The Genetics of Polycystic Ovary Syndrome: An Overview of Candidate Gene Systematic Reviews and Genome-Wide Association Studies
Abstract
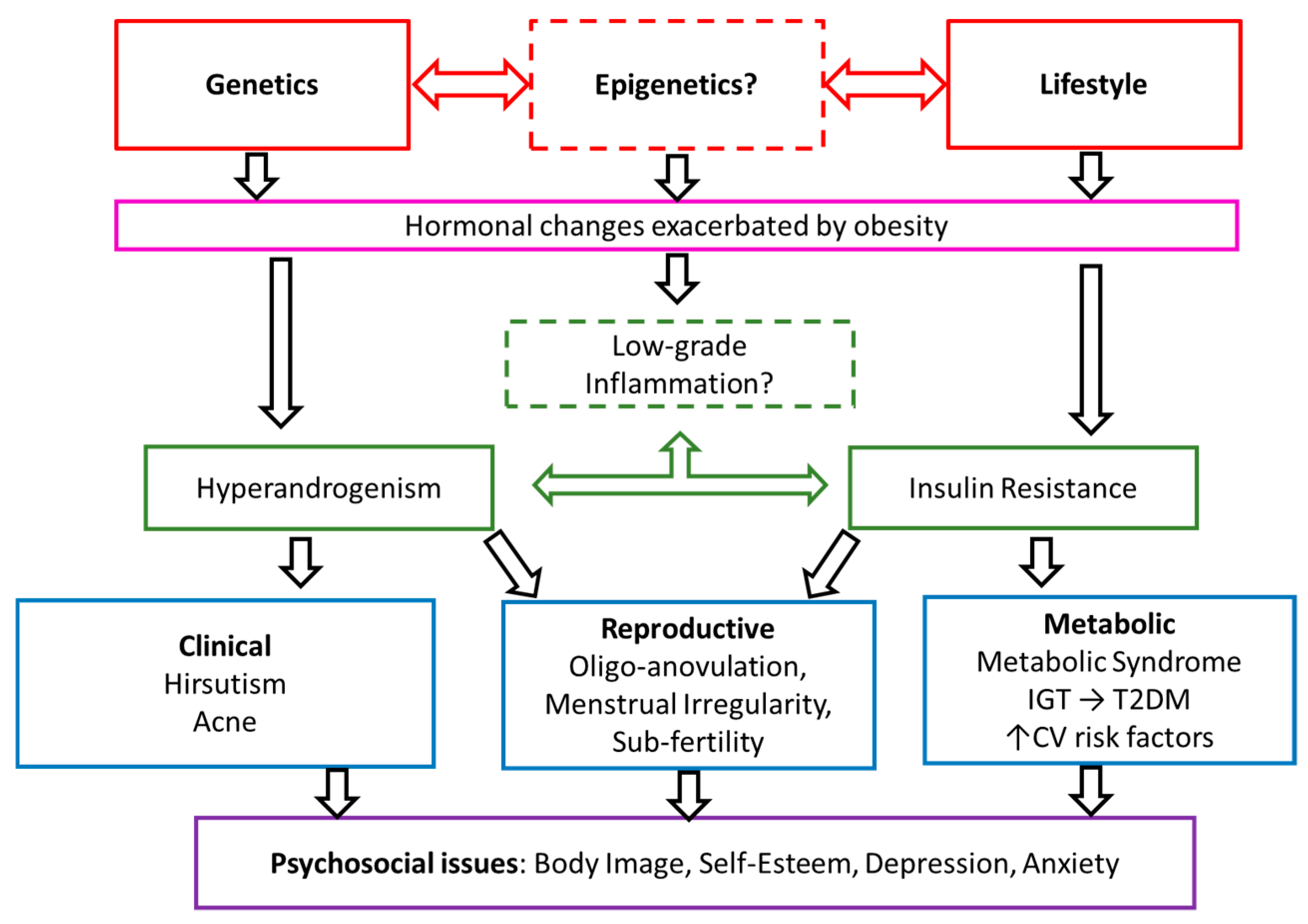
1. Introduction
2. Experimental Section
2.1. Protocol and Registration
