The Impact of Moyamoya Disease and RNF213 Mutations on the Spectrum of Plasma Protein and MicroRNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Patient Information
2.2. Plasma Preparation and Extracellular Vesicle (EV) Isolation
2.3. Isolation of RNA and Small RNA Sequencing Library Construction
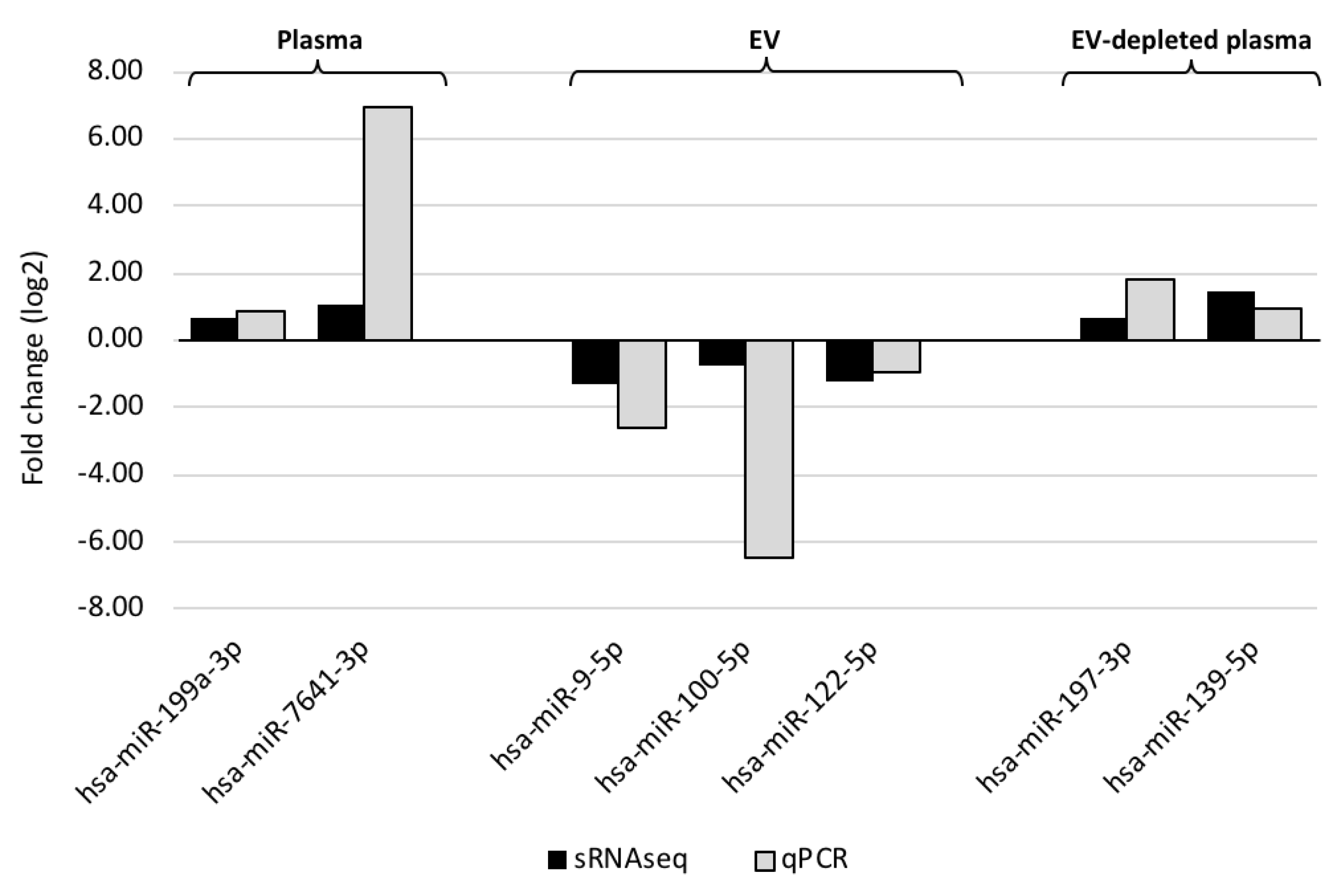
2.4. Small RNAseq Data Analysis and Verification
2.5. Plasma Protein Analysis
2.6. Functional Enrichment and Network Analyses
3. Results
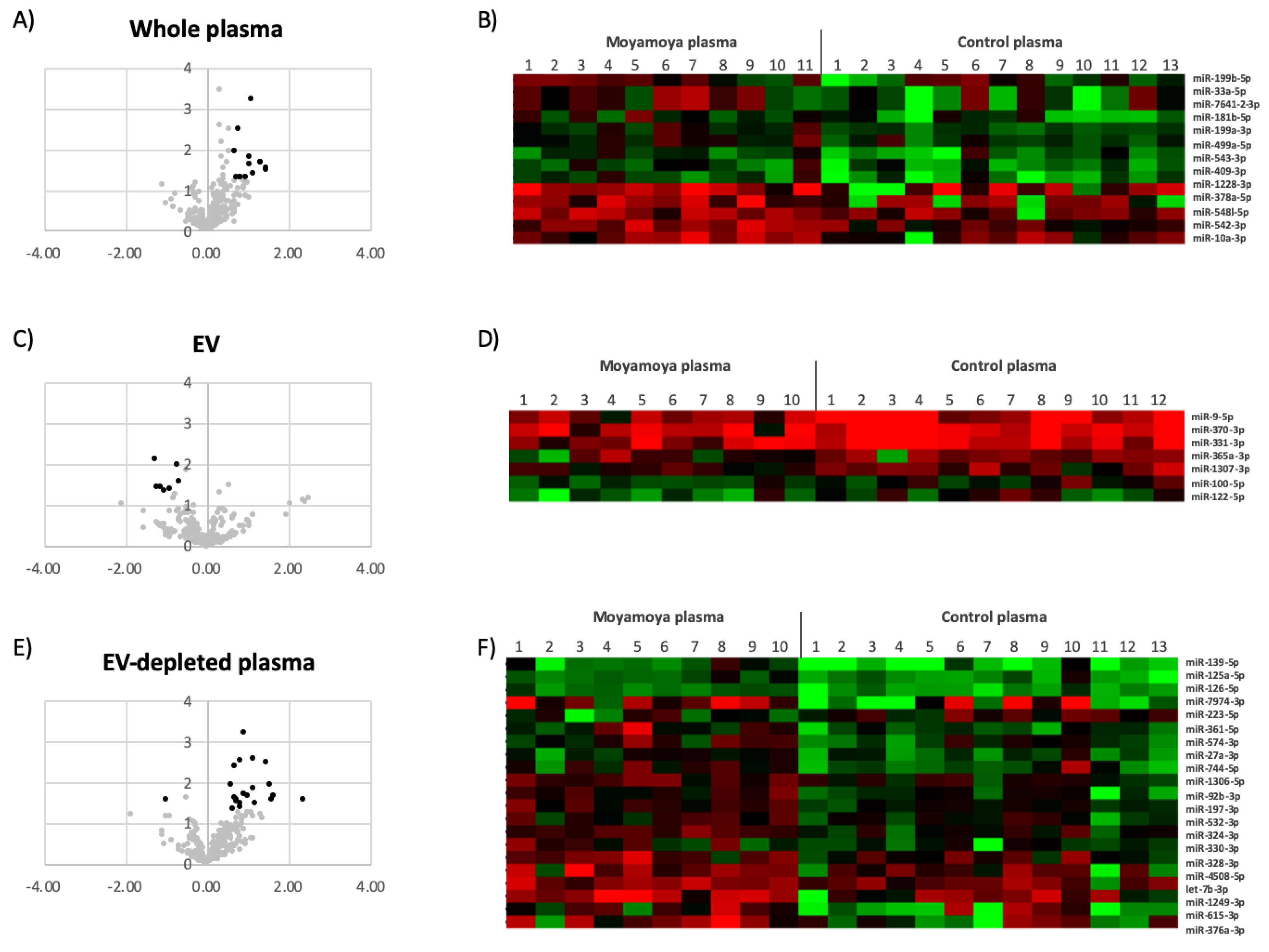
3.1. Characterization of miRNA in Whole Plasma, EVs and EV-Depleted Plasma
3.2. Circulating miRNAs Associated with MMD
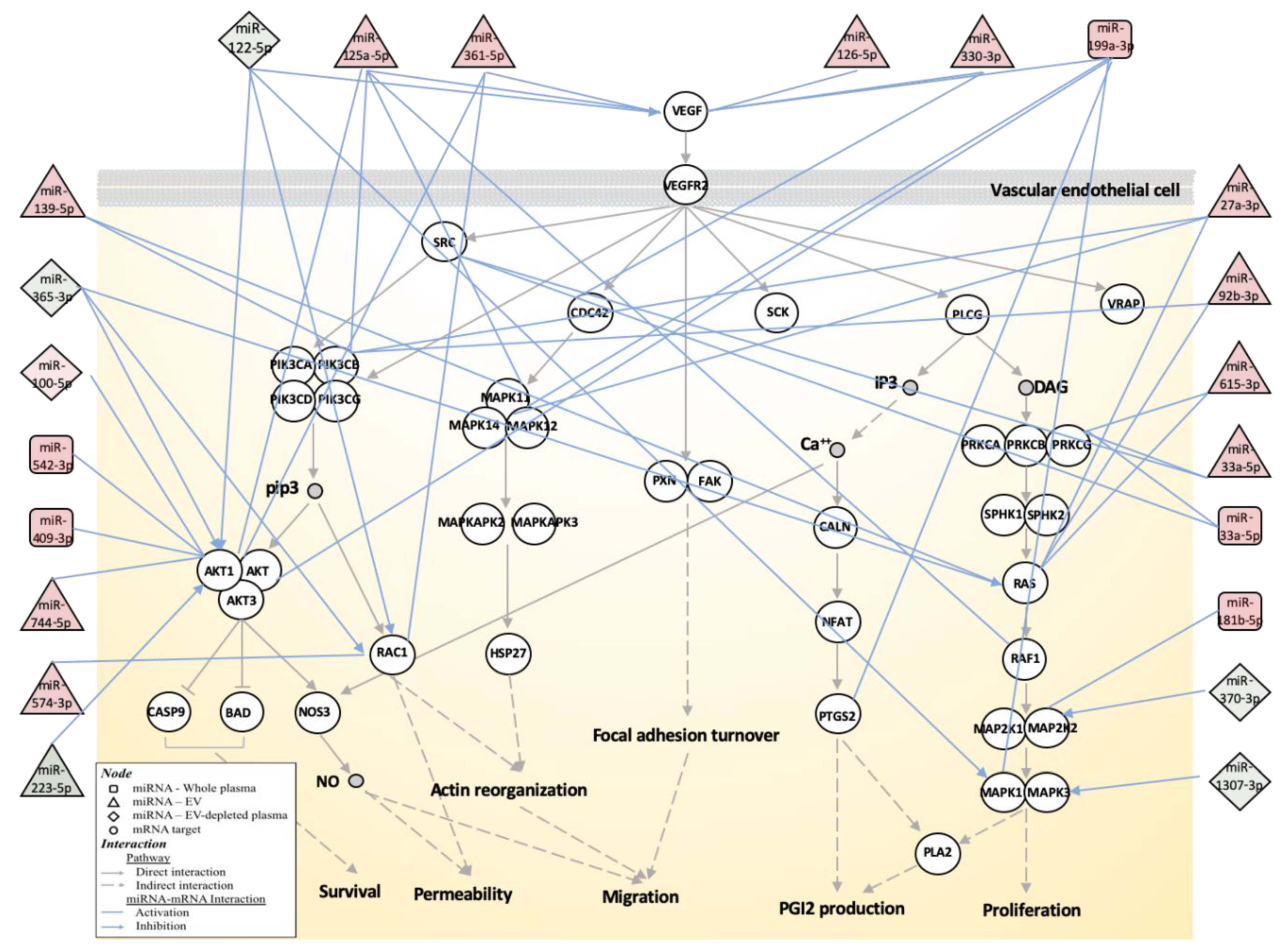
3.3. Biological Function Associated with MMD-Affected miRNAs
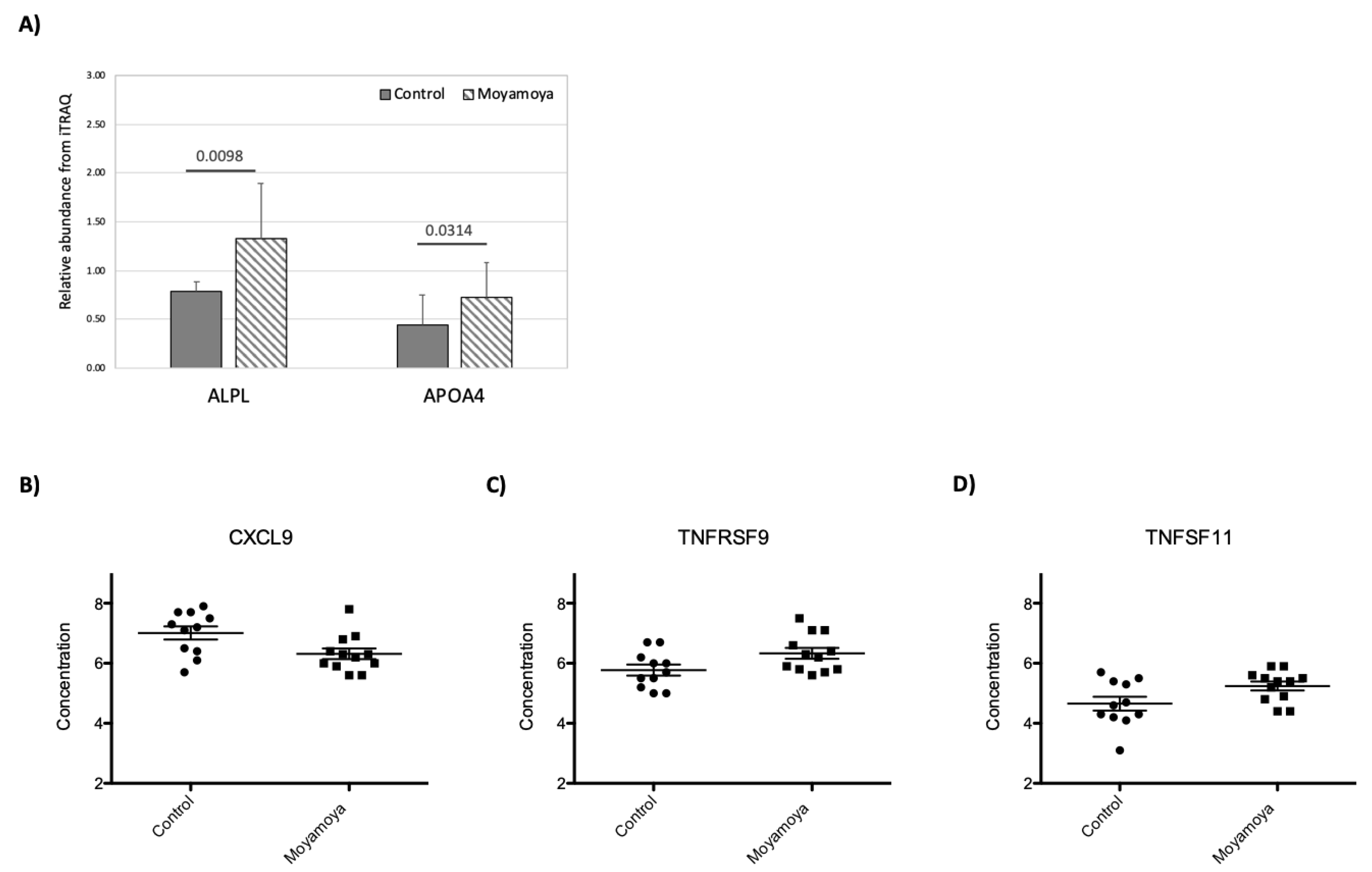
3.4. Impact of MMD on Plasma Proteome
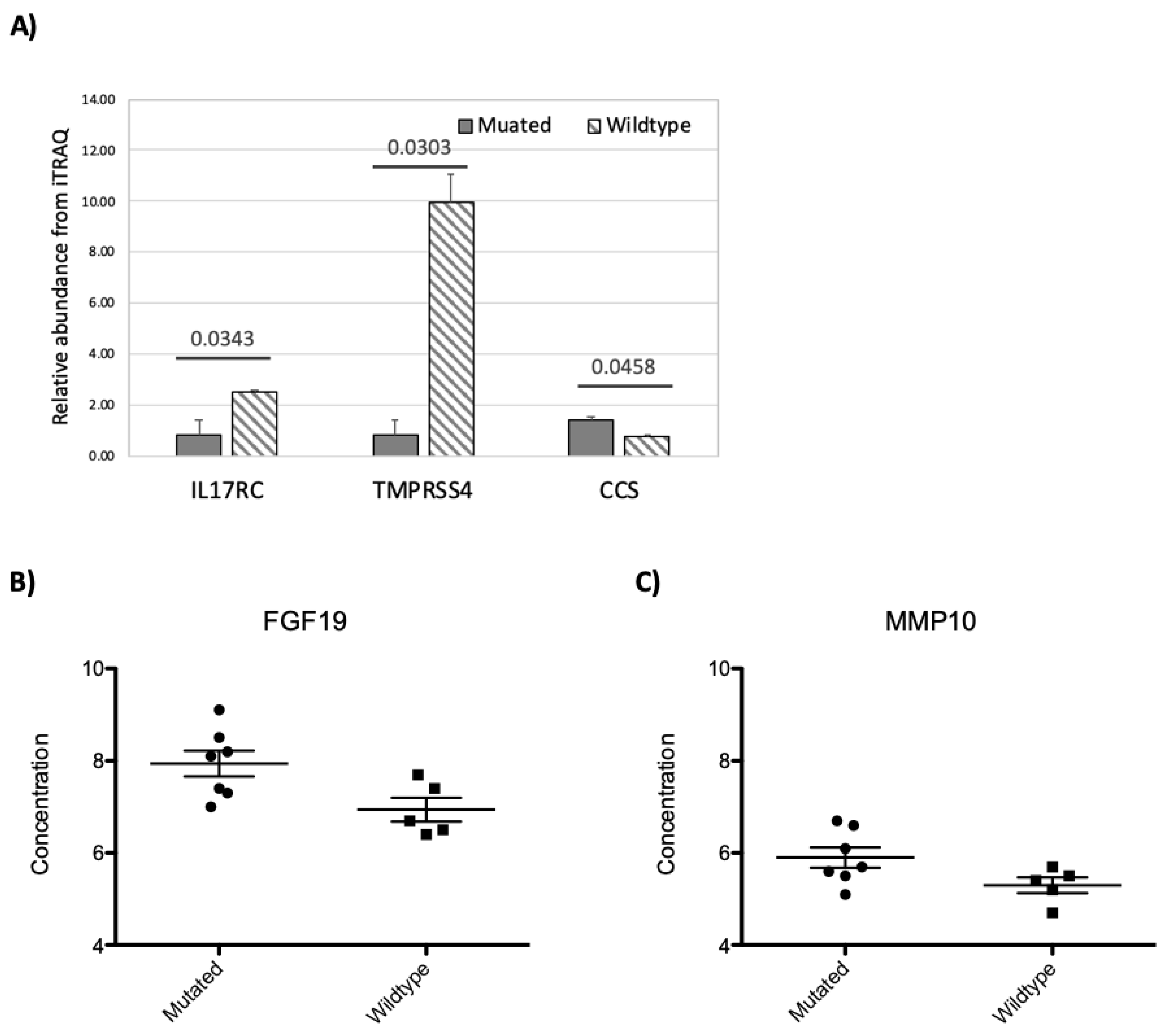
3.5. RNF213 Mutation Affects Plasma Protein and miRNA Concentrations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Suzuki, J.; Takaku, A. Cerebrovascular “moyamoya” disease. Disease showing abnormal net-like vessels in base of brain. Arch. Neurol. 1969, 20, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Masuda, J.; Ogata, J.; Yutani, C. Smooth muscle cell proliferation and localization of macrophages and T cells in the occlusive intracranial major arteries in moyamoya disease. Stroke 1993, 24, 1960–1967. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Oka, K.; Tanaka, K. Histopathology of the brain vascular network in moyamoya disease. Stroke 1983, 14, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, J.; Kodama, N. Moyamoya disease–A review. Stroke 1983, 14, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.C.; Miyamoto, S. Moyamoya disease: Recent progress and outlook. Neurol. Med. Chir. 2010, 50, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Kamada, F.; Aoki, Y.; Narisawa, A.; Abe, Y.; Komatsuzaki, S.; Kikuchi, A.; Kanno, J.; Niihori, T.; Ono, M.; Ishii, N.; et al. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J. Hum. Genet. 