Mapping Driver Mutations to Histopathological Subtypes in Papillary Thyroid Carcinoma: Applying a Deep Convolutional Neural Network
Abstract
1. Introduction
2. Results
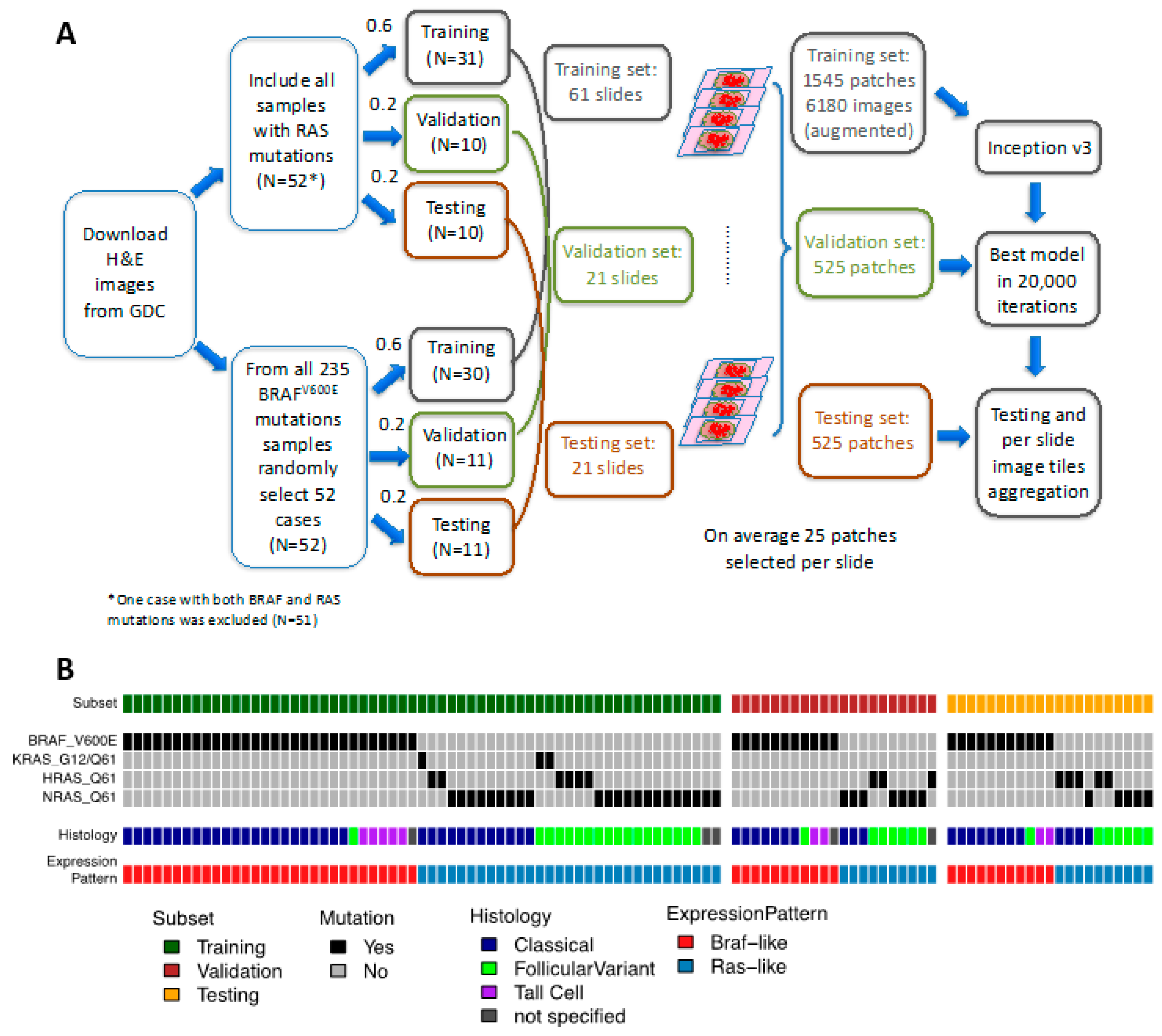
2.1. A CNN Framework for Differentiating Mutations of PTC from Histological Images
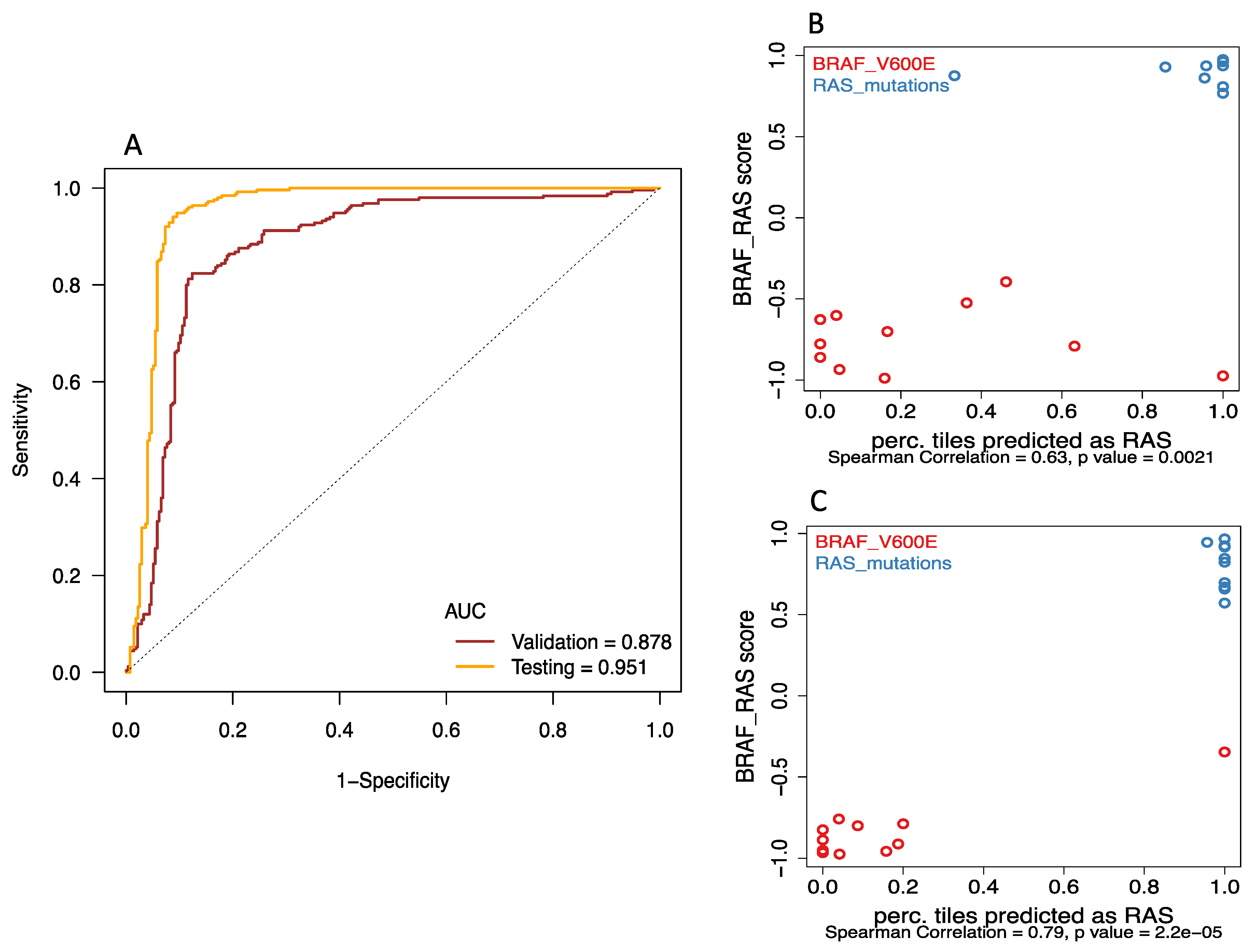
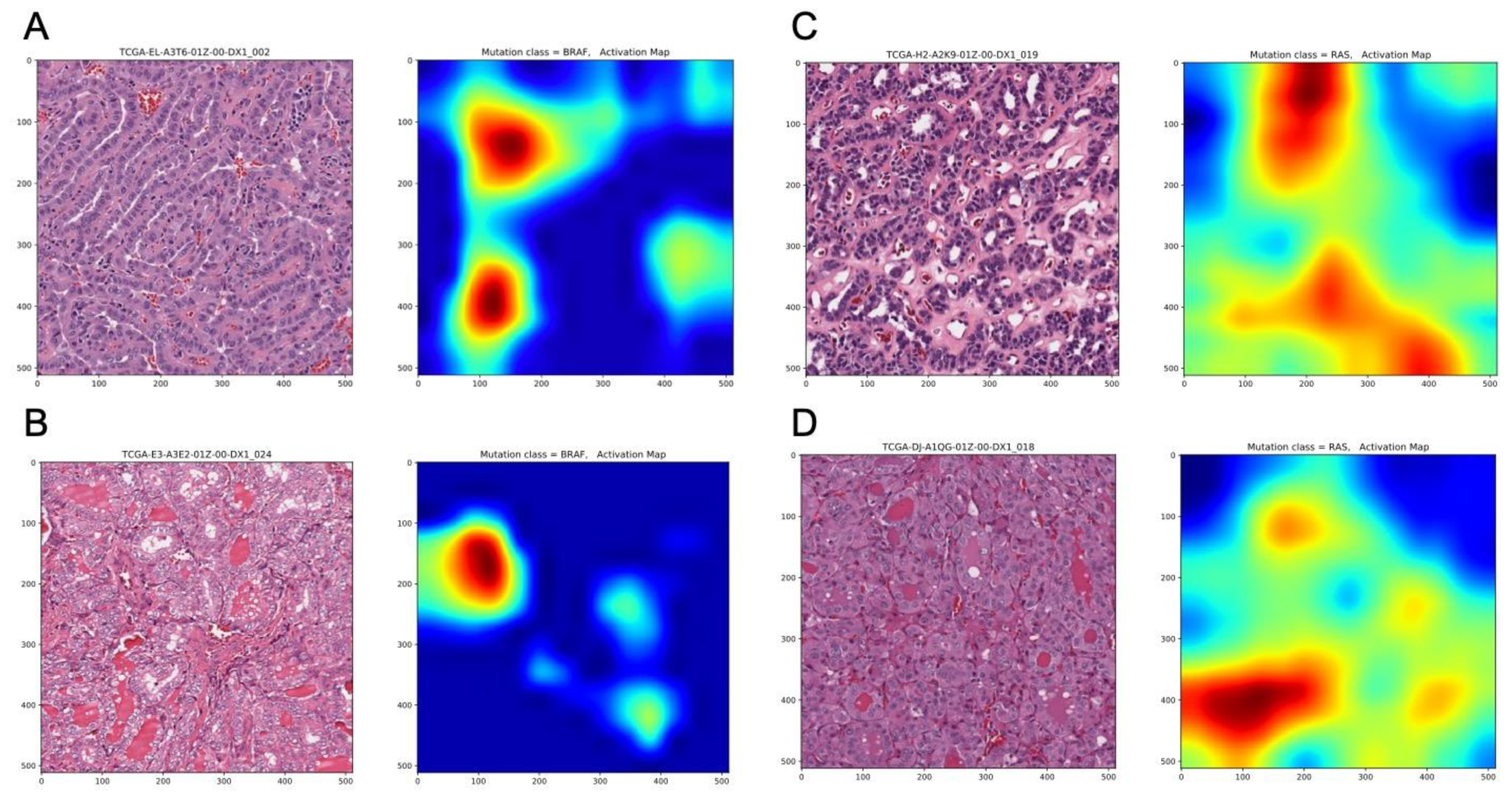
2.2. Deep-Learning Models Successfully Discriminate of BRAFV600E and RAS Mutation of Papillary Thyroid Carcinomas from Histological Images
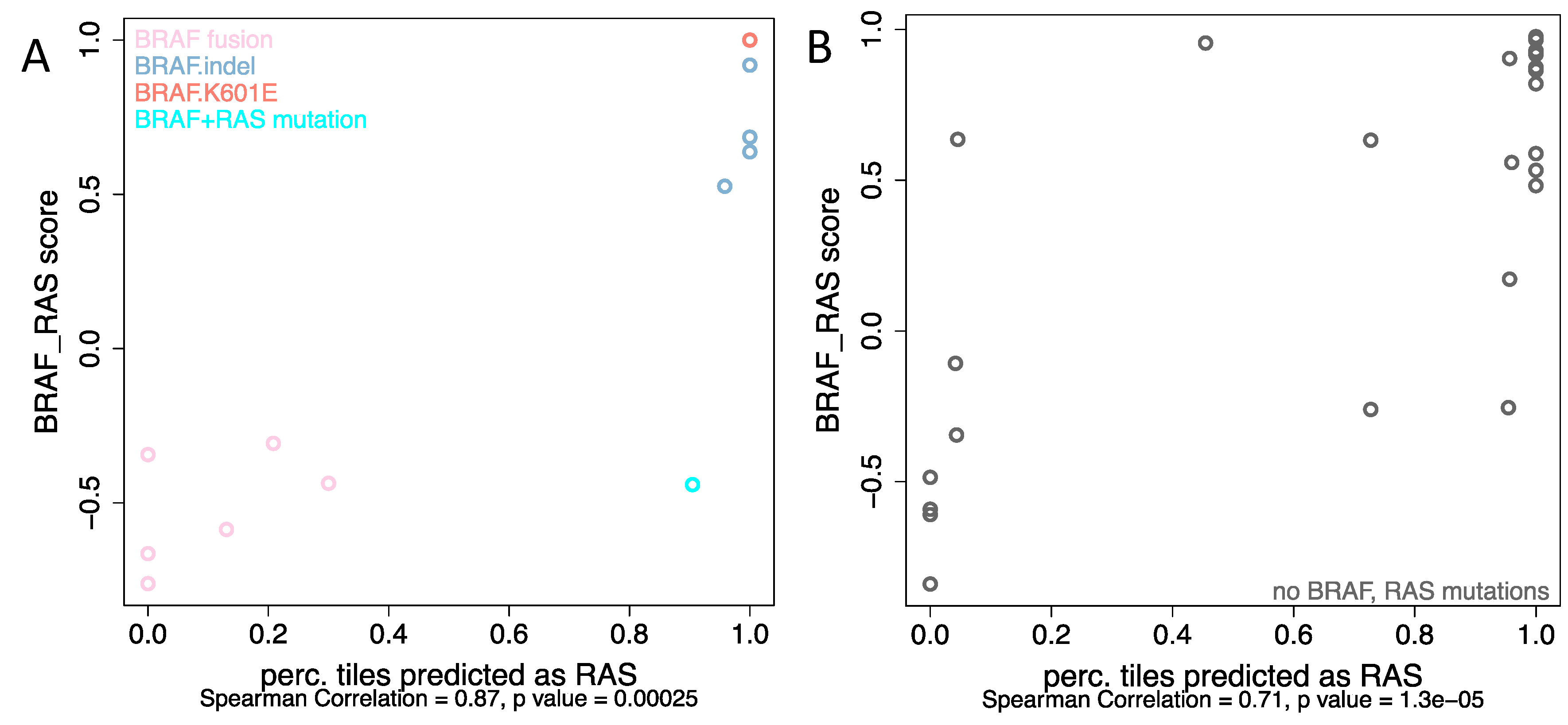
2.3. BRAF Mutations Other Than BRAFV600E Were Classified as RAS-Mutated while BRAF Fusions Were Classified as BRAFV600E Group, Respectively
2.4. Analyses from Histological Images Correlate with Molecular Expression for Tumors without BRAFV600E and RAS Mutations
3. Discussion
4. Methods
4.1. Sample Selection and Image Processing
4.2. Network Architecture and Training Procedures
4.3. Validation and Testing Procedures
4.4. Performance Evaluation
Metrics for Performance Evaluation of Algorithms
4.5. Hardware and Software
4.6. BRS Scores
4.7. Prediction on Additional PTC Tumor Images
4.8. Ethical Statement
