Inflammatory Cell Recruitment in Candida glabrata Biofilm Cell-Infected Mice Receiving Antifungal Chemotherapy
Abstract
1. Introduction
2. Experimental Section
2.1. Ethics Statement
2.2. Organisms and Growth Conditions
2.3. Antifungal Drugs
2.4. Murine Model of Hematogenously Disseminated Infection
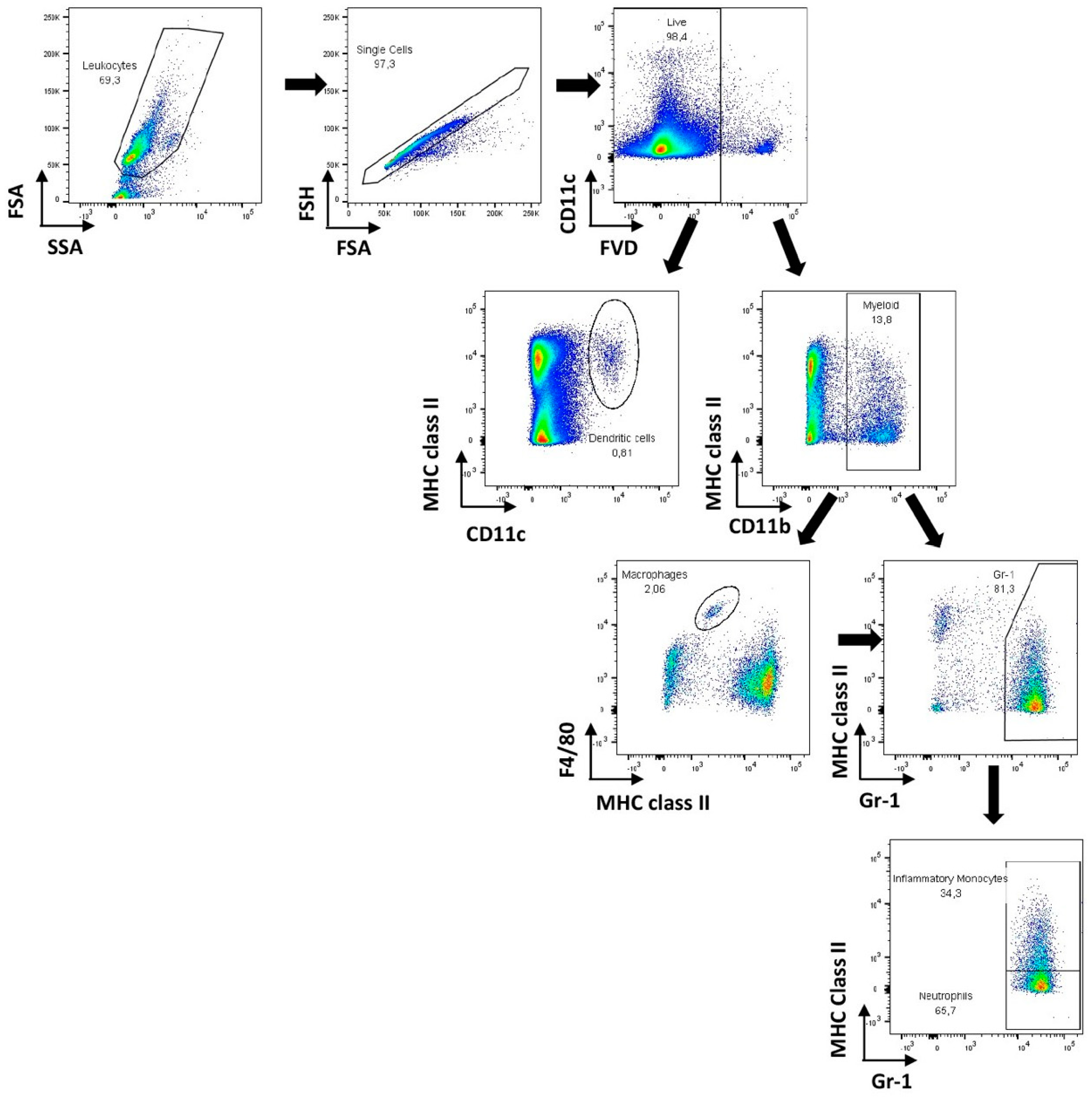
2.5. Flow Cytometry
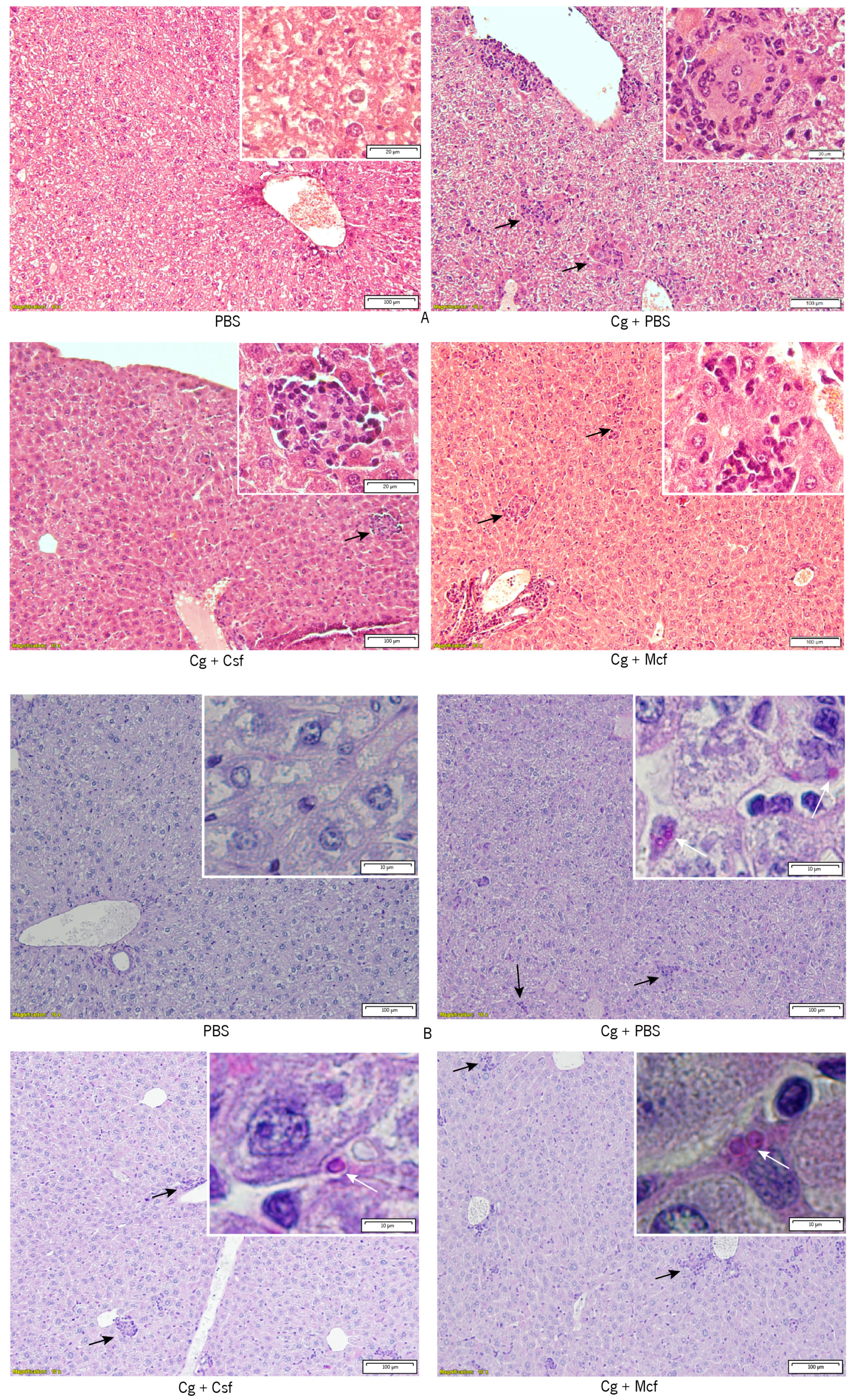
2.6. Histopathologic Examination and Immunohistochemistry
2.7. Statistical Analysis
3. Results and Discussion
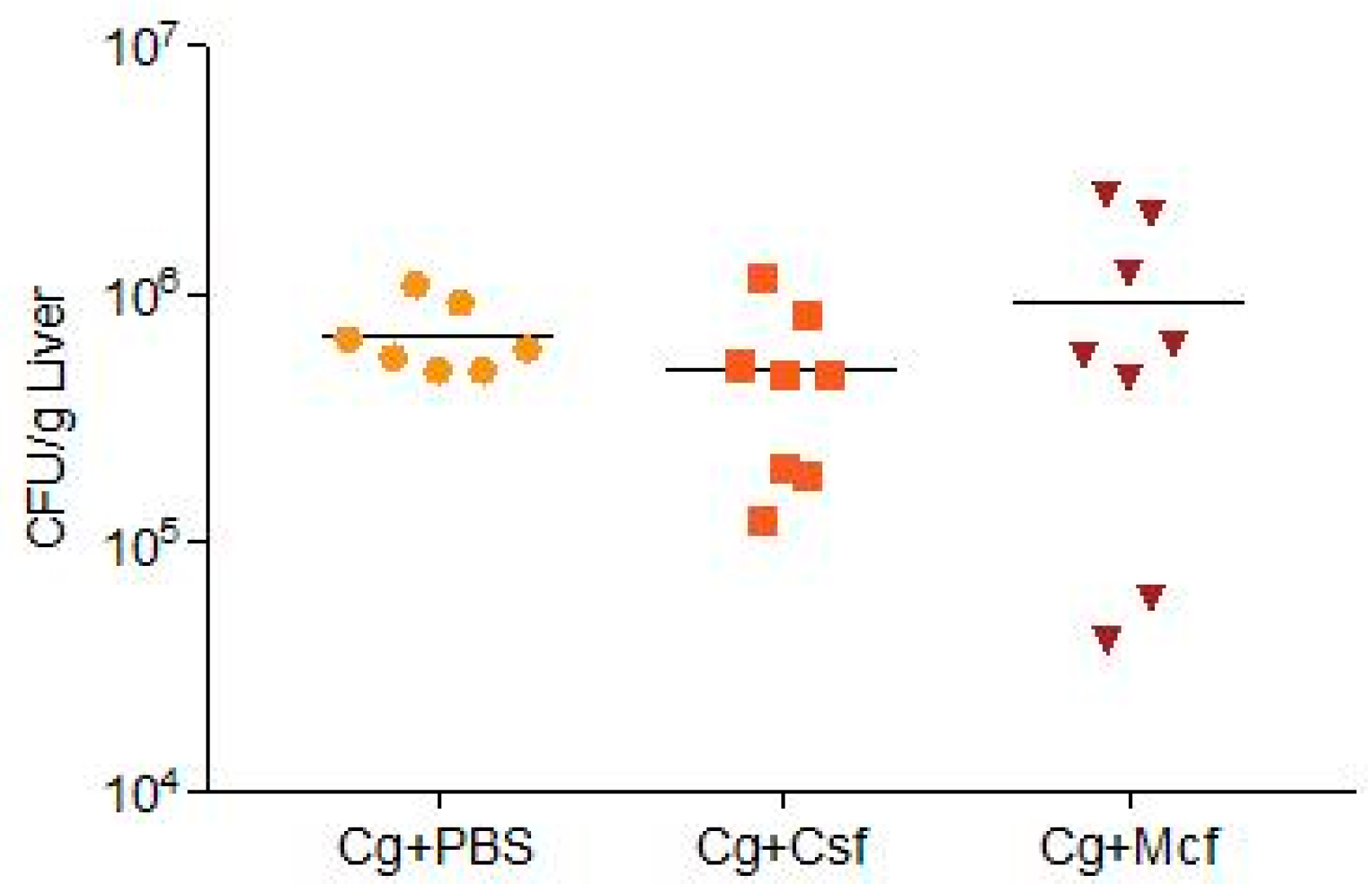
3.1. Fungal Burden Progression Differs Substantially between Liver and Kidneys
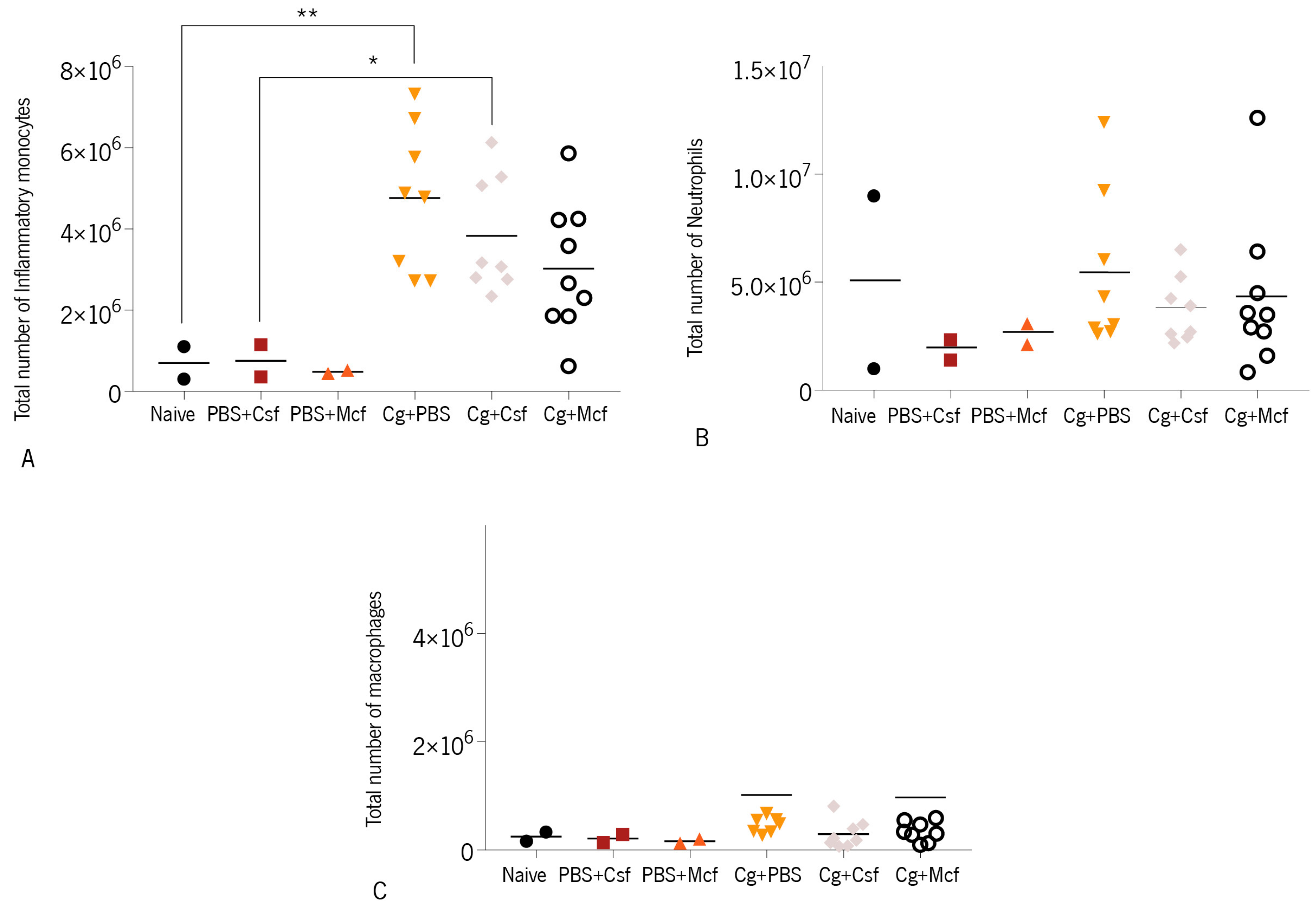
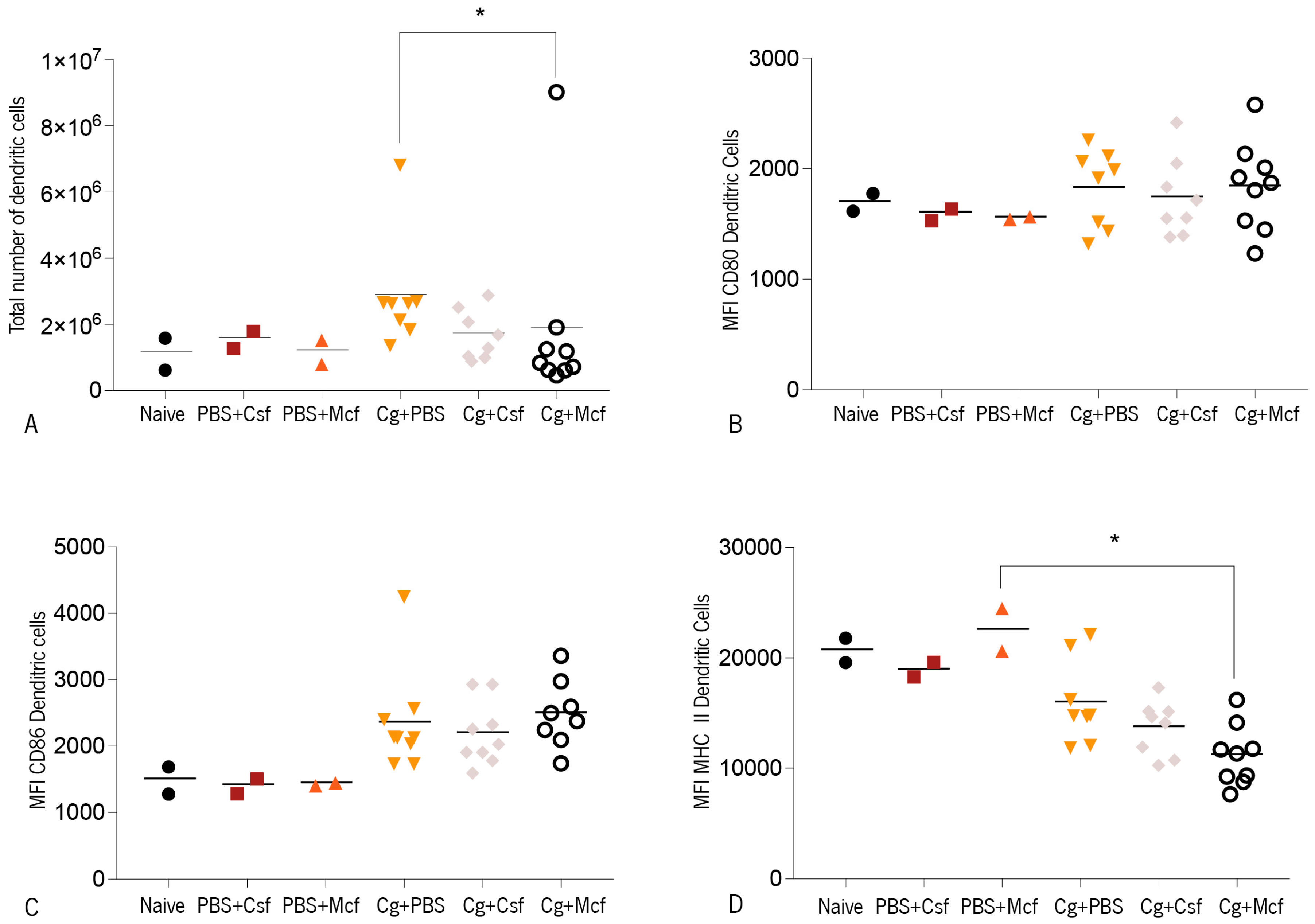
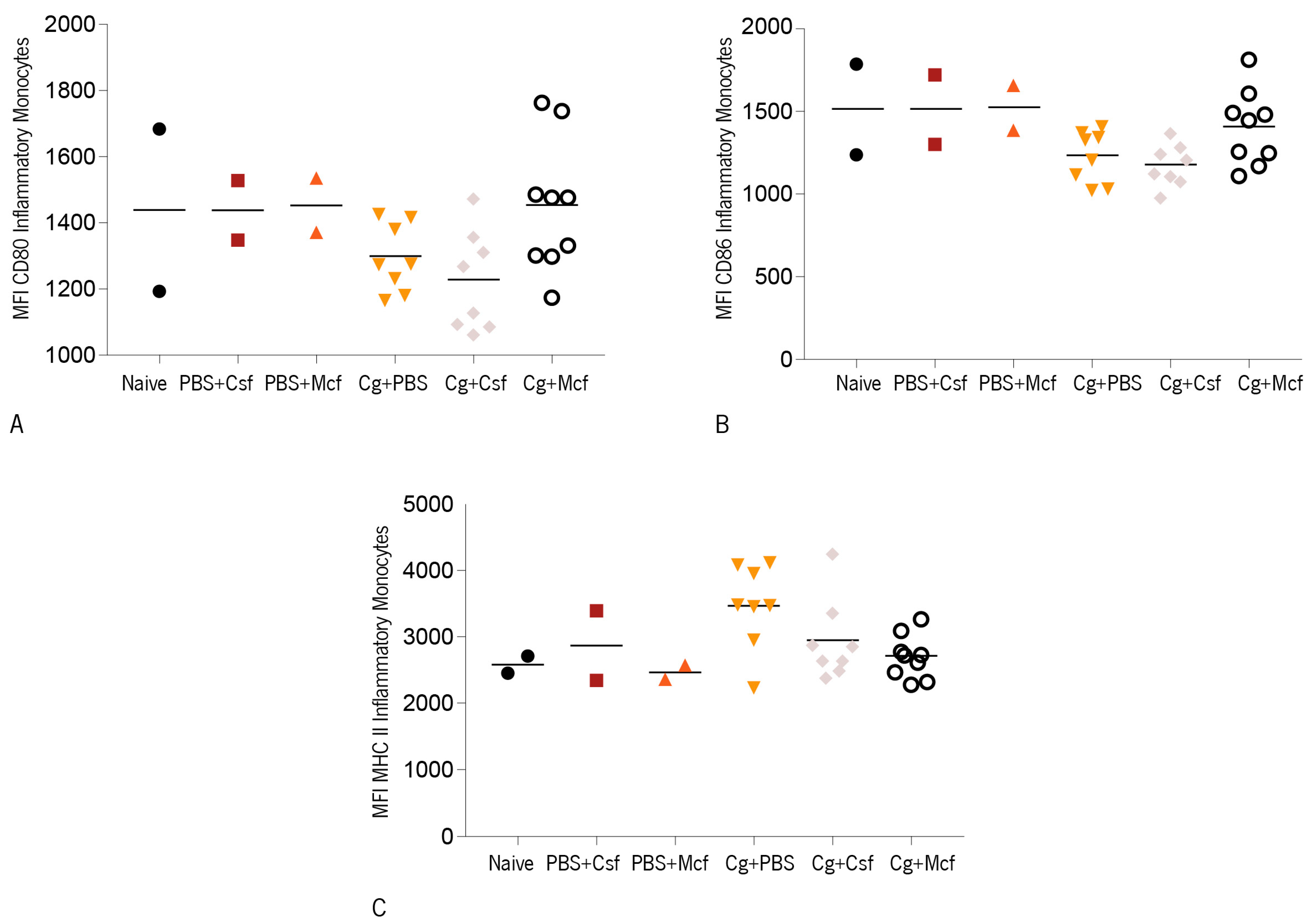
3.2. Host Immune Response to Hematogenously Disseminated Candidiasis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brunke, S.; Seider, K.; Fischer, D.; Jacobsen, I.D.; Kasper, L.; Jablonowski, N.; Wartenberg, A.; Bader, O.; Enache-Angoulvant, A.; Schaller, M.; et al. One Small Step for a Yeast -Microevolution within Macrophages Renders Candida glabrata Hypervirulent Due to a Single Point Mutation. PLoS Pathog. 2014, 10, e1004478. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Rodrigues, M.; Silva, S.; Henriques, M. Candida glabrata Biofilms: How Far Have We Come? J. Fungi 2017, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Archimedes, D.; Carolina, A.; Souza, R.; Colombo, A.L. Revisiting Species Distribution and Antifungal Susceptibility of Candida Bloodstream Isolates from Latin American Medical Centers. J. Fungi 2017, 3, 24. [Google Scholar]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Fidel, P.L.P.; Vazquez, J.A.; Sobel, J. Candida glabrata: Review of Epidemiology, Pathogenesis, and Clinical Disease with Comparison to C. albicans. Clin. Microbiol. Rev. 1999, 12, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C. Candida and candidaemia. Susceptibility and epidemiology. Dan. Med. J. 2013, 60, B4698. [Google Scholar]
- Shin, J.H.; Chae, M.J.; Song, J.W.; Jung, S.-I.; Cho, D.; Kee, S.J.; Kim, S.H.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J. Clin. Microbiol. 2007, 45, 2385–2391. [Google Scholar] [CrossRef]
- Bader, O.; Schwarz, A.; Kraneveld, E.A.; Tangwattanchuleeporn, M.; Schmidt, P.; Jacobsen, M.D.; Gross, U.; De Groot, P.W.J.; Weig, M. Gross Karyotypic and Phenotypic Alterations among Different Progenies of the Candida glabrata CBS138/ ATCC2001 Reference Strain. PLoS ONE 2012, 7, e52218. [Google Scholar] [CrossRef]
- Poláková, S.; Blume, C.; Zárate, J.A.; Mentel, M.; Jørck-Ramberg, D.; Stenderup, J.; Piskur, J. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. USA 2009, 106, 2688–2693. [Google Scholar] [CrossRef]
- Hachem, R.; Hanna, H.; Kontoyiannis, D.; Jiang, Y.; Raad, I. The changing epidemiology of invasive candidiasis: Candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer 2008, 112, 2493–2499. [Google Scholar] [CrossRef]