2011, 56, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Morito, D.; Takashima, S.; Mineharu, Y.; Kobayashi, H.; Hitomi, T.; Hashikata, H.; Matsuura, N.; Yamazaki, S.; Toyoda, A.; et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE 2011, 6, e22542. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.C.; Papke, C.L.; Tran-Fadulu, V.; Regalado, E.S.; Avidan, N.; Johnson, R.J.; Kim, D.H.; Pannu, H.; Willing, M.C.; Sparks, E.; et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease. Am. J. Hum. Genet. 2009, 84, 617–627. [Google Scholar] [CrossRef]
- Hojo, M.; Hoshimaru, M.; Miyamoto, S.; Taki, W.; Nagata, I.; Asahi, M.; Matsuura, N.; Ishizaki, R.; Kikuchi, H.; Hashimoto, N. Role of transforming growth factor-beta1 in the pathogenesis of moyamoya disease. J. Neurosurg. 1998, 89, 623–629. [Google Scholar] [CrossRef]
- Kang, H.S.; Kim, J.H.; Phi, J.H.; Kim, Y.Y.; Kim, J.E.; Wang, K.C.; Cho, B.K.; Kim, S.K. Plasma matrix metalloproteinases, cytokines and angiogenic factors in moyamoya disease. J. Neurol. Neurosurg. Psychiatry 2010, 81, 673–678. [Google Scholar] [CrossRef]
- Malek, A.M.; Connors, S.; Robertson, R.L.; Folkman, J.; Scott, R.M. Elevation of cerebrospinal fluid levels of basic fibroblast growth factor in moyamoya and central nervous system disorders. Pediatr. Neurosurg. 1997, 27, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Rafat, N.; Beck, G.; Pena-Tapia, P.G.; Schmiedek, P.; Vajkoczy, P. Increased levels of circulating endothelial progenitor cells in patients with Moyamoya disease. Stroke 2009, 40, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Takagi, Y.; Kikuta, K.; Nozaki, K.; Fujimoto, M.; Hayashi, J.; Imamura, H.; Hashimoto, N. Expression of hypoxia-inducing factor-1 alpha and endoglin in intimal hyperplasia of the middle cerebral artery of patients with Moyamoya disease. Neurosurgery 2007, 60, 338–345; discussion 345. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, T.; Houkin, K.; Takahashi, A.; Abe, H. Angiogenic factors in moyamoya disease. Stroke 1996, 27, 2160–2165. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407. [Google Scholar] [CrossRef] [PubMed]