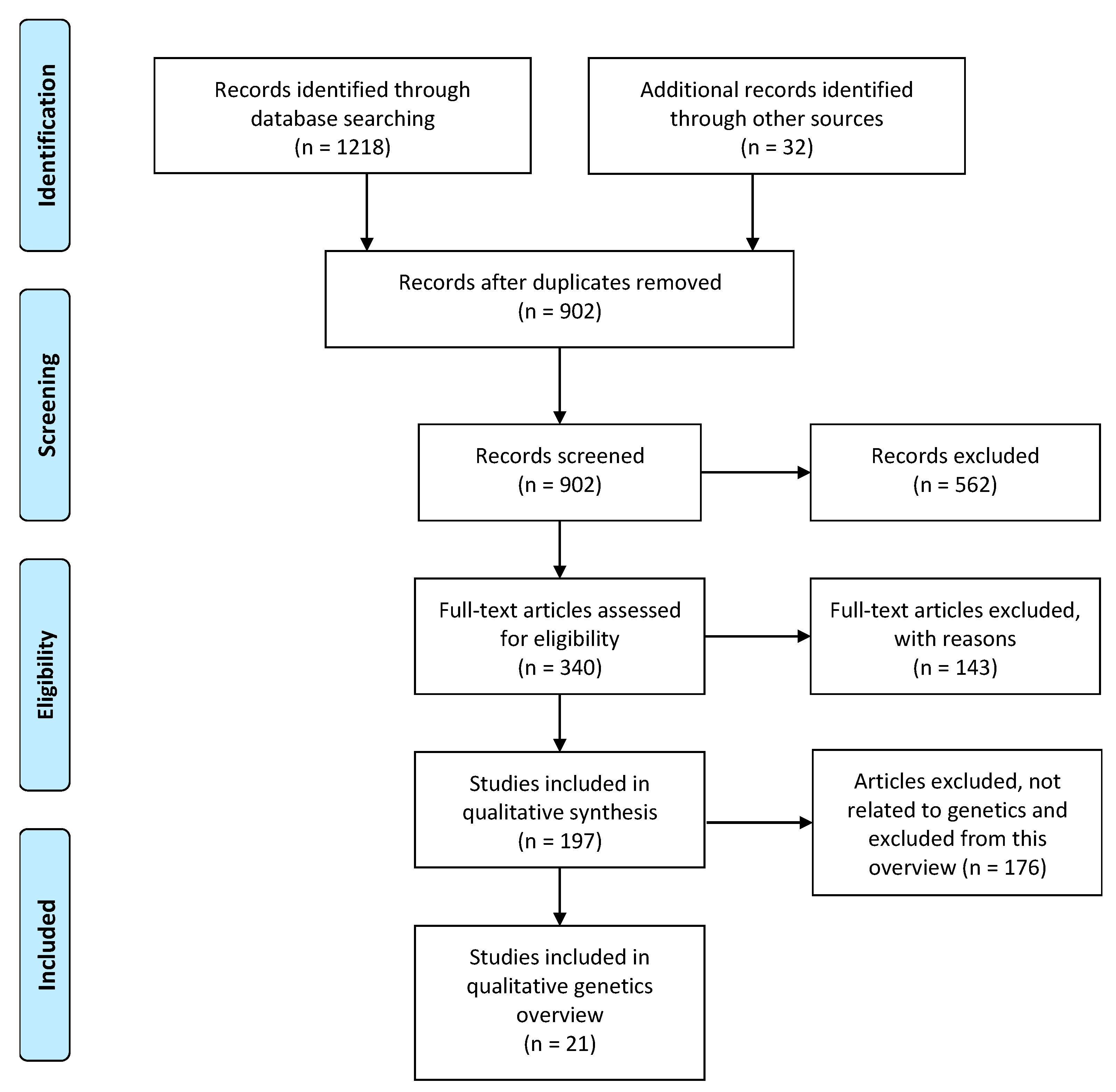
2.2. Systematic Search of Candidate Gene Systematic Review
2.2.1. Literature Search
2.2.2. Eligibility Criteria and Inclusion Criteria
2.2.3. Data Extraction
