Impact of Pregnancy on Intra-Host Genetic Diversity of Influenza A Viruses in Hospitalised Women: A Retrospective Cohort Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. IAV Whole Genome Amplification and Sequencing
2.3. Bioinformatic Analysis
2.4. Statistical Analysis
2.5. Ethical Statement
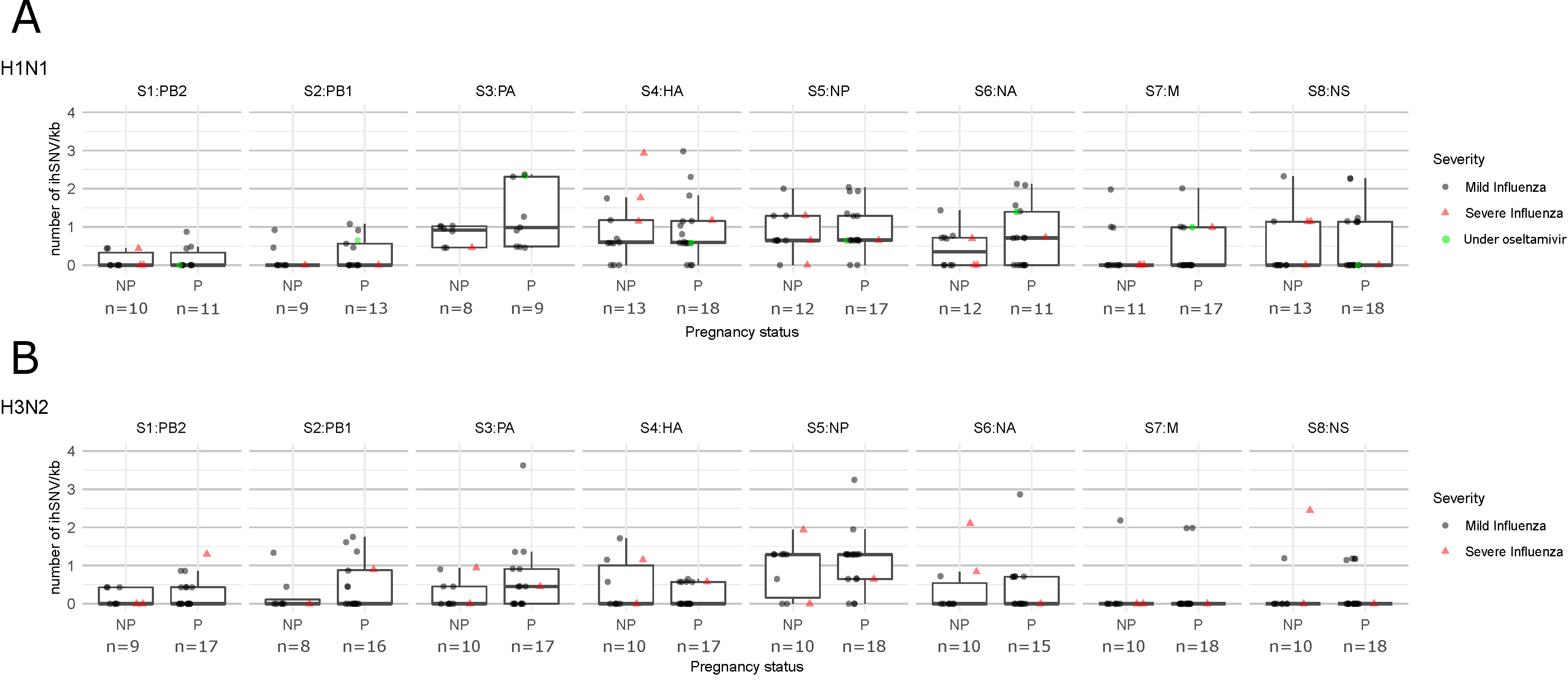
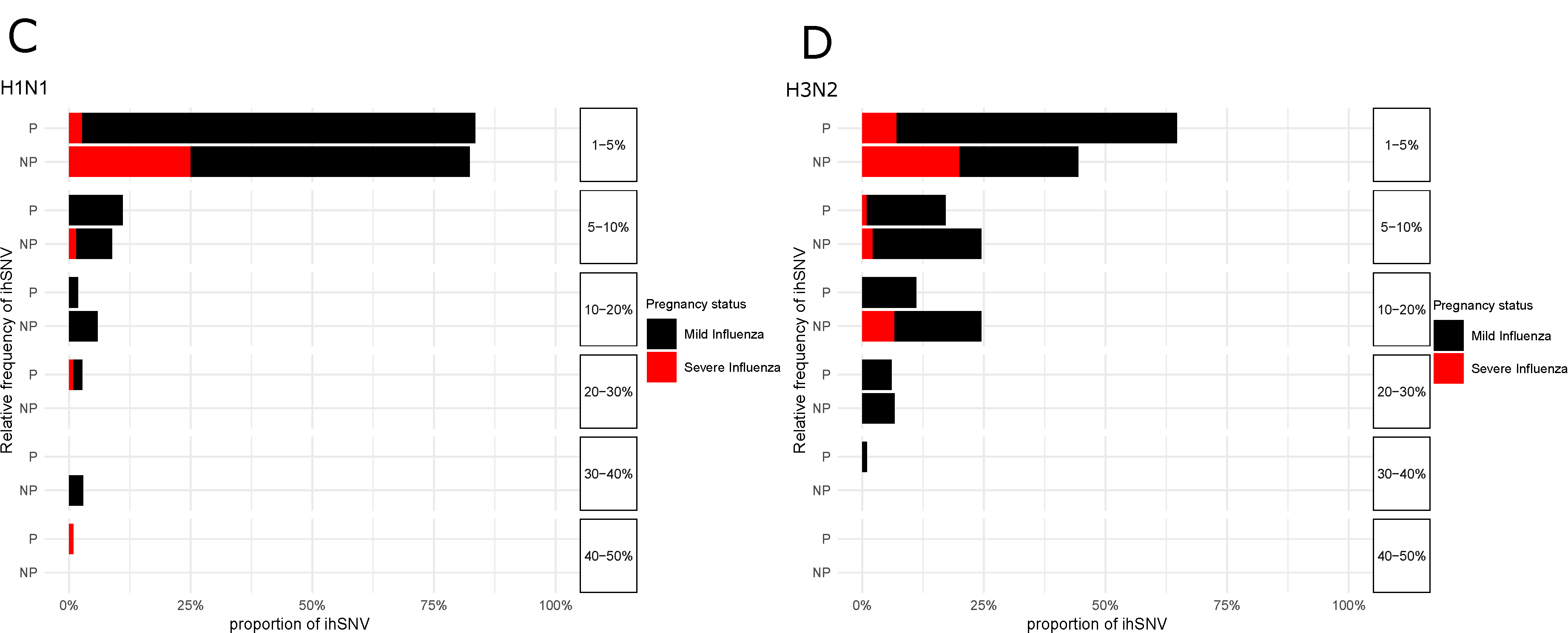
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Domingo, E.; Sheldon, J.; Perales, C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 2012, 76, 159–216. [Google Scholar] [CrossRef] [PubMed]
- Xue, K.S.; Moncla, L.H.; Bedford, T.; Bloom, J.D. Within-Host Evolution of Human Influenza Virus. Trends Microbiol. 2018, 26, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Engels, G.; Hierweger, A.M.; Hoffmann, J.; Thieme, R.; Thiele, S.; Bertram, S.; Dreier, C.; Resa-Infante, P.; Jacobsen, H.; Thiele, K.; et al. Pregnancy-Related Immune Adaptation Promotes the Emergence of Highly Virulent H1N1 Influenza Virus Strains in Allogenically Pregnant Mice. Cell Host Microbe 2017, 21, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Simon, B.; Pichon, M.; Valette, M.; Burfin, G.; Richard, M.; Lina, B.; Josset, L. Whole Genome Sequencing of A(H3N2) Influenza Viruses Reveals Variants Associated with Severity during the 2016–2017 Season. Viruses 2019, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, D.; Von Kuster, G.; Bouvier, E.; Baker, D.; Afgan, E.; Stoler, N.; Galaxy Team; Taylor, J.; Nekrutenko, A. Dissemination of scientific software with Galaxy ToolShed. Genome Biol. 2014, 15, 403. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Internet]. 2018. Available online: https://www.R-project.org/ (accessed on 12 May 2019).
