The GALNT14 Genotype Predicts Postoperative Outcome of Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
2. Experimental Section
3. Results
3.1. Characteristics of Patients
3.2. The GALNT14 Genotype in Association with OS
3.3. Other Clinicopathological Factors in Association with OS and PFS
3.4. The Shapes of Kaplan–Meier Survival Curves for the “GG” and “Non-GG” Genotypes
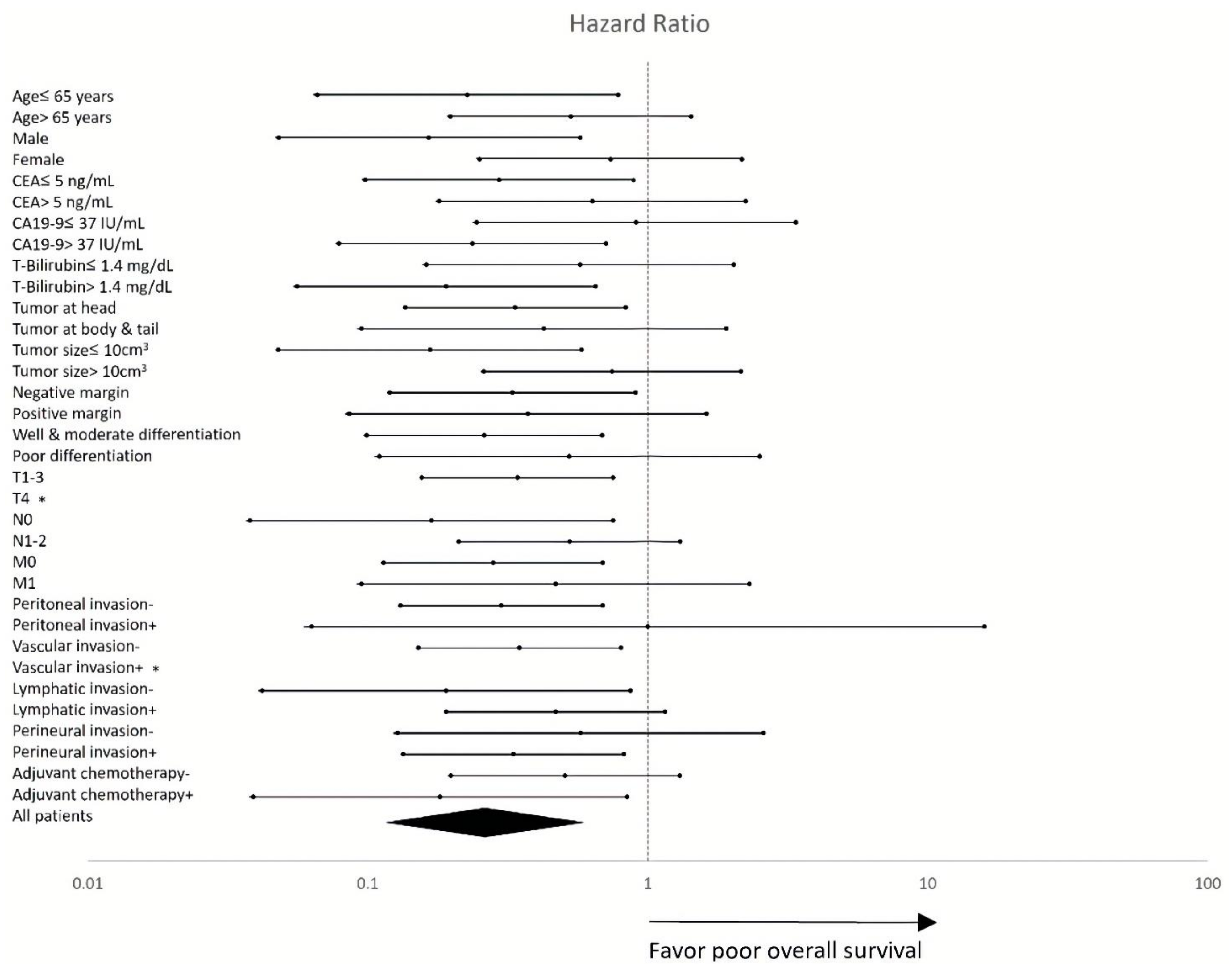
3.5. Subgroup Sensitivity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hasan, S.; Jacob, R.; Manne, U.; Paluri, R. Advances in pancreatic cancer biomarkers. Oncol. Rev. 2019, 13, 410. [Google Scholar] [CrossRef] [PubMed]
- Fong, Z.V.; Winter, J.M. Biomarkers in pancreatic cancer: Diagnostic, prognostic, and predictive. Cancer J. 2012, 18, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Bold, R.J. Re: Ca 19-9 in potentially resectable pancreatic cancer: Perspective to adjust surgical and preoperative therapy. Ann. Surg. Oncol. 2013, 20, 2108–2110. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Loosen, S.H.; Neumann, U.P.; Trautwein, C.; Roderburg, C.; Luedde, T. Current and future biomarkers for pancreatic adenocarcinoma. Tumor Biol. 2017, 39, 1010428317692231. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xie, K.; Wolff, R.; Abbruzzese, J.L. Pancreatic cancer. Lancet 2004, 363, 1049–1057. [Google Scholar] [CrossRef]
- Winter, J.M.; Yeo, C.J.; Brody, J.R. Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer. J. Surg. Oncol. 2013, 107, 15–22. [Google Scholar] [CrossRef]
- Harsha, H.C.; Kandasamy, K.; Ranganathan, P.; Rani, S.; Ramabadran, S.; Gollapudi, S.; Balakrishnan, L.; Dwivedi, S.B.; Telikicherla, D.; Selvan, L.D.; et al. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009, 6, e1000046. [Google Scholar] [CrossRef] [PubMed]
- Brody, J.R.; Witkiewicz, A.K.; Yeo, C.J. The past, present, and future of biomarkers: A need for molecular beacons for the clinical management of pancreatic cancer. Adv. Surg. 2011, 45, 301–321. [Google Scholar] [CrossRef]
- Wang, H.; Tachibana, K.; Zhang, Y.; Iwasaki, H.; Kameyama, A.; Cheng, L.; Guo, J.; Hiruma, T.; Togayachi, A.; Kudo, T.; et al. Cloning and characterization of a novel udp-galnac:Polypeptide n-acetylgalactosaminyltransferase, pp-galnac-t14. Biochem. Biophys. Res. Commun. 2003, 300, 738–744. [Google Scholar] [CrossRef]
- Liang, K.H.; Lin, C.C.; Yeh, C.T. Galnt14 snp as a potential predictor of response to combination chemotherapy using 5-fu, mitoxantrone and cisplatin in advanced hcc. Pharmacogenomics 2011, 12, 1061–1073. [Google Scholar] [CrossRef]