- Malani, A.; Hmoud, J.; Chiu, L.; Carver, P.L.; Bielaczyc, A.; Kauffman, C.A. Candida glabrata Fungemia: Experience in a Tertiary Care Center. Clin. Infect. Dis. 2005, 41, 975–981. [Google Scholar] [CrossRef]
- Playford, E.G.; Marriott, D.; Nguyen, Q.; Chen, S.; Ellis, D.; Slavin, M.; Sorrell, T.C. Candidemia in nonneutropenic critically ill patients: Risk factors for non-albicans Candida spp. Crit. Care Med. 2008, 36, 2034–2039. [Google Scholar] [CrossRef] [PubMed]
- Marriott, D.J.; Playford, E.G.; Chen, S.; Slavin, M.; Nguyen, Q.; Ellis, D.; Sorrell, T.C. Determinants of mortality in non-neutropenic ICU patients with candidaemia for the Australian Candidaemia Study. Crit. Care 2009, 13, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sipsas, N.V.; Lewis, R.E.; Tarrand, J.; Hachem, R.; Rolston, K.V.; Raad, I.I.; Kontoyiannis, D.P. Candidemia in patients with hematologic malignancies in the era of new antifungal agents (2001–2007). Cancer 2009, 115, 4745–4752. [Google Scholar] [CrossRef] [PubMed]
- Di Stasio, D.; Lauritano, D.; Minervini, G.; Paparella, R.S.; Petruzzi, M.; Romano, A.; Candotto, V.; Lucchese, A. Management of denture stomatitis: A narrative review. J. Biol. Regul. Homeost. Agents 2018, 32, 113–116. [Google Scholar] [PubMed]
- Borst, A.; Raimer, M.T.; Warnock, D.W.; Morrison, C.J.; Arthington-Skaggs, B.A. Rapid Acquisition of Stable Azole Resistance by Candida glabrata Isolates Obtained before the Clinical Introduction of Fluconazole. Antimicrob. Agents Chemother. 2005, 49, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Sanglard, D.; Ischer, F.; Calabrese, D.; Majcherczyk, P.A.; Bille, J. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 1999, 43, 2753–2765. [Google Scholar] [CrossRef]
- Akpan, A.; Morgan, R. Oral candidiasis. Postgrad. Med. J. 2002, 78, 455–459. [Google Scholar] [CrossRef]
- Lott, T.J.; Holloway, B.P.; Logan, D.A.; Fundyga, R.; Arnold, J. Towards understanding the evolution of the human commensal yeast Candida albicans. Microbiology 1999, 145, 1137–1143. [Google Scholar] [CrossRef]
- van der Meer, J.W.M.; van de Veerdonk, F.L.; Joosten, L.A.B.; Kullberg, B.-J.; Netea, M.G. Severe Candida spp. infections: New insights into natural immunity. Int. J. Antimicrob. Agents 2010, 36, S58–S62. [Google Scholar] [CrossRef]
- Chandra, J.; Mukherjee, P.K. Candida Biofilms: Development, Architecture, and Resistance. Microbiol. Spectr. 2015, 3, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Kojic, E.M.E.M.; Darouiche, R.O.R.O. Candida infections of medical devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Bjarnsholt, T. The complexity of microbial biofilm research—An introduction to the 3 rd Thematic Issue on Biofilms. Pathog. Dis. 2016. [Google Scholar] [CrossRef] [PubMed]
