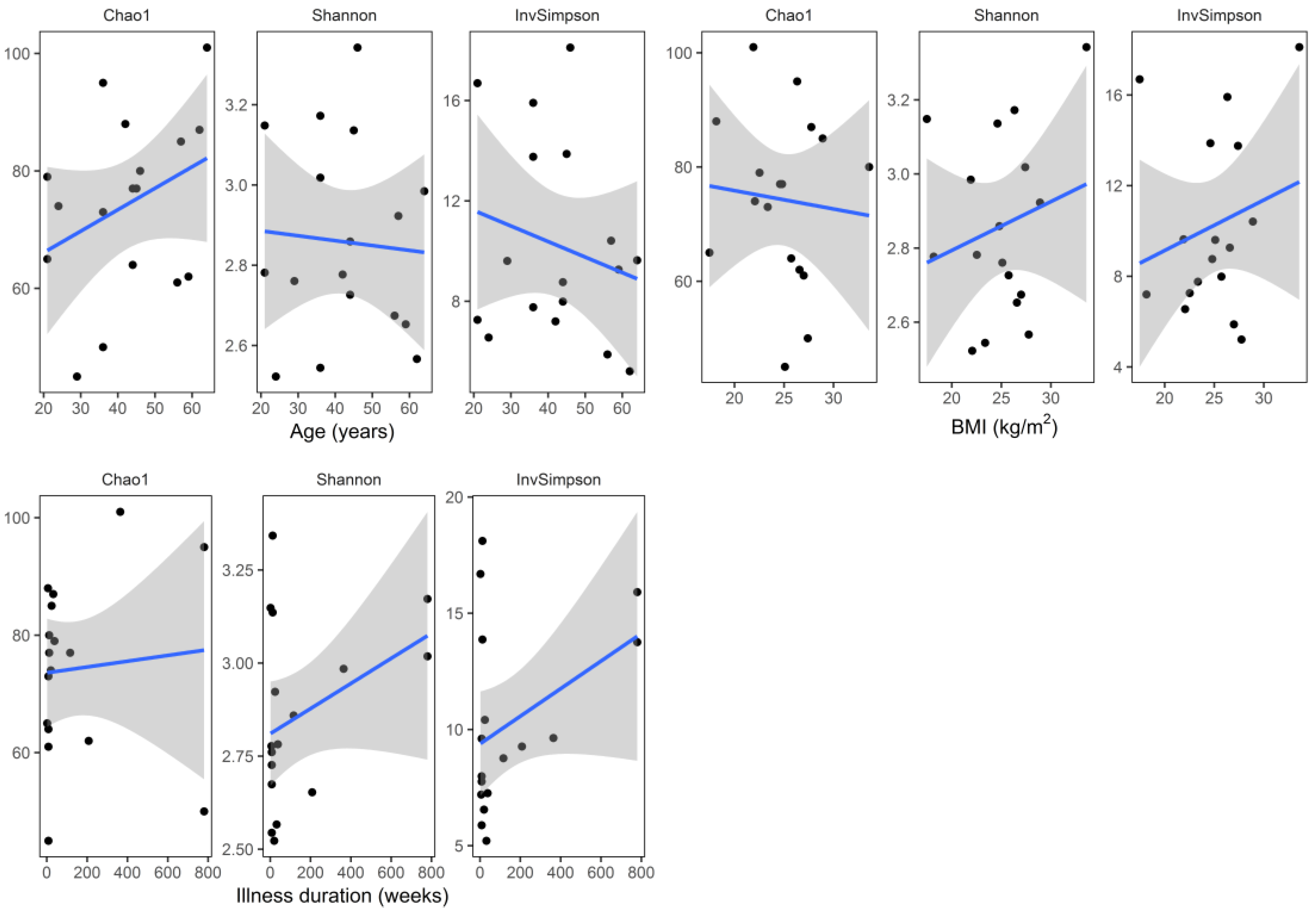
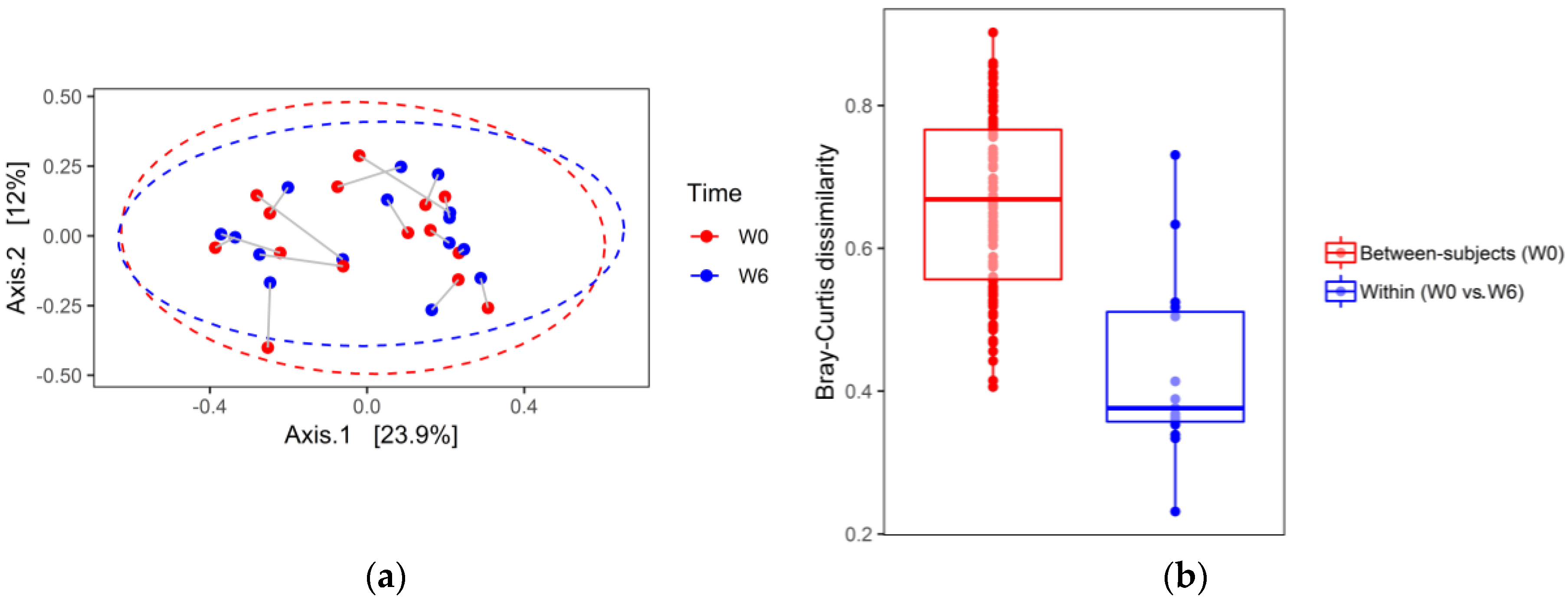
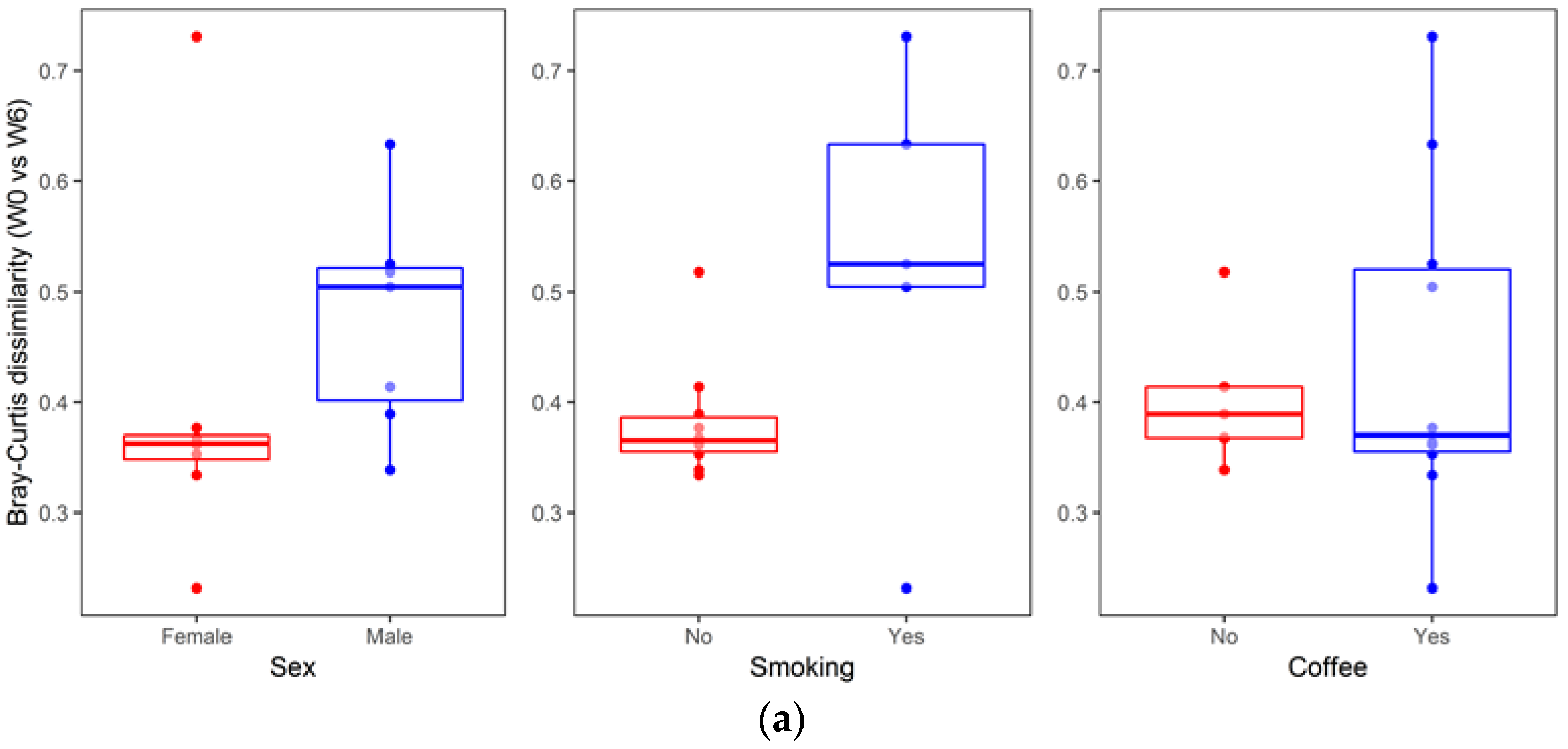
3. Results
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Depression and Other Common Mental Disorders. Available online: http://www.who.int/mental_health/management/depression/prevalence_global_health_estimates/en/ (accessed on 1 January 2019).
- Kiejna, A.; Adamowski, T.; Piotrowski, P.; Moskalewicz, J.; Wojtyniak, B.; Świątkiewicz, G.; Stokwiszewski, J.; Kantorska-Janiec, M.; Zagdańska, M.; Kessler, R. “Epidemiology of mental disorders and access to mental health care. EZOP–Poland”—Research methodology. Psychiatr. Pol. 2015, 49, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Hollon, S.D.; Shelton, R.C.; Wisniewski, S.; Warden, D.; Biggs, M.M.; Friedman, E.S.; Husain, M.; Kupfer, D.J.; Nierenberg, A.A.; Petersen, T.J.; et al. Presenting characteristics of depressed outpatients as a function of recurrence: Preliminary findings from the STAR*D clinical trial. J. Psychiatr. Res. 2006, 40, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Delgado, P.L. Depression: The Case for a Monoamine Deficiency. J. Clin. Psychiatry 2000, 61, 7–11. [Google Scholar] [PubMed]
- Castrén, E.; Rantamäki, T. The role of BDNF and its receptors in depression and antidepressant drug action: Reactivation of developmental plasticity. Dev. Neurobiol. 2010, 70, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Wray, N.R.; Ripke, S.; Mattheisen, M.; Trzaskowski, M.; Byrne, E.M.; Abdellaoui, A.; Adams, M.J.; Agerbo, E.; Air, T.M.; Andlauer, T.M.F.; et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018, 50, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Yamagata, H.; Seki, T.; Watanabe, Y. Epigenetic mechanisms of major depression: Targeting neuronal plasticity. Psychiatry Clin. Neurosci. 2018, 72, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Leonard, B.E. Inflammation and depression: A causal or coincidental link to the pathophysiology? Acta Neuropsychiatr. 2018, 30, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, E.; Wilson, A.A.; Mizrahi, R.; Rusjan, P.M.; Miler, L.; Rajkowska, G.; Suridjan, I.; Kennedy, J.L.; Rekkas, P.V.; Houle, S.; et al. Role of translocator protein density, a marker of neuroinflammation, in the brain during major depressive episodes. JAMA Psychiatry 2015, 72, 268–275. [Google Scholar] [CrossRef]