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chen, A.Y.; Jemal, A.; Ward, E.M. Increasing incidence of differentiated thyroid cancer in the United States, 1988–2005. Cancer 2009, 115, 3801–3807. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.L.; Baloch, Z.; Bernet, V.; Chen, A.; Fahey, T.J., 3rd; Ganly, I.; Hodak, S.P.; Kebebew, E.; Patel, K.N.; Shaha, A.; et al. American Thyroid Association Statement on Surgical Application of Molecular Profiling for Thyroid Nodules: Current Impact on Perioperative Decision Making. Thyroid Off. J. Am. Thyroid Assoc. 2015, 25, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Hay, I.D.; McConahey, W.M.; Goellner, J.R. Managing patients with papillary thyroid carcinoma: Insights gained from the Mayo Clinic’s experience of treating 2,512 consecutive patients during 1940 through 2000. Trans. Am. Clin. Climatol. Assoc. 2002, 113, 241–260. [Google Scholar] [PubMed]
- Huang, Y.; Qu, S.; Zhu, G.; Wang, F.; Liu, R.; Shen, X.; Viola, D.; Elisei, R.; Puxeddu, E.; Fugazzola, L.; et al. BRAF V600E Mutation-Assisted Risk Stratification of Solitary Intrathyroidal Papillary Thyroid Cancer for Precision Treatment. J. Natl. Cancer Inst. 2018, 110, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Tallini, G.; de Biase, D.; Durante, C.; Acquaviva, G.; Bisceglia, M.; Bruno, R.; Bacchi Reggiani, M.L.; Casadei, G.P.; Costante, G.; Cremonini, N.; et al. BRAF V600E and risk stratification of thyroid microcarcinoma: A multicenter pathological and clinical study. Mod. Pathol. 2015, 28, 1343–1359. [Google Scholar] [CrossRef] [PubMed]
- Brose, M.S.; Cabanillas, M.E.; Cohen, E.E.; Wirth, L.J.; Riehl, T.; Yue, H.; Sherman, S.I.; Sherman, E.J. Vemurafenib in patients with BRAF(V600E)-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: A non-randomised, multicentre, open-label, phase 2 trial. Lancet. Oncol. 2016, 17, 1272–1282. [Google Scholar] [CrossRef]
- Jaber, T.; Waguespack, S.G.; Cabanillas, M.E.; Elbanan, M.; Vu, T.; Dadu, R.; Sherman, S.I.; Amit, M.; Santos, E.B.; Zafereo, M.; et al. Targeted Therapy in Advanced Thyroid Cancer to Resensitize Tumors to Radioactive Iodine. J. Clin. Endocrinol. Metab. 2018, 103, 3698–3705. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.B.; Cabanillas, M.E.; Lazar, A.J.; Williams, M.D.; Sanders, D.L.; Ilagan, J.L.; Nolop, K.; Lee, R.J.; Sherman, S.I. Clinical responses to vemurafenib in patients with metastatic papillary thyroid cancer harboring BRAF(V600E) mutation. Thyroid Off. J. Am. Thyroid Assoc. 