- Zarnowski, R.; Westler, W.M.; Lacmbouh, G.A.; Marita, J.M.; Bothe, J.R.; Bernhardt, J.; Sahraoui, A.L.H.; Fontainei, J.; Sanchez, H.; Hatfeld, R.D.; et al. Novel entries in a fungal biofilm matrix encyclopedia. MBio 2014, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- LaFleur, M.D.; Kumamoto, C.A.; Lewis, K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 2006, 50, 3839–3846. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Roussos, N.; Vardakas, K.Z. Relative frequency of albicans and the various non-albicans Candida spp. among candidemia isolates from inpatients in various parts of the world: A systematic review. Int. J. Infect. Dis. 2010, 14, e954–e966. [Google Scholar] [CrossRef] [PubMed]
- Seneviratne, C.J.; Jin, L.; Samaranayake, L.P. Biofilm lifestyle of Candida: A mini review. Oral Dis. 2008, 14, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Douglas, L.J. Candida biofilms and their role in infection. Trends Microbiol. 2003, 11, 30–36. [Google Scholar] [CrossRef]
- Mermel, L.A.; Allon, M.; Bouza, E.; Craven, D.E.; Flynn, P.; O’Grady, N.P.; Raad, I.I.; Rijnders, B.J.A.; Sherertz, R.J.; Warren, D.K. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 49, 1–45. [Google Scholar] [CrossRef]
- Nucci, M.; Anaissie, E.; Betts, R.F.; Dupont, B.F.; Wu, C.; Buell, D.N.; Kovanda, L.; Lortholary, O. Early Removal of Central Venous Catheter in Patients with Candidemia Does Not Improve Outcome: Analysis of 842 Patients from 2 Randomized Clinical Trials. Clin. Infect. Dis. 2010, 51, 295–303. [Google Scholar] [CrossRef]
- Jacobsen, I.D.; Brunke, S.; Seider, K.; Schwarzmüller, T.; Firon, A.; D’enfért, C.; Kuchler, K.; Hube, B. Candida glabrata Persistence in Mice Does Not Depend on Host Immunosuppression and Is Unaffected by Fungal Amino Acid Auxotrophy. Infect. Immun. 2010, 78, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- McCarty, T.P.; Pappas, P.G. Invasive Candidiasis. Infect. Dis. Clin. N. Am. 2016, 30, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Iqbal, N.; Cleveland, A.A.; Farley, M.M.; Harrison, L.H.; Bolden, C.B.; Baughman, W.; Stein, B.; Hollick, R.; Park, B.J.; et al. Species Identification and Antifungal Susceptibility Testing of Candida Bloodstream Isolates from Population-Based Surveillance Studies in Two U.S. Cities from 2008 to 2011. J. Clin. Microbiol. 2012, 50, 3435–3442. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.; Boyken, L.; Hollis, R.; Kroeger, J.; Messer, S.; Tendolkar, S.; Diekema, D. Use of Epidemiological Cutoff Values To Examine 9-Year Trends in Susceptibility of Candida Species to Anidulafungin, Caspofungin, and Micafungin. J. Clin. Microbiol. 2011, 49, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Cleary, J.D.; Garcia-Effron, G.; Chapman, S.W.; Perlin, D.S. Reduced Candida glabrata Susceptibility secondary to an FKS1 Mutation Developed during Candidemia Treatment. Antimicrob. Agents Chemother. 2008, 52, 2263–2265. [Google Scholar] [CrossRef] [PubMed]
- Thompson, G.R.; Wiederhold, N.P.; Vallor, A.C.; Villareal, N.C.; Lewis, J.S.; Patterson, T.F. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob. Agents Chemother. 2008, 52, 3783–3785. [Google Scholar] [CrossRef] [PubMed]