- Miller, A.H.; Raison, C.L. The role of inflammation in depression: From evolutionary imperative to modern treatment target. Nat. Rev. Immunol. 2016, 16, 22–34. [Google Scholar] [CrossRef]
- Hooks, K.B.; Konsman, J.P.; O’Malley, M.A. Microbiota-gut-brain research: A critical analysis. Behav. Brain Sci. 2018, 1–40. [Google Scholar] [CrossRef]
- Zalar, B.; Haslberger, A.; Peterlin, B. The Role of Microbiota in Depression—A brief review. Psychiatr. Danub. 2018, 30, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Huttenhower, C. Chapter 12: Human Microbiome Analysis. PLoS Comput. Biol. 2012, 8, e1002808. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Aizawa, E.; Tsuji, H.; Asahara, T.; Takahashi, T.; Teraishi, T.; Yoshida, S.; Ota, M.; Koga, N.; Hattori, K.; Kunugi, H. Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J. Affect. Disord. 2016, 202, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Kleiman, S.C.; Watson, H.J.; Bulik-Sullivan, E.C.; Huh, E.Y.; Tarantino, L.M.; Bulik, C.M.; Carroll, I.M. The Intestinal Microbiota in Acute Anorexia Nervosa and During Renourishment: Relationship to Depression, Anxiety, and Eating Disorder Psychopathology. Psychosom. Med. 2015, 77, 969–981. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.R.; Borre, Y.; O’ Brien, C.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G.; et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 2016, 82, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Zeng, B.; Zhou, C.; Liu, M.; Fang, Z.; Xu, X.; Zeng, L.; Chen, J.; Fan, S.; Du, X.; et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 2016, 21, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Painold, A.; Mörkl, S.; Kashofer, K.; Halwachs, B.; Dalkner, N.; Bengesser, S.; Birner, A.; Fellendorf, F.; Platzer, M.; Queissner, R.; et al. A step ahead: Exploring the gut microbiota in inpatients with bipolar disorder during a depressive episode. Bipolar Disord. 