- Callis, T.E.; Pandya, K.; Seok, H.Y.; Tang, R.H.; Tatsuguchi, M.; Huang, Z.P.; Chen, J.F.; Deng, Z.; Gunn, B.; Shumate, J.; et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Investig. 2009, 119, 2772–2786. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Jiao, J.Q.; Li, Q.; Long, B.; Wang, K.; Liu, J.P.; Li, Y.R.; Li, P.F. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat. Med. 2011, 17, 71–78. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are Transported in Plasma and Delivered to Recipient Cells by High-Density Lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef]
- Almeida, M.I.; Reis, R.M.; Calin, G.A. MicroRNA history: Discovery, recent applications, and next frontiers. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2011, 717, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Siriwardena, A.K.; Mason, J.M.; Mullamitha, S.; Hancock, H.C.; Jegatheeswaran, S. Management of colorectal cancer presenting with synchronous liver metastases. Nat. Rev. Clin. Oncol. 2014, 11, 446–459. [Google Scholar] [CrossRef] [PubMed]
- Tetta, C.; Ghigo, E.; Silengo, L.; Deregibus, M.C.; Camussi, G. Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine 2013, 44, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, S.; Gullberg, M.; Jarvius, J.; Olsson, C.; Pietras, K.; Gustafsdottir, S.M.; Ostman, A.; Landegren, U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002, 20, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Chen, Y.F.; Fan, P.C.; Wang, K.C.; Wang, K.; Wang, J.; Kuo, M.F. Mutation genotypes of RNF213 gene from moyamoya patients in Taiwan. J. Neurol. Sci. 2015, 353, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Fallen, S.; Baxter, D.; Wu, X.; Kim, T.K.; Shynlova, O.; Lee, M.Y.; Scherler, K.; Lye, S.; Hood, L.; Wang, K. Extracellular vesicle RNAs reflect placenta dysfunction and are a biomarker source for preterm labour. J. Cell. Mol. Med. 2018, 22, 2760–2773. [Google Scholar] [CrossRef] [PubMed]