- Chapeland-Leclerc, F.; Hennequin, C.; Papon, N.; Noël, T.; Girard, A.; Socié, G.; Ribaud, P.; Lacroix, C. Acquisition of Flucytosine, Azole, and Caspofungin Resistance in Candida glabrata Bloodstream Isolates Serially Obtained from a Hematopoietic Stem Cell Transplant Recipient. Antimicrob. Agents Chemother. 2010, 54, 1360–1362. [Google Scholar] [CrossRef] [PubMed]
- Durán-Valle, M.T.; Gago, S.; Gómez-López, A.; Cuenca-Estrella, M.; Jiménez Díez-Canseco, L.; Gómez-Garcés, J.L.; Zaragoza, O. Recurrent Episodes of Candidemia Due to Candida glabrata with a Mutation in Hot Spot 1 of the FKS2 Gene Developed after Prolonged Therapy with Caspofungin. Antimcrob. Agents Chemoter. 2012, 56, 3417–3419. [Google Scholar] [CrossRef]
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Kwa, A.L.; Cheng, S.; Du, C.; Clancy, C.J. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob. Agents Chemother. 2012, 56, 4862–4869. [Google Scholar] [CrossRef]
- Pfeiffer, C.D.; Garcia-Effron, G.; Zaas, A.K.; Perfect, J.R.; Perlin, D.S.; Alexander, B.D. Breakthrough Invasive Candidiasis in Patients on Micafungin. J. Clin. Microbiol. 2010, 48, 2373–2380. [Google Scholar] [CrossRef]
- Bizerra, F.C.; Jimenez-Ortigosa, C.; Souza, A.C.R.; Breda, G.L.; Queiroz-Telles, F.; Perlin, D.S.; Colombo, A.L. Breakthrough candidemia due to multidrug-resistant Candida glabrata during prophylaxis with a low dose of micafungin. Antimcrob. Agents Chemoter. 2014, 58, 2438–2440. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.D.; Iqbal, N.; Bolden, C.B.; Kuykendall, R.J.; Harrison, L.H.; Farley, M.M.; Schaffner, W.; Beldavs, Z.G.; Chiller, T.M.; Park, B.J.; et al. Role of FKS mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob. Agents Chemother. 2014, 58, 4690–4696. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kelly, R.; Kahn, J.N.N.; Robles, J.; Hsu, M.J.M.-J.; Register, E.; Li, W.; Vyas, V.; Fan, H.; Abruzzo, G.; et al. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 2005, 49, 3264–3273. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S.; Teppler, H.; Donowitz, G.R.; Maertens, J.A.; Baden, L.R.; Milne, S.; Brown, A.J.; Gow, N.A. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 2007, 10, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Effron, G.; Chua, D.J.; Tomada, J.R.; Dipersio, J.; Perlin, D.S.; Ghannoum, M.; Bonilla, H. Novel FKS Mutations Associated with Echinocandin Resistance in Candida Species. Antimicrob. Agents Chemother. 2010, 54, 2225–2227. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, E.; Silva, S.; Rodrigues, C.F.; Alves, C.; Azeredo, J.; Henriques, M. Effects of fluconazole on Candida glabrata biofilms and its relationship with ABC transporter gene expression. Biofouling 2014, 30, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Gonçalves, B.; Rodrigues, M.E.; Silva, S.; Azeredo, J.; Henriques, M. The Effectiveness of Voriconazole in Therapy of Candida glabrata’s Biofilms Oral Infections and Its Influence on the Matrix Composition and Gene Expression. Mycopathologia 2017, 182, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Henriques, M. Oral mucositis caused by Candida glabrata biofilms: Failure of the concomitant use of fluconazole and ascorbic acid. Ther. Adv. Infect. Dis. 2017, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.; Perlin, D.; Jensen, R.; Howard, S.; Goodwin, J.; Hopec, W. Differential in vivo activities of anidulafungin, caspofungin, and micafungin against Candida glabrata isolates with and without FSK resistance mutations. Antimicrob. Agents Chemoter. 2012, 2435–2442. [Google Scholar] [CrossRef]