2013, 23, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Howell, G.M.; Hodak, S.P.; Yip, L. RAS mutations in thyroid cancer. Oncologist 2013, 18, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Pratilas, C.A.; Taylor, B.S.; Ye, Q.; Viale, A.; Sander, C.; Solit, D.B.; Rosen, N. (V600E) BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. USA 2009, 106, 4519–4524. [Google Scholar] [CrossRef] [PubMed]
- Xing, M. BRAF mutation in thyroid cancer. Endocr. Relat. Cancer 2005, 12, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Durante, C.; Puxeddu, E.; Ferretti, E.; Morisi, R.; Moretti, S.; Bruno, R.; Barbi, F.; Avenia, N.; Scipioni, A.; Verrienti, A.; et al. BRAF mutations in papillary thyroid carcinomas inhibit genes involved in iodine metabolism. J. Clin. Endocrinol. Metab. 2007, 92, 2840–2843. [Google Scholar] [CrossRef] [PubMed]
- Elisei, R.; Ugolini, C.; Viola, D.; Lupi, C.; Biagini, A.; Giannini, R.; Romei, C.; Miccoli, P.; Pinchera, A.; Basolo, F. BRAF (V600E) mutation and outcome of patients with papillary thyroid carcinoma: A 15-year median follow-up study. J. Clin. Endocrinol. Metab. 