- Etheridge, A.; Wang, K.; Baxter, D.; Galas, D. Preparation of Small RNA NGS Libraries from Biofluids. Methods Mol. Biol. 2018, 1740, 163–175. [Google Scholar] [PubMed]
- Wu, X.; Kim, T.K.; Baxter, D.; Scherler, K.; Gordon, A.; Fong, O.; Etheridge, A.; Galas, D.J.; Wang, K. sRNAnalyzer-a flexible and customizable small RNA sequencing data analysis pipeline. Nucleic Acids Res. 2017, 45, 12140–12151. [Google Scholar] [CrossRef]
- Geer, L.Y.; Markey, S.P.; Kowalak, J.A.; Wagner, L.; Xu, M.; Maynard, D.M.; Yang, X.; Shi, W.; Bryant, S.H. Open mass spectrometry search algorithm. J. Proteome Res. 2004, 3, 958–964. [Google Scholar] [CrossRef]
- Chou, C.H.; Chang, N.W.; Shrestha, S.; Hsu, S.D.; Lin, Y.L.; Lee, W.H.; Yang, C.D.; Hong, H.C.; Wei, T.Y.; Tu, S.J.; et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016, 44, D239–D247. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G. Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef] [PubMed]
- Morito, D.; Nishikawa, K.; Hoseki, J.; Kitamura, A.; Kotani, Y.; Kiso, K.; Kinjo, M.; Fujiyoshi, Y.; Nagata, K. Moyamoya disease-associated protein mysterin/RNF213 is a novel AAA+ ATPase, which dynamically changes its oligomeric state. Sci. Rep. 2014, 4, 4442. [Google Scholar] [CrossRef]
- Scholz, B.; Korn, C.; Wojtarowicz, J.; Mogler, C.; Augustin, I.; Boutros, M.; Niehrs, C.; Augustin, H.G. Endothelial RSPO3 Controls Vascular Stability and Pruning through Non-canonical WNT/Ca(2+)/NFAT Signaling. Dev. Cell 2016, 36, 79–93. [Google Scholar] [CrossRef]
- Ohkubo, K.; Sakai, Y.; Inoue, H.; Akamine, S.; Ishizaki, Y.; Matsushita, Y.; Sanefuji, M.; Torisu, H.; Ihara, K.; Sardiello, M.; et al. Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells. Sci. Rep. 2015, 5, 13191. [Google Scholar] [CrossRef]
- Dai, D.; Lu, Q.; Huang, Q.; Yang, P.; Hong, B.; Xu, Y.; Zhao, W.; Liu, J.; Li, Q. Serum miRNA signature in Moyamoya disease. PLoS ONE 2014, 9, e102382. [Google Scholar] [CrossRef]