- Andes, D.; Diekema, D.J.; Pfaller, M.A.; Bohrmuller, J.; Marchillo, K.; Lepak, A.; Andes, D.; Diekema, D.J.; Pfaller, M.A.; Bohrmuller, J.; et al. In Vivo comparison of the pharmacodynamic targets for echinocandin drugs against Candida species. Antimicrob. Agents Chemother. 2010, 54, 2497–2506. [Google Scholar] [CrossRef]
- Teixeira, L.; Moreira, J.; Melo, J.; Bezerra, F.; Marques, R.M.; Fer-Reirinha, P.; Correia, A.; Monteiro, M.P.; Ferreira, P.G.; Vilanova, M. Immune response in the adipose tissue of lean mice infected with the protozoan parasite Neospora caninum. Immunology 2014, 145, 242–257. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Saraswat, D.; Tati, S.; Edgerton, M. Novel Aggregation Properties of Candida albicans Secreted Aspartyl Proteinase Sap6 Mediate Virulence in Oral Candidiasis. Infect. Immun. 2015, 83, 2614–2626. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R. Current Epidemiology of Candida Infection. Clin. Microbiol. Newsl. 2014, 36, 131–136. [Google Scholar] [CrossRef]
- Silva, S.; Rodrigues, C.F.; Araújo, D.; Rodrigues, M.E.; Henriques, M. Candida Species Biofilms’ Antifungal Resistance. J. Fungi 2017, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Safdar, A.; Bannister, T.W.; Safdar, Z.; Ellis, M.; Ain, A. The predictors of outcome in immunocompetent patients with hematogenous candidasis. Int. J. Infect. Dis. 2004, 8, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Van Dijck, P.; Sjollema, J.; Camue, B.P.A.; Lagrou, K.; Berman, J.; d’Enfert, C.; Andes, D.R.; Arendrup, M.C.; Brakhage, A.A.; Calderone, R.; et al. Methodologies for in vitro and in vivo evaluation of efficacy of antifungal and antibiofilm agents and surface coatings against fungal biofilms. Microb. Cell 2018, 5, 300–326. [Google Scholar] [CrossRef] [PubMed]
- Correia, A.; Lermann, U.; Teixeira, L.; Cerca, F.; Botelho, S.; Gil Da Costa, R.M.; Sampaio, P.; Gärtner, F.; Morschhäuser, J.; Vilanova, M.; et al. Limited Role of Secreted Aspartyl Proteinases Sap1 to Sap6 in Candida albicans Virulence and Host Immune Response in Murine Hematogenously Disseminated Candidiasis. Infect. Immun. 2010, 78, 4839–4849. [Google Scholar] [CrossRef]
- Brieland, J.; Essig, D.; Jackson, C.; Frank, D.; Loebenberg, D.; Menzel, F.; Arnold, B.; DiDomenico, B.; Hare, R. Comparison of pathogenesis and host immune responses to Candida glabrata and Candida albicans in systemically infected immunocompetent mice. Infect. Immun. 2001, 69, 5046–5055. [Google Scholar] [CrossRef]
- Kaur, R.; Ma, B.; Cormack, B.P. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. USA 2007, 104, 7628–7633. [Google Scholar] [CrossRef]