- Tsou, Y.K.; Liang, K.H.; Lin, W.R.; Chang, H.K.; Tseng, C.K.; Yeh, C.T. Galnt14 genotype as a response predictor for concurrent chemoradiotherapy in advanced esophageal squamous cell carcinoma. Oncotarget 2017, 8, 29151–29160. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.R.; Hsu, C.W.; Chen, Y.C.; Chang, M.L.; Liang, K.H.; Huang, Y.H.; Yeh, C.T. Galnt14 genotype, alpha-fetoprotein and therapeutic side effects predict post-chemotherapy survival in patients with advanced hepatocellular carcinoma. Mol. Clin. Oncol. 2014, 2, 630–640. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, W.R.; Chiang, J.M.; Liang, K.H.; Lim, S.N.; Lai, M.W.; Tsou, Y.K.; Hsieh, T.Y.; Hsu, C.K.; Yeh, C.T. Galnt14 genotype predicts postoperative outcome of stage iii colorectal cancer with oxaliplatin as adjuvant chemotherapy. Medicine 2016, 95, e3487. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.H.; Yeh, T.S.; Wu, R.C.; Yeh, C.N.; Yeh, C.T. Galnt14 genotype is associated with perineural invasion, lymph node metastasis and overall survival in resected cholangiocarcinoma. Oncol. Lett. 2017, 13, 4215–4223. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, T.H.; Lin, W.R.; Lee, C.; Chiu, C.T.; Hsu, J.T.; Yeh, T.S.; Lin, K.H.; Le, P.H.; Yeh, C.T. Prognostic stratification of advanced gastric signet ring cell carcinoma by clinicopathological factors and galnt14 genotype. J. Cancer 2018, 9, 3540–3547. [Google Scholar] [CrossRef]
- Yeh, C.T.; Liang, K.H.; Lin, C.C.; Chang, M.L.; Hsu, C.L.; Hung, C.F. A single nucleotide polymorphism on the galnt14 gene as an effective predictor of response to chemotherapy in advanced hepatocellular carcinoma. Int. J. Cancer 2014, 134, 1214–1224. [Google Scholar] [CrossRef]
- Afanasyeva, M.A.; Putlyaeva, L.V.; Demin, D.E.; Kulakovskiy, I.V.; Vorontsov, I.E.; Fridman, M.V.; Makeev, V.J.; Kuprash, D.V.; Schwartz, A.M. The single nucleotide variant rs12722489 determines differential estrogen receptor binding and enhancer properties of an il2ra intronic region. PLoS ONE 2017, 12, e0172681. [Google Scholar] [CrossRef]
- Yasuda, H.; Oh, C.D.; Chen, D.; de Crombrugghe, B.; Kim, J.H. A novel regulatory mechanism of type ii collagen expression via a sox9-dependent enhancer in intron 6. J. Biol. Chem. 2017, 292, 528–538. [Google Scholar] [CrossRef]
- Pattison, J.M.; Posternak, V.; Cole, M.D. Transcription factor klf5 binds a cyclin e1 polymorphic intronic enhancer to confer increased bladder cancer risk. Mol. Cancer Res. 2016, 14, 1078–1086. [Google Scholar] [CrossRef]