2018. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, J.; Gui, S.; Zhou, C.; Chen, J.; Yang, C.; Hu, Z.; Wang, H.; Zhong, X.; Zeng, L.; et al. Comparative metaproteomics analysis shows altered fecal microbiota signatures in patients with major depressive disorder. Neuroreport 2018, 29, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.; Hart, R.A.; Charlesworth, R.P.G.; Sharpley, C.F. Gut microbiome and depression: What we know and what we need to know. Rev. Neurosci. 2018, 29, 629–643. [Google Scholar] [CrossRef] [PubMed]
- López-Muñoz, F.; Alamo, C. Monoaminergic neurotransmission: The history of the discovery of antidepressants from 1950s until today. Curr. Pharm. Des. 2009, 15, 1563–1586. [Google Scholar] [CrossRef]
- Lieb, J. The immunostimulating and antimicrobial properties of lithium and antidepressants. J. Infect. 2004, 49, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Bellido, J.L.; Munoz-Criado, S.; Garcìa-Rodrìguez, J.A. Antimicrobial activity of psychotropic drugs: Selective serotonin reuptake inhibitors. Int. J. Antimicrob. Agents 2000, 14, 177–180. [Google Scholar] [CrossRef]
- Csiszar, K.; Molnar, J. Mechanism of action of tricyclic drugs on Escherichia coli and Yersinia enterocolitica plasmid maintenance and replication. Anticancer Res. 1992, 12, 2267–2272. [Google Scholar] [PubMed]
- Weinbach, E.C.; Levenbook, L.; Alling, D.W. Binding of tricyclic antidepressant drugs to trophozoites of giardia lamblia. Comp. Biochem. Physiol. C: Comp. Pharmacol. Toxicol. 1992, 102, 391–396. [Google Scholar] [CrossRef]
- Molnár, J. Antiplasmid activity of tricyclic compounds. Methods Find Exp. Clin. Pharmacol. 1988, 10, 467–474. [Google Scholar] [PubMed]
- Ayaz, M.; Subhan, F.; Ahmed, J.; Khan, A.-U.; Ullah, F.; Ullah, I.; Ali, G.; Syed, N.-I.-H.; Hussain, S. Sertraline enhances the activity of antimicrobial agents against pathogens of clinical relevance. J. Biol. Res. 2015, 22, 4. [Google Scholar] [CrossRef] [PubMed]
- Coban, A.Y.; Tanriverdi Cayci, Y.; Keleş Uludağ, S.; Durupinar, B. Investigation of antibacterial activity of sertralin. Mikrobiyol. Bul. 2009, 43, 651–656. [Google Scholar]