- Srikantha, T.; Daniels, K.J.; Wu, W.; Lockhart, S.R.; Yi, S.; Sahni, N.; Ma, N.; Soll Correspondence, D.R.; Soll, D.R. Dark brown is the more virulent of the switch phenotypes of Candida glabrata. Microbiology 2008, 154, 3309–3318. [Google Scholar] [CrossRef]
- Atkinson, B.A.; Bouthet, C.; Bocanegra, R.; Correa, A.; Luther, M.F.; Graybill, J.R. Comparison of fluconazole, amphotericin B and flucytosine in treatment of a murine model of disseminated infection with Candida glabrata in immunocompromised mice. J. Antimicrob. Chemother. 1995, 35, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Moet, G.J.; Messer, S.A.; Jones, R.N.; Castanheira, M. Candida bloodstream infections: Comparison of species distributions and antifungal resistance patterns in community-onset and nosocomial isolates in the SENTRY Antimicrobial Surveillance Program, 2008-2009. Antimicrob. Agents Chemother. 2011, 55, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Ortigosa, C.J.; Shor, E.; Perlin, D.S. Genetic Drivers of Multidrug Resistance in Candida glabrata. Front. Microbiol. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Morio, F.; Jensen, R.H.; Le Pape, P.; Arendrup, M.C. Molecular basis of antifungal drug resistance in yeasts. Int. J. Antimicrob. Agents 2017, 17, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S. Mechanisms of echinocandin antifungal drug resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Sanguinetti, M.; De Bernardis, F.; Torelli, R.; Posteraro, B.; Vandeputte, P.; Sanglard, D. Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice. Antimicrob. Agents Chemother. 2011, 55, 1852–1860. [Google Scholar] [CrossRef] [PubMed]
- Al-fattani, M.A.; Douglas, L.J. Penetration of Candida Biofilms by Antifungal Agents. Antimicrob. Agents Chemother. 2004, 48, 3291–3297. [Google Scholar] [CrossRef]
- De Luca, C.; Guglielminetti, M.; Ferrario, A.; Calabrὸ, M.; Casari, E. Candidemia: Species involved, virulence factors and antimycotic susceptibility. New Microbiol. 2012, 35, 459–468. [Google Scholar]
- Grandesso, S.; Sapino, B.; Mazzuccato, S.; Solinas, M.; Bedin, M.; D’Angelo, M.; Gion, M. Study on in vitro susceptibility of Candida spp. isolated from blood culture. Infect. Med. 2012, 20, 25–30. [Google Scholar]
- Lewis, R.; Kontoyiannis, D.; Darouiche, R.; Raad, I.; Prince, R. Antifungal activity of amphotericin B, fluconazole, and voriconazole in an in vitro model of Candida catheter-related bloodstream infection. Antimicrob. Agents Chemother. 2002, 46, 3499–3505. [Google Scholar] [CrossRef]
- Donlan, R.; Costerton, J. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Basso, L.R.; Gast, C.E.; Mao, Y.; Wong, B. Fluconazole Transport into Candida albicans Secretory Vesicles by the Membrane Proteins Cdr1p, Cdr2p, and Mdr1p. Eukaryot. Cell 2010, 9, 960–970. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Joosten, L.A.; van der Meer, J.W.; Kullberg, B.J.; van de Veerdonk, F.L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 2015, 15, 630. [Google Scholar] [CrossRef] [PubMed]