- Henson, D.M.; Chou, C.; Sakurai, N.; Egawa, T. A silencer-proximal intronic region is required for sustained cd4 expression in postselection thymocytes. J. Immunol. 2014, 192, 4620–4627. [Google Scholar] [CrossRef]
- Seo, S.; Takayama, K.; Uno, K.; Ohi, K.; Hashimoto, R.; Nishizawa, D.; Ikeda, K.; Ozaki, N.; Nabeshima, T.; Miyamoto, Y.; et al. Functional analysis of deep intronic snp rs13438494 in intron 24 of pclo gene. PLoS ONE 2013, 8, e76960. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.-H.; Lin, C.-L.; Chen, S.-F.; Chiu, C.-W.; Yang, P.-C.; Chang, M.-L.; Lin, C.-C.; Sung, K.-F.; Yeh, C.; Hung, C.-F.; et al. Galnt14 genotype effectively predicts the therapeutic response in unresectable hepatocellular carcinoma treated with transcatheter arterial chemoembolization. Pharmacogenomics 2016, 17, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Chia, J.; Goh, G.; Bard, F. Short o-galnac glycans: Regulation and role in tumor development and clinical perspectives. Biochim. Biophys. Acta 2016, 1860, 1623–1639. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.W.; Punnoose, E.A.; Januario, T.; Lawrence, D.A.; Pitti, R.M.; Lancaster, K.; Lee, D.; von Goetz, M.; Yee, S.F.; Totpal, K.; et al. Death-receptor o-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand apo2l/trail. Nat. Med. 2007, 13, 1070–1077. [Google Scholar] [CrossRef]
- Jeon, M.Y.; Seo, S.U.; Woo, S.M.; Min, K.J.; Byun, H.S.; Hur, G.M.; Kang, S.C.; Kwon, T.K. Oridonin enhances trail-induced apoptosis through galnt14-mediated dr5 glycosylation. Biochimie 2019, 165, 108–114. [Google Scholar] [CrossRef]
- Wang, R.; Yu, C.; Zhao, D.; Wu, M.; Yang, Z. The mucin-type glycosylating enzyme polypeptide n-acetylgalactosaminyltransferase 14 promotes the migration of ovarian cancer by modifying mucin 13. Oncol. Rep. 2013, 30, 667–676. [Google Scholar] [CrossRef]
- Song, K.H.; Park, M.S.; Nandu, T.S.; Gadad, S.; Kim, S.C.; Kim, M.Y. Galnt14 promotes lung-specific breast cancer metastasis by modulating self-renewal and interaction with the lung microenvironment. Nat. Commun. 2016, 7, 13796. [Google Scholar] [CrossRef]
- Tempero, M.A.; Malafa, M.P.; Chiorean, E.G.; Czito, B.; Scaife, C.; Narang, A.K.; Fountzilas, C.; Wolpin, B.M.; Al-Hawary, M.; Asbun, H.; et al. Pancreatic adenocarcinoma, version 1.2019. J. Natl. Compr. Cancer Netw. 2019, 17, 202–210. [Google Scholar] [CrossRef]