- Kruszewska, H.; Zareba, T.; Tyski, S. Examination of antimicrobial activity of selected non-antibiotic medicinal preparations. Acta Pol. Pharm. 2012, 69, 1368–1371. [Google Scholar] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Mello, B.S.F.; Monte, A.S.; McIntyre, R.S.; Soczynska, J.K.; Custódio, C.S.; Cordeiro, R.C.; Chaves, J.H.; Vasconcelos, S.M.M.; Nobre, H.V.; Florenço de Sousa, F.C.; et al. Effects of doxycycline on depressive-like behavior in mice after lipopolysaccharide (LPS) administration. J. Psychiatr. Res. 2013, 47, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Miyaoka, T.; Wake, R.; Furuya, M.; Liaury, K.; Ieda, M.; Kawakami, K.; Tsuchie, K.; Taki, M.; Ishihara, K.; Araki, T.; et al. Minocycline as adjunctive therapy for patients with unipolar psychotic depression: An open-label study. Prog. Neuropsychopharmacol. Biol. Psychiatry 2012, 37, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Akilandeswari, K.; Ruckmani, K.; Ranjith, M. Efficacy of antibacterial activity of antibiotics ciprofloxacin and gentamycin improved with anti-depressant drug, escitalopram. Int. J. Pharm. Sci. Rev. Res. 2013, 21, 71–74. [Google Scholar]
- Cussotto, S.; Strain, C.R.; Fouhy, F.; Strain, R.G.; Peterson, V.L.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Differential effects of psychotropic drugs on microbiome composition and gastrointestinal function. Psychopharmacology 2018, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Jarosz, M.; Rychlik, E.; Stoś, K.; Wierzejska, R.; Wojtasik, A.; Charzewska, J.; Mojska, H.; Szponar, L.; Sajór, I.; Kłosiewicz-Latoszek, L.; et al. Normy żywienia dla populacji Polski; Instytut Żywności i Żywienia: Warszawa, Poland, 2017; ISBN 978-83-86060-89-4. [Google Scholar]
- Almonacid, D.E.; Kraal, L.; Ossandon, F.J.; Budovskaya, Y.V.; Cardenas, J.P.; Bik, E.M.; Goddard, A.D.; Richman, J.; Apte, Z.S. 16S rRNA gene sequencing and healthy reference ranges for 28 clinically relevant microbial taxa from the human gut microbiome. PLoS ONE 2017, 12, e0176555. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Morris, E.K.; Caruso, T.; Buscot, F.; Fischer, M.; Hancock, C.; Maier, T.S.; Meiners, T.; Müller, C.; Obermaier, E.; Prati, D.; et al. Choosing and using diversity indices: Insights for ecological applications from the German Biodiversity Exploratories. Ecol. Evol. 2014, 4, 3514–3524. [Google Scholar] [CrossRef]
- Kim, B.-R.; Shin, J.; Guevarra, R.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R. Deciphering diversity indices for better understanding of the microbial communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Naseribafrouei, A.; Hestad, K.; Avershina, E.; Sekelja, M.; Linløkken, A.; Wilson, R.; Rudi, K. Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil. 2014, 26, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Shi, X.; Li, Z.; Shen, Y.; Shi, X.; Wang, L.; Li, G.; Yuan, Y.; Wang, J.; Zhang, Y.; et al. Possible association of Firmicutes in the gut microbiota of patients with major depressive disorder. Neuropsychiatr. Dis. Treat. 2018, 14, 3329–3337. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed]
- Shade, A. Diversity is the question, not the answer. ISME J. 2017, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Skonieczna-Żydecka, K.; Grochans, E.; Maciejewska, D.; Szkup, M.; Schneider-Matyka, D.; Jurczak, A.; Łoniewski, I.; Kaczmarczyk, M.; Marlicz, W.; Czerwińska-Rogowska, M.; et al. Faecal Short Chain Fatty Acids Profile is Changed in Polish Depressive Women. Nutrients 2018, 10, 1939. [Google Scholar] [CrossRef] [PubMed]
- Bishehsari, F.; Engen, P.A.; Preite, N.Z.; Tuncil, Y.E.; Naqib, A.; Shaikh, M.; Rossi, M.; Wilber, S.; Green, S.J.; Hamaker, B.R.; et al. Dietary Fiber Treatment Corrects the Composition of Gut Microbiota, Promotes SCFA Production, and Suppresses Colon Carcinogenesis. Genes 2018, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 3654. [Google Scholar] [CrossRef]
- So, D.; Whelan, K.; Rossi, M.; Morrison, M.; Holtmann, G.; Kelly, J.T.; Shanahan, E.R.; Staudacher, H.M.; Campbell, K.L. Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2018, 107, 965–983. [Google Scholar] [CrossRef]