- Shoham, S.; Levitz, S.M. The immune response to fungal infections. Br. J. Haematol. 2005, 129, 569–582. [Google Scholar] [CrossRef] [PubMed]
- V Zquez-torres, A.S.; Balish, E. Macrophages in Resistance to Candidiasis. Microbiol. Mol. Biol. Rev. 1997, 61, 170–192. [Google Scholar]
- Ehrensaft, D.V.; Epstein, R.B.; Sarpel, S.; Andersen, B.R. Disseminated candidiasis in leukopenic dogs. Proc. Soc. Exp. Biol. Med. 1979, 160, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Elin, R.J.; Edelin, J.B.; Wolff, S.M. Infection and Immunoglobulin Concentrations in Chediak-Higashi Mice. Infect. Immun. 1974, 10, 88–91. [Google Scholar]
- Holm, H.W.; Marwin, R.M. Effects of surface active agents on the susceptibility of Swiss mice to Candida albicans. Mycopathol. Mycol. Appl. 1967, 33, 186–192. [Google Scholar] [CrossRef]
- Sharpe, H.; Romani Arlene Cenci, L.; Pitzurra, L.; Spreca, A.; Kopf, M.; Mencacci, A.; Montagnoli, C.; Bacci, A.; Cenci, E.; Sharpe, A.H.; et al. CD80+ Gr-1+ Myeloid Cells Inhibit Development of Antifungal Th1 Immunity in Mice with Candidiasis. J. Immunol. Ref. 2017, 169, 3180–3190. [Google Scholar]
- Taylor, P.R.; Tsoni, S.V.; Willment, J.A.; Dennehy, K.M.; Rosas, M.; Findon, H.; Haynes, K.; Steele, C.; Botto, M.; Gordon, S.; et al. Dectin-1 is required for β-glucan recognition and control of fungal infection. Nat. Immunol. 2007, 8, 31–38. [Google Scholar] [CrossRef]
- Taylor, P.R.; Brown, G.D.; Geldhof, A.B.; Martinez-Pomares, L.; Gordon, S. Pattern recognition receptors and differentiation antigens define murine myeloid cell heterogeneity ex vivo. Eur. J. Immunol. 2003, 33, 2090–2097. [Google Scholar] [CrossRef] [PubMed]
- Westwater, C.; Schofield, D.A.; Nicholas, P.J.; Paulling, E.E.; Balish, E. Candida glabrata and Candida albicans; dissimilar tissue tropism and infectivity in a gnotobiotic model of mucosal candidiasis. FEMS Immunol. Med. Microbiol. 2007, 51, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Brunke, S.; Hube, B. Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell. Microbiol. 2013, 15, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Keppler-Ross, S.; Douglas, L.; Konopka, J.B.; Dean, N. Recognition of Yeast by Murine Macrophages Requires Mannan but Not Glucan. Eukaryot. Cell 2010, 9, 1776–1787. [Google Scholar] [CrossRef] [PubMed]
- Seider, K.; Brunke, S.; Schild, L.; Jablonowski, N.; Wilson, D.; Majer, O.; Dagmar, B.; Haas, A.; Kuchler, K.; Schaller, M.; et al. The Facultative Intracellular Pathogen Candida glabrata Subverts Macrophage Cytokine Production and Phagolysosome Maturation. J. Immunol. 2011, 187, 3072–3086. [Google Scholar] [CrossRef] [PubMed]
- Roetzer, A.; Gratz, N.; Kovarik, P.; Schüller, C. Autophagy supports Candida glabrata survival during phagocytosis. Cell. Microbiol. 2010, 12, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Jandric, Z.; Schuller, C. Stress response in Candida glabrata: Pieces of a fragmented picture. Future Mirobiol. 2011, 6, 1475–1484. [Google Scholar] [CrossRef]