- Conroy, T.; Ducreux, M. Adjuvant treatment of pancreatic cancer. Curr. Opin. Oncol. 2019, 31, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Klaiber, U.; Leonhardt, C.S.; Strobel, O.; Tjaden, C.; Hackert, T.; Neoptolemos, J.P. Neoadjuvant and adjuvant chemotherapy in pancreatic cancer. Langenbeck Arch. Surg. 2018, 403, 917–932. [Google Scholar] [CrossRef] [PubMed]
- Chaulagain, C.P.; Rothschild, J.; Saif, M.W. Is s-1 a potential game changer in adjuvant therapy of pancreatic cancer? JOP 2013, 14, 329–333. [Google Scholar] [PubMed]
- Visser, B.C.; Ma, Y.; Zak, Y.; Poultsides, G.A.; Norton, J.A.; Rhoads, K.F. Failure to comply with nccn guidelines for the management of pancreatic cancer compromises outcomes. HPB 2012, 14, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Cabana, M.D.; Rand, C.S.; Powe, N.R.; Wu, A.W.; Wilson, M.H.; Abboud, P.A.; Rubin, H.R. Why don’t physicians follow clinical practice guidelines? A framework for improvement. JAMA 1999, 282, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Ghaneh, P.; Kleeff, J.; Halloran, C.M.; Raraty, M.; Jackson, R.; Melling, J.; Jones, O.; Palmer, D.H.; Cox, T.F.; Smith, C.J.; et al. The impact of positive resection margins on survival and recurrence following resection and adjuvant chemotherapy for pancreatic ductal adenocarcinoma. Ann. Surg. 2019, 269, 520–529. [Google Scholar] [CrossRef]
- Tummers, W.S.; Groen, J.V.; Sibinga Mulder, B.G.; Farina-Sarasqueta, A.; Morreau, J.; Putter, H.; van de Velde, C.J.; Vahrmeijer, A.L.; Bonsing, B.A.; Mieog, J.S.; et al. Impact of resection margin status on recurrence and survival in pancreatic cancer surgery. Br. J. Surg. 2019, 106, 1055–1065. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Agarwal, B.; Correa, A.M.; Ho, L. Survival in pancreatic carcinoma based on tumor size. Pancreas 2008, 36, e15–e20. [Google Scholar] [CrossRef]
- Takahashi, C.; Shridhar, R.; Huston, J.; Meredith, K. Correlation of tumor size and survival in pancreatic cancer. J. Gastrointest. Oncol. 2018, 9, 910–921. [Google Scholar] [CrossRef]
- Marchegiani, G.; Andrianello, S.; Malleo, G.; De Gregorio, L.; Scarpa, A.; Mino-Kenudson, M.; Maggino, L.; Ferrone, C.R.; Lillemoe, K.D.; Bassi, C.; et al. Does size matter in pancreatic cancer?: Reappraisal of tumour dimension as a predictor of outcome beyond the tnm. Ann. Surg. 2017, 266, 142–148. [Google Scholar] [CrossRef]

| Variable * | All Patients (n = 103) | GALNT14 “GG” (n = 21) | GALNT14 “Non-GG” (n = 82) | p-Value † |
|---|---|---|---|---|
| Gender | 0.816 | |||
| Male | 66 (64%) | 13 (61.9%) | 53 (64.6%) | |
| Female | 37 (36%) | 8 (38.1%) | 29 (35.4%) | |
| Ages of resection (y) | 64.0 ± 10.5 | 62.5 ± 11.7 | 64.4 ± 10.2 | 0.460 |