- Rao, T.S.S.; Asha, M.R.; Ramesh, B.N.; Rao, K.S.J. Understanding nutrition, depression and mental illnesses. Indian J. Psychiatry 2008, 50, 77–82. [Google Scholar]



| Variable | Recommended Daily Intake * | Result | |
|---|---|---|---|
| Mean | Range | ||
| Energy, kcal | 2995 | 2865–3165 | |
| Protein, g | 106 | 89–122 | |
| Fat, g | 102 | 91–116 | |
| Carbohydrates, g | 420 | 387–442 | |
| Fiber, g | >25 | 31 | 28–33 |
| % Energy from protein | 10–20 | 14 | 12–15 |
| % Energy from fat | 20–35 | 31 | 29–33 |
| % Energy from carbohydrates | 45–65 | 56 | 54–56 |
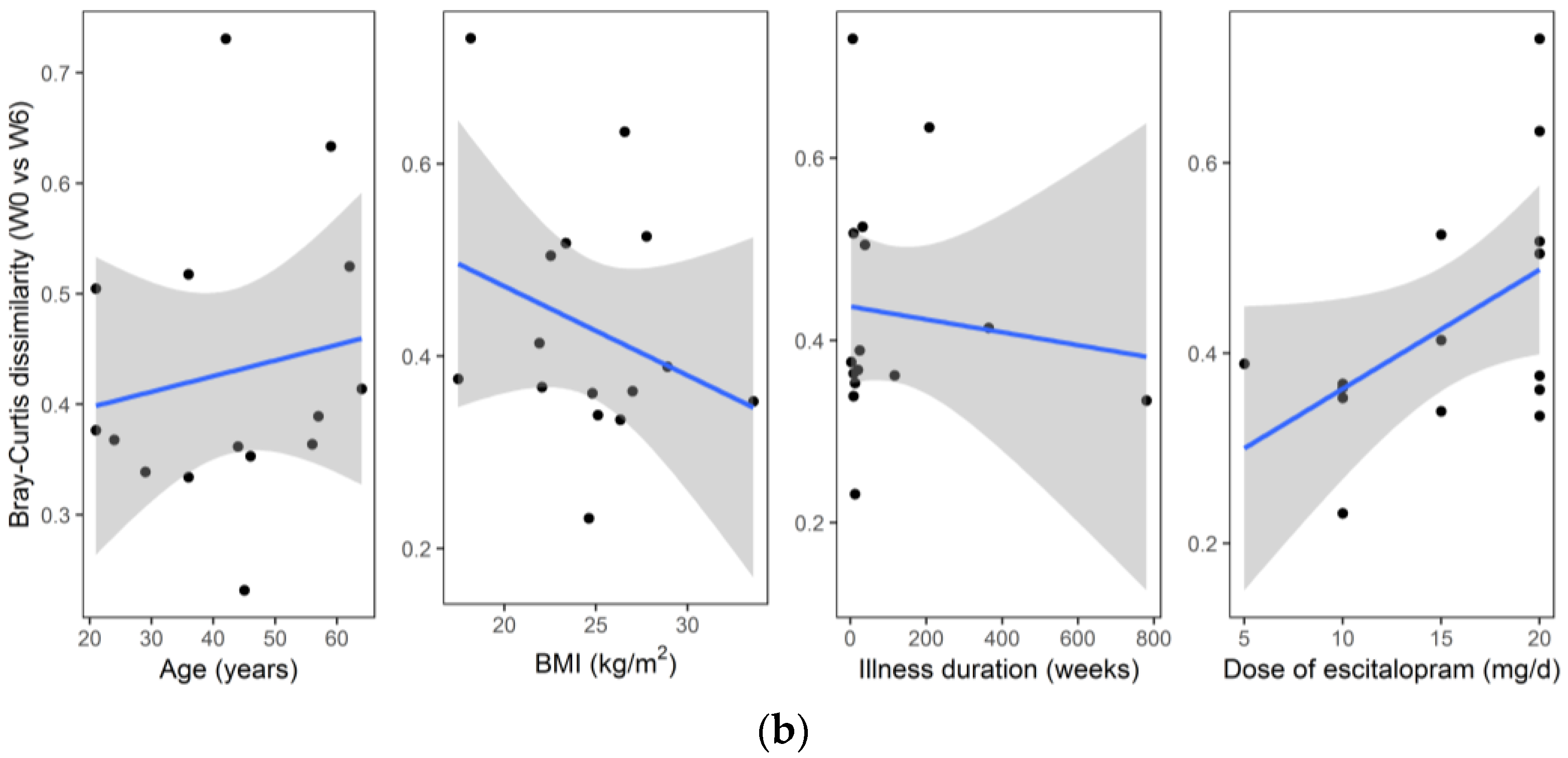
| Characteristic | n = 17 |
|---|---|
| Age, years | 42.5 ± 13.9 (21.0–64.0) |
| Sex (M/F), n (%) | 8/9 (47/53) |
| BMI, kg/m2 | 24.9 ± 3.9 (17.5–33.6) |
| Smoking (Y/N), n (%) | 6/11 (35/65) |
| Coffee (Y/N), n (%) | 12/5 (71/29) |
| Age of diagnosis of depression, years | 40.1 ± 14.6 (21.0–62.0) |
| Illness duration, weeks | 143 ± 258 (2–780) |
| Age of onset of psychiatric treatment, years | 38.6 ± 14.4 (19.0–62.0) |
| Current episode duration, weeks | 11.5 ± 7.9 (2.0–32.0) |
| Phylum | W0 (%) | W6 (%) | Raw p † | p1 | p2 |
|---|---|---|---|---|---|
| Firmicutes | 46.9 | 42.7 | 0.526 | 0.792 | 0.658 |
| Bacteroidetes | 40.0 | 47.4 | 0.239 | 0.667 | 0.650 |
| Actinobacteria | 5.5 | 4.6 | 0.661 | 0.792 | 0.661 |
| Proteobacteria | 3.5 | 4.2 | 0.390 | 0.792 | 0.650 |
| Verrucomicrobia | 4.1 | 1.0 | 0.348 | 0.792 | 0.650 |
| Class | W0 (%) | W6 (%) | Raw p † | p1 | p2 |
|---|---|---|---|---|---|
| Bacteroidia | 40.0 | 47.4 | 0.239 | 0.877 | 0.902 |
| Clostridia | 43.0 | 39.7 | 0.620 | 0.996 | 0.902 |
| Actinobacteria | 5.5 | 4.6 | 0.661 | 0.996 | 0.902 |
| Verrucomicrobiae | 4.1 | 1.0 | 0.348 | 0.952 | 0.902 |
| Negativicutes | 2.4 | 2.3 | 0.902 | 0.998 | 0.902 |
| Betaproteobacteria | 1.6 | 2.1 | 0.393 | 0.963 | 0.902 |
| Gammaproteobacteria | 1.3 | 1.4 | 0.852 | 0.998 | 0.902 |
| Bacilli | 1.2 | 0.3 | 0.009 | 0.076 | 0.085 |
| Deltaproteobacteria | 0.7 | 0.8 | 0.610 | 0.996 | 0.902 |
| Erysipelotrychia | 0.3 | 0.4 | 0.893 | 0.998 | 0.902 |
| Order | W0 (%) | W6 (%) | Raw p † | p1 | p2 |
|---|---|---|---|---|---|