- Bonifazi, P.; Zelante, T.; D’Angelo, C.; De Luca, A.; Moretti, S.; Bozza, S.; Perruccio, K.; Iannitti, R.G.; Giovannini, G.; Volpi, C.; et al. Balancing inflammation and tolerance in vivo through dendritic cells by the commensal Candida albicans. Mucosal Immunol. 2009, 2, 362–374. [Google Scholar] [CrossRef]
- Moraes Nicola, A.; Casadevall, A.; Goldman, D.L.; Nicola, A.M.; Casadevall, A.; Goldman, D.L. Fungal killing by mammalian phagocytic cells. Curr. Opin. Microbiol. 2008, 11, 313–317. [Google Scholar] [CrossRef]
- Romani, L.; Montagnoli, C.; Bozza, S.; Perruccio, K.; Spreca, A.; Allavena, P.; Verbeek, S.; Calderone, R.A.; Bistoni, F.; Puccetti, P. The exploitation of distinct recognition receptors in dendritic cells determines the full range of host immune relationships with Candida albicans. Int. Immunol. 2004, 16, 149–161. [Google Scholar] [CrossRef]
- Shi, D.; Li, D.; Qingxin, Y.; Qiu, Y.; Yan, H.; Shen, Y.; Guixia, L.; Liu, W. Silenced suppressor of cytokine signaling 1 (SOCS1) enhances the maturation and antifungal immunity of dendritic cells in response to Candida albicans in vitro. Immunol. Res. 2015, 61, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Ohno, N.; Uchiyama, M.; Tsuzuki, A.; Tokunaka, K.; Miura, N.N.; Adachi, Y.; Aizawa, M.W.; Tamura, H.; Tanaka, S.; Yadomae, T. Solubilization of yeast cell-wall β-(1→3)-D-glucan by sodium hypochlorite oxidation and dimethyl sulfoxide extraction. Carbohydr. Res. 1999, 316, 161–172. [Google Scholar] [CrossRef]
- Rodrigues, C.F.; Boas, D.; Haynes, K.; Henriques, M. The MNN2 Gene Knockout Modulates the Antifungal Resistance of Biofilms of Candida glabrata. Biomolecules 2018, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Rodrigues, M.E.; Henriques, M. Susceptibility of Candida glabrata biofilms to echinocandins: Alterations in the matrix composition. Biofouling 2018, 34, 892–7014. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, C.F.; Correia, A.; Vilanova, M.; Henriques, M. Inflammatory Cell Recruitment in Candida glabrata Biofilm Cell-Infected Mice Receiving Antifungal Chemotherapy. J. Clin. Med. 2019, 8, 142. https://doi.org/10.3390/jcm8020142
Rodrigues CF, Correia A, Vilanova M, Henriques M. Inflammatory Cell Recruitment in Candida glabrata Biofilm Cell-Infected Mice Receiving Antifungal Chemotherapy. Journal of Clinical Medicine. 2019; 8(2):142. https://doi.org/10.3390/jcm8020142
Chicago/Turabian StyleRodrigues, Célia F., Alexandra Correia, Manuel Vilanova, and Mariana Henriques. 2019. "Inflammatory Cell Recruitment in Candida glabrata Biofilm Cell-Infected Mice Receiving Antifungal Chemotherapy" Journal of Clinical Medicine 8, no. 2: 142. https://doi.org/10.3390/jcm8020142
APA StyleRodrigues, C. F., Correia, A., Vilanova, M., & Henriques, M. (2019). Inflammatory Cell Recruitment in Candida glabrata Biofilm Cell-Infected Mice Receiving Antifungal Chemotherapy. Journal of Clinical Medicine, 8(2), 142. https://doi.org/10.3390/jcm8020142