| CEA | 3.1 (0.9–65.2) | 3.1 (0.9–65.2) | 3.1 (0.9–64.3) | 0.577 |
| CA19-9 | 198.7 (2–20310.4) | 162.7 (2–8515.9) | 205 (2–20310.4) | 0.716 |
| T-bilirubin | 2.6 (0.3–23.7) | 2.9 (0.5–12.1) | 2.4 (0.3–23.7) | 0.923 |
| Tumor location | 0.491 | |||
| Head | 73 (72.3%) | 16 (76.2%) | 57 (71.3%) | |
| Body | 15 (14.9%) | 4 (19.0%) | 11 (13.8%) | |
| Tail | 13 (12.9%) | 1 (4.8%) | 12 (15%) | |
| Tumor size (cm3) | 14.1 (0.5–2535) | 6 (0.5–130.6) | 14.3 (0.8–2535) | 0.145 |
| Free margin | 0.374 | |||
| Positive (0 mm) | 27 (30.7%) | 5 (26.3%) | 22 (31.9%) | |
| Very close (0–3 mm) | 14 (15.9%) | 5 (26.3%) | 9 (13.0%) | |
| Negative (>3 mm) | 47 (53.4%) | 9 (47.4%) | 38 (55.1%) | |
| Differentiation | 0.881 | |||
| Well | 18 (17.6%) | 4 (19.0%) | 14 (17.3%) | |
| Moderate | 66 (64.7%) | 13 (61.9%) | 53 (65.4%) | |
| Poor | 18 (17.6%) | 4 (19.0%) | 14 (17.3%) | |
| Tumor invasion | 0.198 | |||
| pT1–3 | 94 (91.3%) | 21 (100%) | 73 (89%) | |
| pT4 | 9 (8.7%) | 0 (0%) | 9 (11%) | |
| Regional LN | 0.814 | |||
| N0 | 43 (41.7%) | 8 (38.1%) | 35 (42.7%) | |
| N1 | 53 (51.5%) | 12 (57.1%) | 41 (50%) | |
| N2 | 7 (6.8%) | 1 (4.8%) | 6 (7.3%) | |
| Metastasis | 0.501 | |||
| M0 | 88 (85.4%) | 17 (81%) | 71 (86.6%) | |
| M1 | 15 (14.6%) | 4 (19%) | 11 (13.4%) | |
| Peritoneal invasion | 1.000 | |||
| No | 99 (96.1%) | 20 (95.2%) | 79 (96.3%) | |
| Yes | 4 (3.9%) | 1 (4.8%) | 3 (3.7%) | |
| Vascular invasion | 0.232 | |||
| No | 78 (75.7%) | 18 (85.7%) | 60 (73.2%) | |
| Yes | 25 (24.3%) | 3 (14.3%) | 22 (26.8%) | |
| Lymphatic invasion | 0.705 | |||
| No | 38 (36.9%) | 7 (33.3%) | 31 (37.8%) | |
| Yes | 65 (63.1%) | 14 (66.7%) | 51 (62.2%) | |
| Perineural invasion | 0.136 | |||
| No | 28 (27.2%) | 3 (14.3%) | 25 (30.5%) | |
| Yes | 75 (72.8%) | 18 (85.7%) | 57 (69.5%) | |
| Adjuvant C/T | 0.153 | |||
| No | 72 (69.9%) | 12 (57.1%) | 60 (73.2%) | |
| Yes | 31 (30.1%) | 9 (42.9%) | 22 (26.8%) |
| Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|
| N | HR | 95%CI | p-Value | HR | 95%CI | p-Value | |
| GALNT14 | 0.342 | 0.156–0.748 | 0.007 | 0.273 | 0.114–0.652 | 0.003 | |
| GG | 21 | ||||||
| Non-GG | 82 | ||||||
| Age | 0.934 | 0.562–1.552 | 0.793 | ||||
| ≤65 y | 57 | ||||||
| >65 y | 46 | ||||||
| Gender | 0.864 | 0.512–1.457 | 0.584 | ||||
| Male | 66 | ||||||
| Female | 37 | ||||||
| CEA | 0.724 | 0.404–1.298 | 0.279 | ||||
| ≤5 ng/mL | 54 | ||||||
| >5 ng/mL | 32 | ||||||
| CA19-9 | 1.354 | 0.717–2.555 | 0.35 | ||||
| ≤37 IU/mL | 19 | ||||||
| >37 IU/mL | 67 | ||||||
| T-bilirubin | 0.82 | 0.459–1.463 | 0.502 | ||||