- Uchino, H.; Ito, M.; Kazumata, K.; Hama, Y.; Hamauchi, S.; Terasaka, S.; Sasaki, H.; Houkin, K. Circulating miRNome profiling in Moyamoya disease-discordant monozygotic twins and endothelial microRNA expression analysis using iPS cell line. BMC Med. Genom. 2018, 11, 72. [Google Scholar] [CrossRef]
- Dewdney, B.; Trollope, A.; Moxon, J.; Thomas Manapurathe, D.; Biros, E.; Golledge, J. Circulating MicroRNAs as Biomarkers for Acute Ischemic Stroke: A Systematic Review. J. Stroke Cereb. Dis. 2018, 27, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Sepramaniam, S.; Tan, J.R.; Tan, K.S.; DeSilva, D.A.; Tavintharan, S.; Woon, F.P.; Wang, C.W.; Yong, F.L.; Karolina, D.S.; Kaur, P.; et al. Circulating microRNAs as biomarkers of acute stroke. Int. J. Mol. Sci. 2014, 15, 1418–1432. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Morimoto, T.; Kamatani, Y.; Kamimura, T.; Kobayashi, H.; Harada, K.; Tomita, T.; Higashiyama, A.; Takahashi, J.C.; Nakagawara, J.; et al. Moyamoya Disease Susceptibility Variant RNF213 p.R4810K Increases the Risk of Ischemic Stroke Attributable to Large-Artery Atherosclerosis. Circulation 2019, 139, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Yasmeen, S.; Kaur, S.; Mirza, A.H.; Brodin, B.; Pociot, F.; Kruuse, C.J.M.N. miRNA-27a-3p and miRNA-222-3p as Novel Modulators of Phosphodiesterase 3a (PDE3A) in Cerebral Microvascular Endothelial Cells. Mol. Neurobiol. 2019, 56, 5304–5314. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Ji, Y.; Peng, J.; Zhou, X.; Chen, X.; Zhao, H.; Xu, T.; Chen, L.; Xu, Y. Increased Brain-Specific MiR-9 and MiR-124 in the Serum Exosomes of Acute Ischemic Stroke Patients. PLoS ONE 2016, 11, e0163645. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, S.S.; Nygaard, A.B.; Carlsen, A.L.; Heegaard, N.H.H.; Bak, M.; Christensen, T.J.B.R. Elevation of brain-enriched miRNAs in cerebrospinal fluid of patients with acute ischemic stroke. Biomark. Res. 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.; Cao, X.; Han, L.; Zhao, H.; Qiu, S.; Yan, Y.; Wang, X.; Chen, X.; Zheng, W.; Xu, X.; et al. Association of increased Treg and Th17 with pathogenesis of moyamoya disease. Sci. Rep. 2017, 7, 3071. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, A.; Kobayashi, H.; Liu, W.; Fujii, Y.; Senevirathna, S.T.; Nanayakkara, S.; Okuda, H.; Hitomi, T.; Harada, K.H.; Takenaka, K.; et al. P.R4810K, a polymorphism of RNF213, the susceptibility gene for moyamoya disease, is associated with blood pressure. Environ. Health Prev. Med. 2013, 18, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Nam, T.M.; Jo, K.I.; Yeon, J.Y.; Hong, S.C.; Kim, J.S. Coronary heart disease in moyamoya disease: Are they concomitant or coincidence? J. Korean Med. Sci. 2015, 30, 470–474. [Google Scholar] [CrossRef]
- Morimoto, T.; Mineharu, Y.; Ono, K.; Nakatochi, M.; Ichihara, S.; Kabata, R.; Takagi, Y.; Cao, Y.; Zhao, L.; Kobayashi, H.; et al. Significant association of RNF213 p.R4810K, a moyamoya susceptibility variant, with coronary artery disease. PLoS ONE 2017, 12, e0175649. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [PubMed]
- Frank, F.; Sonenberg, N.; Nagar, B. Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010, 465, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.J.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
| Category (mean) | Whole Plasma | Extracellular Vesicle (EV) | EV-Depleted Plasma | |||
|---|---|---|---|---|---|---|
| Moyamoya | Control | Moyamoya | Control | Moyamoya | Control | |
| Raw read count | 8,275,547 | 8,843,102 | 7,771,542 | 8,907,214 | 8,160,659 | 7,942,143 |
| Trimmed read count | 4,962,493 | 5,229,659 | 4,134,795 | 4,165,114 | 4,232,272 | 5,505,643 |
| Mapped reads | 686,951 | 672,717 | 64,844 | 68,817 | 191,987 | 497,845 |
| Observed miRNA (at least one mapped read) | 598 | 574 | 384 | 366 | 467 | 497 |
| Detected miRNA (at least 10 mapped reads) | 333 | 333 | 225 | 233 | 234 | 276 |
| Sample Type | miRNA ID | Moyamoya/Control | |
|---|---|---|---|
| Concentration Change (log 2) | p-Value | ||
| Whole Plasma | hsa-miR-10a-3p | 1.12 | 0.039138 |