| Bacteroidales | 40.0 | 47.4 | 0.239 | 0.954 | 0.998 |
| Clostridiales | 43.0 | 39.7 | 0.620 | 0.999 | 0.998 |
| Lactobacillales | 1.2 | 0.3 | 0.009 | 0.096 | 0.119 |
| Selenomonadales | 2.4 | 2.3 | 0.902 | 0.999 | 0.998 |
| Bifidobacteriales | 3.7 | 2.7 | 0.594 | 0.999 | 0.998 |
| Coriobacteriales | 1.8 | 1.9 | 0.700 | 0.999 | 0.998 |
| Actinomycetales | 0.02 | 0.02 | 0.385 | 0.992 | 0.998 |
| Enterobacteriales | 1.2 | 1.2 | 0.927 | 0.999 | 0.998 |
| Erysipelotrichales | 0.3 | 0.4 | 0.893 | 0.999 | 0.998 |
| Burkholderiales | 1.6 | 2.1 | 0.393 | 0.992 | 0.998 |
| Desulfovibrionales | 0.7 | 0.8 | 0.610 | 0.999 | 0.998 |
| Pasteurellales | 0.1 | 0.1 | 0.844 | 0.999 | 0.998 |
| Neisseriales | 0.0006 | 0.0008 | 1.0 | 1.0 | 1.0 |
| Verrucomicrobiales | 4.1 | 1.0 | 0.348 | 0.988 | 0.998 |
| Family | W0 (%) | W6 (%) | Raw p † | p1 | p2 |
|---|---|---|---|---|---|
| Bacteroidaceae | 29.9 | 33.4 | 0.484 | 1.0 | 0.987 |
| Lachnospiraceae | 26.9 | 24.6 | 0.617 | 1.0 | 0.987 |
| Streptococcaceae | 0.6 | 0.3 | 0.191 | 0.992 | 0.987 |
| Lactobacillaceae | 0.4 | 0.06 | 0.110 | 0.934 | 0.987 |
| Porphyromonadaceae | 6.7 | 8.7 | 0.160 | 0.986 | 0.987 |
| Bifidobacteriaceae | 3.7 | 2.7 | 0.594 | 1.0 | 0.987 |
| Veillonellaceae | 0.9 | 0.7 | 0.694 | 1.0 | 0.987 |
| Coriobacteriaceae | 1.7 | 1.9 | 0.700 | 1.0 | 0.987 |
| Rikenellaceae | 2.8 | 4.8 | 0.015 | 0.282 | 0.508 |
| Actinomycetaceae | 0.02 | 0.02 | 0.423 | 1.0 | 0.987 |
| Ruminococcaceae | 15.5 | 14.4 | 0.753 | 1.0 | 0.987 |
| Erysipelotrichaceae | 0.3 | 0.4 | 0.893 | 1.0 | 0.987 |
| Enterobacteriaceae | 1.2 | 1.2 | 0.927 | 1.0 | 0.987 |
| Prevotellaceae | 0.5 | 0.5 | 0.888 | 1.0 | 0.987 |
| Acidaminococcaceae | 1.5 | 1.6 | 0.907 | 1.0 | 0.987 |
| Sutterellaceae | 1.6 | 2.0 | 0.403 | 1.0 | 0.987 |
| Desulfovibrionaceae | 0.7 | 0.8 | 0.610 | 1.0 | 0.987 |
| Clostridiales Family XI. IncertaeSedis | 0.001 | 0.001 | 0.645 | 1.0 | 0.987 |
| Peptostreptococcaceae | 0.3 | 0.5 | 0.504 | 1.0 | 0.987 |
| Enterococcaceae | 0.2 | 0.01 | 0.252 | 0.999 | 0.987 |
| Pasteurellaceae | 0.1 | 0.1 | 0.844 | 1.0 | 0.987 |
| Eubacteriaceae | 0.007 | 0.002 | 0.083 | 0.869 | 0.987 |