2008, 93, 3943–3949. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.; Alzahrani, A.S.; Carson, K.A.; Viola, D.; Elisei, R.; Bendlova, B.; Yip, L.; Mian, C.; Vianello, F.; Tuttle, R.M.; et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA 2013, 309, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Velazquez Vega, J.E.; Brat, D.J.; Cooper, L.A.D. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979. [Google Scholar] [CrossRef] [PubMed]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyo, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Kim, W.Y.; Hwang, T.S.; Lee, S.S.; Kim, H.; Han, H.S.; Lim, S.D.; Kim, W.S.; Yoo, Y.B.; Park, K.S. BRAF and RAS mutations in follicular variants of papillary thyroid carcinoma. Endocr. Pathol. 2013, 24, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.; Weigelt, B.; Wen, H.C.; Pareja, F.; Raghavendra, A.; Martelotto, L.G.; Burke, K.A.; Basili, T.; Li, A.; Geyer, F.C.; et al. IDH2 Mutations Define a Unique Subtype of Breast Cancer with Altered Nuclear Polarity. Cancer Res. 2016, 76, 7118–7129. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, P.; Kazemi, E.; Imielinski, M.; Elemento, O.; Hajirasouliha, I. Deep Convolutional Neural Networks Enable Discrimination of Heterogeneous Digital Pathology Images. EBioMedicine 2018, 27, 317–328. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsou, P.; Wu, C.-J. Mapping Driver Mutations to Histopathological Subtypes in Papillary Thyroid Carcinoma: Applying a Deep Convolutional Neural Network. J. Clin. Med. 2019, 8, 1675. https://doi.org/10.3390/jcm8101675
Tsou P, Wu C-J. Mapping Driver Mutations to Histopathological Subtypes in Papillary Thyroid Carcinoma: Applying a Deep Convolutional Neural Network. Journal of Clinical Medicine. 2019; 8(10):1675. https://doi.org/10.3390/jcm8101675
Chicago/Turabian StyleTsou, Peiling, and Chang-Jiun Wu. 2019. "Mapping Driver Mutations to Histopathological Subtypes in Papillary Thyroid Carcinoma: Applying a Deep Convolutional Neural Network" Journal of Clinical Medicine 8, no. 10: 1675. https://doi.org/10.3390/jcm8101675
APA StyleTsou, P., & Wu, C.-J. (2019). Mapping Driver Mutations to Histopathological Subtypes in Papillary Thyroid Carcinoma: Applying a Deep Convolutional Neural Network. Journal of Clinical Medicine, 8(10), 1675. https://doi.org/10.3390/jcm8101675