- Pichon, M.; Simon, B.; Valette, M.; Bal, A.; Picard, C.; Escuret, V.; Ottmann, M.; Gillet, Y.; Ader, F.; Lina, B.; et al. Evolution of influenza genome diversity during infection in immunocompetent patients. bioRxiv 2018, 435263. [Google Scholar]
- Debbink, K.; McCrone, J.T.; Petrie, J.G.; Truscon, R.; Johnson, E.; Mantlo, E.K.; Monto, A.S.; Lauring, A.S. Vaccination has minimal impact on the intrahost diversity of H3N2 influenza viruses. PLoS Pathog. 2017, 13, e1006194. [Google Scholar] [CrossRef] [PubMed]
- Sobel Leonard, A.; McClain, M.T.; Smith, G.J.D.; Wentworth, D.E.; Halpin, R.A.; Lin, X.; Ransier, A.; Stockwell, T.B.; Das, S.R.; Gilbert, A.S.; et al. Deep Sequencing of Influenza a Virus from a Human Challenge Study Reveals a Selective Bottleneck and Only Limited Intrahost Genetic Diversification. J. Virol. 2016, 90, 11247–11258. [Google Scholar] [CrossRef] [PubMed]
- Dinis, J.M.; Florek, N.W.; Fatola, O.O.; Moncla, L.H.; Mutschler, J.P.; Charlier, O.K.; Meece, J.K.; Belongia, E.A.; Friedrich, T.C. Deep Sequencing Reveals Potential Antigenic Variants at Low Frequencies in Influenza a Virus-Infected Humans. J. Virol. 2016, 90, 3355–3365. [Google Scholar] [CrossRef] [PubMed]
- Xue, K.S.; Bloom, J.D. Reconciling disparate estimates of viral genetic diversity during human influenza infections. Nat. Genet. 2019, 51, 1298–1301. [Google Scholar] [CrossRef] [PubMed]
- Pichon, M.; Picard, C.; Simon, B.; Gaymard, A.; Renard, C.; Massenavette, B.; Malcus, C.; Monneret, G.; Morfin-Sherpa, F.; Valette, M.; et al. Clinical management and viral genomic diversity analysis of a child’s influenza A(H1N1)pdm09 infection in the context of a severe combined immunodeficiency. Antivir. Res. 2018, 160, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Thangavel, R.R.; Bouvier, N.M. Animal models for influenza virus pathogenesis, transmission, and immunology. J. Immunol. Methods 2014, 410, 60–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Castelán-Vega, J.A.; Jiménez-Alberto, A.; Vassell, R.; Ye, Z.; Weiss, C.D. A mutation in the receptor binding site enhances infectivity of 2009 H1N1 influenza hemagglutinin pseudotypes without changing antigenicity. Virology 2010, 407, 374–380. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oliveira, M.J.; Motta Fdo, C.; Siqueira, M.M.; Resende, P.C.; Born Pda, S.; Souza, T.M.; Mesquita, M.; Oliveira Mde, L.; Carney, S.; Mello, W.A.; et al. Molecular findings from influenza A(H1N1)pdm09 detected in patients from a Brazilian equatorial region during the pandemic period. Mem. Inst. Oswaldo Cruz 2014, 109, 912–917. [Google Scholar] [CrossRef] [PubMed]
| Pregnant Women n = 36 | Non-Pregnant Women n = 23 | p | |
|---|---|---|---|
| Age, mean years (SD) | 30.6 (0.96) | 34.7 (1.5) | 0.03 |
| Trimesters, n (%) | |||
| Trimester 1 | 2 (5.6) | ||
| Trimester 2 | 9 (25.0) | ||
| Trimester 3 | 25 (69.4 | ||
| With underlying condition, n (%) | 10 (30.3) | 10 (43.5) | 0.21 |