| ≤1.4 mg/dL | 33 | ||||||
| >1.4 mg/dL | 57 | ||||||
| Tumor location | 0.837 | 0.468–1.496 | 0.548 | ||||
| Head | 73 | ||||||
| Body & Tail | 28 | ||||||
| Tumor size | 0.961 | 0.573–1.611 | 0.88 | ||||
| ≤10 cm3 | 46 | ||||||
| >10 cm3 | 57 | ||||||
| Free margin | 0.528 | 0.294–0.950 | 0.033 | 0.434 | 0.198–0.949 | 0.037 | |
| Negative | 61 | ||||||
| Positive | 27 | ||||||
| Differentiation | 0.9 | 0.455–1.781 | 0.762 | ||||
| Well & moderate | 84 | ||||||
| Poor | 18 | ||||||
| Tumor invasion | 0.764 | 0.275–2.128 | 0.607 | ||||
| pT1–3 | 94 | ||||||
| pT4 | 9 | ||||||
| Regional LN | 0.754 | 0.449–1.267 | 0.287 | ||||
| N0 | 43 | ||||||
| N1-2 | 60 | ||||||
| Metastasis | 0.402 | 0.191–0.847 | 0.017 | 0.302 | 0.109–0.833 | 0.021 | |
| M0 | 88 | ||||||
| M1 | 15 | ||||||
| Peritoneal invasion | 0.925 | 0.222–3.845 | 0.914 | ||||
| No | 99 | ||||||
| Yes | 4 | ||||||
| Vascular invasion | 1.324 | 0.670–2.616 | 0.42 | ||||
| No | 78 | ||||||
| Yes | 25 | ||||||
| Lymphatic invasion | 0.668 | 0.389–1.145 | 0.142 | ||||
| No | 38 | ||||||
| Yes | 65 | ||||||
| Perineural invasion | 1.385 | 0.808–2.373 | 0.236 | ||||
| No | 28 | ||||||
| Yes | 75 | ||||||
| Adjuvant C/T | 1.988 | 1.131–3.492 | 0.017 | 0.861 | 0.386–1.924 | 0.716 | |
| No | 72 | ||||||
| Yes | 31 | ||||||
| Univariate Analysis | ||||
|---|---|---|---|---|
| N | HR | 95%CI | p-value | |
| GALNT14 | 0.632 | 0.321–1.245 | 0.185 | |
| GG | 17 | |||
| Non-GG | 68 | |||
| Age | 1.022 | 0.611–1.709 | 0.934 | |
| ≤65 y | 46 | |||
| >65 y | 39 | |||
| Gender | 0.836 | 0.493–1.416 | 0.505 | |
| Male | 56 | |||
| Female | 29 | |||
| CEA | 1.048 | 0.578–1.901 | 0.877 | |
| ≤5 ng/mL | 50 | |||
| >5 ng/mL | 24 | |||
| CA19-9 | 0.998 | 0.512–1.947 | 0.996 | |
| ≤37 IU/mL | 17 | |||
| >37 IU/mL | 56 | |||
| T-bilirubin | 1.08 | 0.622–1.875 | 0.784 | |
| ≤1.4 mg/dL | 26 | |||
| >1.4 mg/dL | 50 | |||
| Tumor location | 0.654 | 0.356–1.203 | 0.172 | |
| Head | 67 | |||
| Body & Tail | 17 | |||
| Tumor size | 0.601 | 0.357–1.011 | 0.055 | |
| ≤10 cm3 | 41 | |||
| >10 cm3 | 44 | |||
| Free margin | 0.657 | 0.371–1.163 | 0.149 | |
| Negative | 50 | |||
| Positive | 23 | |||
| Differentiation | 1.262 | 0.571–2.789 | 0.565 | |
| Well & moderate | 71 | |||
| Poor | 13 | |||
| Tumor invasion | 0.914 | 0.282–2.959 | 0.881 | |
| pT1–3 | 79 | |||
| pT4 | 6 | |||
| Regional LN | 0.644 | 0.383–1.085 | 0.099 | |
| N0 | 39 | |||
| N1-2 | 46 | |||
| Metastasis | - | - | - | |
| M0 | 85 | |||
| M1 | 0 | |||
| Peritoneal invasion | - | - | - | |
| No | 85 | |||
| Yes | 0 | |||