| hsa-miR-1228-3p | 1.44 | 0.034762 | |
| hsa-miR-181b-5p | 0.76 | 0.003383 | |
| hsa-miR-199a-3p | 0.70 | 0.011899 | |
| hsa-miR-199b-5p | 0.80 | 0.049975 | |
| hsa-miR-33a-5p | 1.32 | 0.022562 | |
| hsa-miR-378a-5p | 1.47 | 0.030569 | |
| hsa-miR-409-3p | 1.03 | 0.016035 | |
| hsa-miR-499a-5p | 0.95 | 0.049400 | |
| hsa-miR-542-3p | 1.09 | 0.000614 | |
| hsa-miR-543-3p | 0.73 | 0.049719 | |
| hsa-miR-548l-5p | 0.84 | 0.049396 | |
| hsa-miR-7641-3p | 1.06 | 0.024955 | |
| EV | hsa-miR-100-5p | −0.73 | 0.009984 |
| hsa-miR-122-5p | −1.21 | 0.037025 | |
| hsa-miR-1307-3p | −0.70 | 0.027819 | |
| hsa-miR-331-3p | −0.91 | 0.042423 | |
| hsa-miR-365a-3p | −1.03 | 0.044760 | |
| hsa-miR-370-3p | −1.13 | 0.036237 | |
| hsa-miR-9-5p | −1.28 | 0.007390 | |
| EV-Depleted Plasma | hsa-let-7b-3p | 0.71 | 0.032790 |
| hsa-miR-1249-3p | 1.53 | 0.012610 | |
| hsa-miR-125a-5p | 0.71 | 0.024496 | |
| hsa-miR-126-5p | 0.60 | 0.012774 | |
| hsa-miR-1306-5p | 0.82 | 0.003148 | |
| hsa-miR-139-5p | 1.47 | 0.003326 | |
| hsa-miR-197-3p | 0.69 | 0.004247 | |
| hsa-miR-27a-3p | 0.74 | 0.028641 | |
| hsa-miR-324-3p | 0.90 | 0.000670 | |
| hsa-miR-328-3p | 0.81 | 0.036318 | |
| hsa-miR-33a-5p | 2.06 | 0.022378 | |
| hsa-miR-330-3p | 0.85 | 0.045555 | |
| hsa-miR-361-5p | 1.12 | 0.014715 | |
| hsa-miR-376a-3p | 1.64 | 0.023245 | |
| hsa-miR-4508-5p | 1.21 | 0.034114 | |
| hsa-miR-532-3p | 0.64 | 0.048844 | |
| hsa-miR-574-3p | 0.93 | 0.020531 | |
| hsa-miR-615-3p | 1.59 | 0.027183 | |
| hsa-miR-744-5p | 1.01 | 0.023024 | |
| hsa-miR-7974-3p | 2.37 | 0.028771 | |
| hsa-miR-92b-3p | 1.13 | 0.002922 | |
| hsa-miR-223-5p | −1.01 | 0.027949 | |
| Sample Type | miRNA ID | Mutated/Wildtype | |
|---|---|---|---|
| Concentration Difference (log 2) | p-Value | ||
| Whole Plasma | hsa-miR-15b-5p | 0.68 | 0.021039 |
| hsa-miR-17-3p | 0.66 | 0.023532 | |
| hsa-miR-101-5p | 0.74 | 0.006891 | |
| hsa-miR-576-5p | 0.70 | 0.027591 | |
| hsa-miR-766-3p | 1.28 | 0.000960 | |
| hsa-miR-628-3p | −0.88 | 0.036486 | |
| EV | hsa-miR-25-3p | 0.93 | 0.034067 |