| Time since onset of symptoms, median days (IQR) | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | 0.80 |
| Oseltamivir treatment before sampling, n (%) | 1 (2.8) | 0 (0.0) | 1.00 |
| Hospitalisation for influenza, n (%) | 11 (30.6) | 11 (47.8) | 0.18 |
| Severe influenza, n (%) | 2 (5.6) | 5 (21.7) | 0.10 |
| IAV viral load, mean Ct (SD) | 26.01 (4.2) | 26.35 (4.2) | 0.80 |
| IAV sub-type and clades, n (%) | |||
| H1N1 6B1 | 18 (50) | 13 (56.5) | 0.96 |
| H3N2 | 18 (50) | 10 (43.5) | 0.82 |
| H3N2 3c2a1 | 13 (36.1) | 9 (39.1) | 0.65 |
| H3N2 3c2a2 | 3 (8.3) | 0 (0.0) | 0.40 |
| H3N2 3c2a3 | 2 (5.5) | 1 (4.3) | 0.83 |
| H3N2 3c3b | 1 (2.8) | 0 (0.0) | 1.00 |
| Pregnant Women n = 36 | Non-Pregnant Women n = 23 | p Occurrence of NS-ihSNV | p Relative Frequencies | |
|---|---|---|---|---|
| H1N1, women ratio* (min–max of ihSNV relative frequencies) | ||||
| S2-PB1 | ||||
| K279Q | 4/6 (2.3–8.6%) | 1/6 (1.8%) | 0.24 | |
| Q460K | 1/6 (1.6%) | 0/6 | 1.00 | |
| S3-PA | ||||
| E493A | 9/9 (1.3–14.3%) | 8/8 (1.7–10.5%) | 1.00 | 0.55 |
| S4-HA** | ||||
| P182Q | 1/16 (4.6%) | 4/12 (1.3–3.6%) | 0.13 | |
| T342A | 2/13 (1.2–1.4%) | 2/8 (1.2–1.7%) | 0.53 | >0.999 |
| E356K | 9/15 (1.2–7.6%) | 3/12 (1.2–6.2%) | 0.12 | >0.999 |
| S5-NP | ||||
| T130K | 13/17 (1.1–1.8%) | 9/12 (1.1–3.5%) | >0.99 | 0.07 |
| H3N2, women ratio* (min–max of ihSNV relative frequencies) | ||||
| S2-PB1 | ||||
| K279Q | 4/13 (1.5–3.4%) | 0/6 | 0.25 | |
| Q460K | 3/13 (1.3–2.0%) | 0/6 | 0.52 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Destras, G.; Pichon, M.; Simon, B.; Valette, M.; Escuret, V.; Bolze, P.-A.; Dubernard, G.; Gaucherand, P.; Lina, B.; Josset, L. Impact of Pregnancy on Intra-Host Genetic Diversity of Influenza A Viruses in Hospitalised Women: A Retrospective Cohort Study. J. Clin. Med. 2019, 8, 1974. https://doi.org/10.3390/jcm8111974
Destras G, Pichon M, Simon B, Valette M, Escuret V, Bolze P-A, Dubernard G, Gaucherand P, Lina B, Josset L. Impact of Pregnancy on Intra-Host Genetic Diversity of Influenza A Viruses in Hospitalised Women: A Retrospective Cohort Study. Journal of Clinical Medicine. 2019; 8(11):1974. https://doi.org/10.3390/jcm8111974
Chicago/Turabian StyleDestras, Gregory, Maxime Pichon, Bruno Simon, Martine Valette, Vanessa Escuret, Pierre-Adrien Bolze, Gil Dubernard, Pascal Gaucherand, Bruno Lina, and Laurence Josset. 2019. "Impact of Pregnancy on Intra-Host Genetic Diversity of Influenza A Viruses in Hospitalised Women: A Retrospective Cohort Study" Journal of Clinical Medicine 8, no. 11: 1974. https://doi.org/10.3390/jcm8111974
APA StyleDestras, G., Pichon, M., Simon, B., Valette, M., Escuret, V., Bolze, P.-A., Dubernard, G., Gaucherand, P., Lina, B., & Josset, L. (2019). Impact of Pregnancy on Intra-Host Genetic Diversity of Influenza A Viruses in Hospitalised Women: A Retrospective Cohort Study. Journal of Clinical Medicine, 8(11), 1974. https://doi.org/10.3390/jcm8111974