| Vascular invasion | 0.959 | 0.518–1.776 | 0.895 | |
| No | 66 | |||
| Yes | 19 | |||
| Lymphatic invasion | 0.648 | 0.378–1.108 | 0.113 | |
| No | 34 | |||
| Yes | 51 | |||
| Perineural invasion | 0.93 | 0.517–1.673 | 0.81 | |
| No | 22 | |||
| Yes | 63 | |||
| Adjuvant C/T | 1.323 | 0.789–2.219 | 0.289 | |
| No | 55 | |||
| Yes | 30 | |||
| Variable * | Death Within 1 Y (n = 34) | Death More Than 1 Y (n = 26) | p-Value |
|---|---|---|---|
| Gender | 0.276 | ||
| Male | 23 (67.6%) | 14 (53.8%) | |
| Female | 11 (32.4%) | 12 (46.2%) | |
| Ages of resection (y) | 64.2 ± 11.4 | 63.0 ± 9.3 | 0.652 |
| CEA | 4.1 (1.0-28.9) | 2.9 (0.9-65.2) | 0.435 |
| CA19-9 | 222.5 (2.0–20310.4) | 155.0 (2.0–2388.2) | 0.200 |
| T-bilirubin | 4.3 (0.3–23.7) | 4.2 (0.5–21.1) | 0.950 |
| Tumor location | 1.000 | ||
| Head | 25 (73.5%) | 19 (73.1%) | |
| Body | 6 (17.6%) | 4 (15.4%) | |
| Tail | 3 (8.8%) | 3 (11.5%) | |
| Tumor size (cm3) | 12.4 (0.5–432.0) | 6.8 (0.8–161.0) | 0.546 |
| Free margin | 0.543 | ||
| Positive (0 mm) | 12 (44.4%) | 7 (30.4%) | |
| Very close (0–3 mm) | 4 (14.8%) | 3 (13.0%) | |
| Negative (>3 mm) | 11 (40.7%) | 13 (56.5%) | |
| Differentiation | 0.188 | ||
| Well | 5 (14.7%) | 7 (26.9%) | |
| Moderate | 21 (61.8%) | 17 (65.4%) | |
| Poor | 8 (23.5%) | 2 (7.7%) | |
| Tumor invasion | 0.626 | ||
| pT1–3 | 31 (91.2%) | 25 (96.2%) | |
| pT4 | 3 (8.8%) | 1 (3.8%) | |
| Regional LN | 0.288 | ||
| N0 | 11 (32.4%) | 13 (50.0%) | |
| N1 | 22 (64.7%) | 13 (50.0%) | |
| N2 | 1 (2.9%) | 0 (0%) | |
| Metastasis | 0.719 | ||
| M0 | 28 (82.4%) | 23 (88.5%) | |
| M1 | 6 (17.6%) | 3 (11.5%) | |
| Peritoneal invasion | 0.184 | ||
| No | 34 (100.0%) | 24 (92.3%) | |
| Yes | 0 (0%) | 2 (7.7%) | |
| Vascular invasion | 0.163 | ||
| No | 26 (76.5%) | 24 (92.3%) | |
| Yes | 8 (23.5%) | 2 (7.7%) | |
| Lymphatic invasion | 0.065 | ||
| No | 8 (23.5%) | 12 (46.2%) | |
| Yes | 26 (76.5%) | 14 (53.8%) | |
| Perineural invasion | 0.461 | ||
| No | 10 (29.4%) | 10 (38.5%) | |
| Yes | 24 (70.6%) | 16 (61.5%) | |
| Adjuvant C/T | 0.017 | ||
| No | 28 (82.4%) | 14 (53.8%) | |
| Yes | 6 (17.6%) | 12 (46.2%) |
| Variable * | Progression Within 1 Y (n = 37) | Progression More Than 1 Y (n = 23) | p-Value |
|---|---|---|---|
| Gender | 0.755 | ||
| Male | 24 (64.9%) | 14 (60.9%) | |
| Female | 13 (35.1%) | 9 (39.1%) | |
| Ages of resection (y) | 63.7 ± 10.6 | 63.4 ± 9.6 | 0.889 |
| CEA | 2.5 (0.9–64.3) | 2.8 (1.1–22.5) | 0.851 |
| CA19-9 | 192.4 (2.0–4271.4) | 265.8 (6.4–7551.0) | 0.198 |
| T-bilirubin | 2.2 (0.5–17.4) | 4.2 (0.6–21.1) | 0.211 |