| hsa-miR-145-5p | 2.35 | 0.018981 | |
| hsa-miR-148a-3p | 0.76 | 0.024250 | |
| hsa-miR-186-5p | 0.70 | 0.033598 | |
| hsa-miR-188-5p | 0.96 | 0.044339 | |
| hsa-miR-200c-3p | 0.89 | 0.043478 | |
| hsa-miR-330-3p | 0.71 | 0.001653 | |
| hsa-miR-423-3p | 1.04 | 0.033586 | |
| hsa-miR-484-5p | 0.78 | 0.032059 | |
| hsa-miR-486-5p | 0.83 | 0.042868 | |
| hsa-miR-652-3p | 0.94 | 0.022147 | |
| hsa-miR-144-3p | −0.94 | 0.003342 | |
| hsa-miR-190a-5p | −1.45 | 0.031924 | |
| EV-Depleted Plasma | hsa-miR-22-3p | 0.76 | 0.008363 |
| hsa-miR-320b-3p | 0.94 | 0.009862 | |
| hsa-miR-7641-3p | 2.70 | 0.027610 | |
| hsa-miR-30a-5p | −0.68 | 0.016327 | |
| hsa-miR-125b-5p | −1.03 | 0.017852 | |
| hsa-miR-224-5p | −1.43 | 0.004563 | |
| hsa-miR-421-3p | −1.85 | 0.047318 | |
| hsa-miR-433-3p | −2.43 | 0.046585 | |
| hsa-miR-4532-5p | −2.45 | 0.049418 | |
| hsa-miR-574-3p | −0.92 | 0.048682 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.-J.; Fallen, S.; Zhou, Y.; Baxter, D.; Scherler, K.; Kuo, M.-F.; Wang, K. The Impact of Moyamoya Disease and RNF213 Mutations on the Spectrum of Plasma Protein and MicroRNA. J. Clin. Med. 2019, 8, 1648. https://doi.org/10.3390/jcm8101648
Lee M-J, Fallen S, Zhou Y, Baxter D, Scherler K, Kuo M-F, Wang K. The Impact of Moyamoya Disease and RNF213 Mutations on the Spectrum of Plasma Protein and MicroRNA. Journal of Clinical Medicine. 2019; 8(10):1648. https://doi.org/10.3390/jcm8101648
Chicago/Turabian StyleLee, Ming-Jen, Shannon Fallen, Yong Zhou, David Baxter, Kelsey Scherler, Meng-Fai Kuo, and Kai Wang. 2019. "The Impact of Moyamoya Disease and RNF213 Mutations on the Spectrum of Plasma Protein and MicroRNA" Journal of Clinical Medicine 8, no. 10: 1648. https://doi.org/10.3390/jcm8101648
APA StyleLee, M.-J., Fallen, S., Zhou, Y., Baxter, D., Scherler, K., Kuo, M.-F., & Wang, K. (2019). The Impact of Moyamoya Disease and RNF213 Mutations on the Spectrum of Plasma Protein and MicroRNA. Journal of Clinical Medicine, 8(10), 1648. https://doi.org/10.3390/jcm8101648