| Clostridiaceae | 0.005 | 0.004 | 0.922 | 1.0 | 0.987 |
| Verrucomicrobiaceae | 4.1 | 1.0 | 0.348 | 1.0 | 0.987 |
| Oxalobacteraceae | 0.02 | 0.03 | 0.449 | 1.0 | 0.987 |
| Oscillospiraceae | 0.0003 | 0.002 | 0.499 | 1.0 | 0.987 |
| Neisseriaceae | 0.001 | 0.001 | 1.0 | 1.0 | 1.0 |
| Leuconostocaceae | 0.001 | 0.002 | 0.879 | 1.0 | 0.987 |
| Corynebacteriaceae | 0.001 | 0.001 | 0.506 | 1.0 | 0.987 |
| Christensenellaceae | 0.0006 | 0.002 | 0.503 | 1.0 | 0.987 |
| Catabacteriaceae | 0.0003 | 0.0002 | 1.0 | 1.0 | 1.0 |
| Carnobacteriaceae | 0.003 | 0.001 | 0.254 | 0.999 | 0.987 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liśkiewicz, P.; Pełka-Wysiecka, J.; Kaczmarczyk, M.; Łoniewski, I.; Wroński, M.; Bąba-Kubiś, A.; Skonieczna-Żydecka, K.; Marlicz, W.; Misiak, B.; Samochowiec, J. Fecal Microbiota Analysis in Patients Going through a Depressive Episode during Treatment in a Psychiatric Hospital Setting. J. Clin. Med. 2019, 8, 164. https://doi.org/10.3390/jcm8020164
Liśkiewicz P, Pełka-Wysiecka J, Kaczmarczyk M, Łoniewski I, Wroński M, Bąba-Kubiś A, Skonieczna-Żydecka K, Marlicz W, Misiak B, Samochowiec J. Fecal Microbiota Analysis in Patients Going through a Depressive Episode during Treatment in a Psychiatric Hospital Setting. Journal of Clinical Medicine. 2019; 8(2):164. https://doi.org/10.3390/jcm8020164
Chicago/Turabian StyleLiśkiewicz, Paweł, Justyna Pełka-Wysiecka, Mariusz Kaczmarczyk, Igor Łoniewski, Michał Wroński, Agata Bąba-Kubiś, Karolina Skonieczna-Żydecka, Wojciech Marlicz, Błażej Misiak, and Jerzy Samochowiec. 2019. "Fecal Microbiota Analysis in Patients Going through a Depressive Episode during Treatment in a Psychiatric Hospital Setting" Journal of Clinical Medicine 8, no. 2: 164. https://doi.org/10.3390/jcm8020164
APA StyleLiśkiewicz, P., Pełka-Wysiecka, J., Kaczmarczyk, M., Łoniewski, I., Wroński, M., Bąba-Kubiś, A., Skonieczna-Żydecka, K., Marlicz, W., Misiak, B., & Samochowiec, J. (2019). Fecal Microbiota Analysis in Patients Going through a Depressive Episode during Treatment in a Psychiatric Hospital Setting. Journal of Clinical Medicine, 8(2), 164. https://doi.org/10.3390/jcm8020164