| Tumor location | 0.654 | ||
| Head | 27 (73.0%) | 19 (82.6%) | |
| Body | 7 (18.9%) | 2 (8.7%) | |
| Tail | 3 (8.1%) | 2 (8.7%) | |
| Tumor size (cm3) | 14.3 (0.5–130.6) | 14.1 (0.8–280.0) | 0.738 |
| Free margin | 0.164 | ||
| Positive (0 mm) | 14 (42.4%) | 4 (18.2%) | |
| Very close (0–3 mm) | 5 (15.2%) | 4 (18.2%) | |
| Negative (>3 mm) | 14 (42.4%) | 14 (63.6%) | |
| Differentiation | 0.285 | ||
| Well | 8 (21.6%) | 3 (13.6%) | |
| Moderate | 23 (62.2%) | 18 (81.8%) | |
| Poor | 6 (16.2%) | 1 (4.5%) | |
| Tumor invasion | 1.000 | ||
| pT1–3 | 35 (94.6%) | 22 (95.7%) | |
| pT4 | 2 (5.4%) | 1 (4.3%) | |
| Regional LN | 0.233 | ||
| N0 | 12 (32.4%) | 12 (52.2%) | |
| N1 | 19 (51.4%) | 10 (43.5%) | |
| N2 | 6 (16.2%) | 1 (4.3%) | |
| Metastasis † | |||
| M0 | - | - | |
| M1 | - | - | |
| Peritoneal invasion† | |||
| No | - | - | |
| Yes | - | - | |
| Vascular invasion | 1.000 | ||
| No | 29 (78.4%) | 18 (78.3%) | |
| Yes | 8 (21.6%) | 5 (21.7%) | |
| Lymphatic invasion | 0.101 | ||
| No | 10 (27.0%) | 11 (47.8%) | |
| Yes | 27 (73.0%) | 12 (52.2%) | |
| Perineural invasion | 0.646 | ||
| No | 10 (27.0%) | 5 (21.7%) | |
| Yes | 27 (73.0%) | 18 (78.3%) | |
| Adjuvant C/T | 0.051 | ||
| No | 24 (64.9%) | 9 (39.1%) | |
| Yes | 13 (35.1%) | 14 (60.9%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiang, C.-C.; Yeh, C.-T.; Hwang, T.-L.; Chu, Y.-D.; Lim, S.-N.; Chen, C.-W.; Kuo, C.-J.; Le, P.-H.; Chen, T.-H.; Lin, W.-R. The GALNT14 Genotype Predicts Postoperative Outcome of Pancreatic Ductal Adenocarcinoma. J. Clin. Med. 2019, 8, 2225. https://doi.org/10.3390/jcm8122225
Chiang C-C, Yeh C-T, Hwang T-L, Chu Y-D, Lim S-N, Chen C-W, Kuo C-J, Le P-H, Chen T-H, Lin W-R. The GALNT14 Genotype Predicts Postoperative Outcome of Pancreatic Ductal Adenocarcinoma. Journal of Clinical Medicine. 2019; 8(12):2225. https://doi.org/10.3390/jcm8122225
Chicago/Turabian StyleChiang, Chun-Cheng, Chau-Ting Yeh, Tsann-Long Hwang, Yu-De Chu, Siew-Na Lim, Chun-Wei Chen, Chia-Jung Kuo, Puo-Hsien Le, Tsung-Hsing Chen, and Wey-Ran Lin. 2019. "The GALNT14 Genotype Predicts Postoperative Outcome of Pancreatic Ductal Adenocarcinoma" Journal of Clinical Medicine 8, no. 12: 2225. https://doi.org/10.3390/jcm8122225
APA StyleChiang, C.-C., Yeh, C.-T., Hwang, T.-L., Chu, Y.-D., Lim, S.-N., Chen, C.-W., Kuo, C.-J., Le, P.-H., Chen, T.-H., & Lin, W.-R. (2019). The GALNT14 Genotype Predicts Postoperative Outcome of Pancreatic Ductal Adenocarcinoma. Journal of Clinical Medicine, 8(12), 2225. https://doi.org/10.3390/jcm8122225