2.2.4. Quality Assessment of Systematic Reviews
2.3. Semi-Structured Search of Genome-Wide Association Studies (GWAS)
3. Results
3.1. Candidate Gene Systematic Reviews
3.1.1. Eligibility Assessment
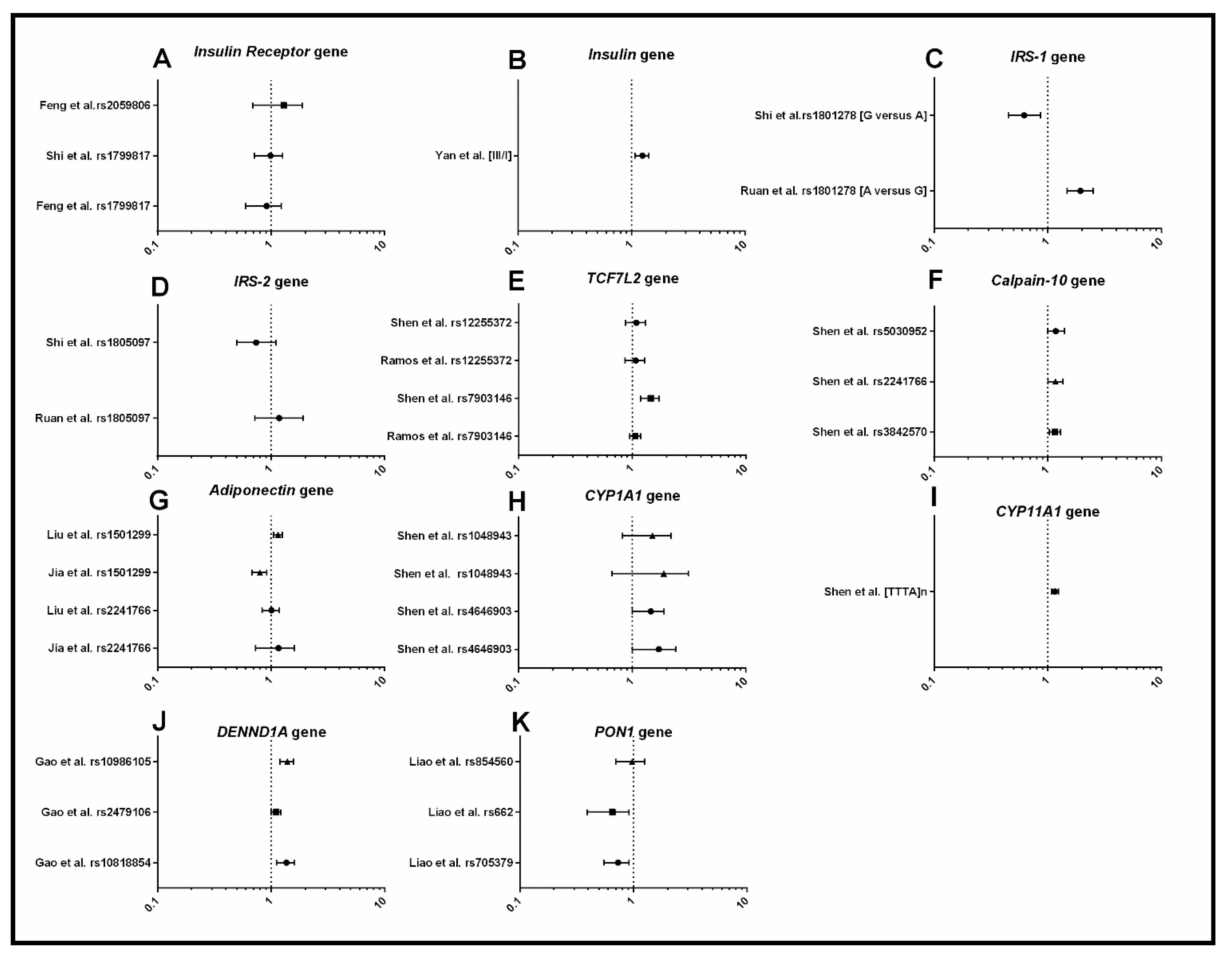
3.1.2. Metabolism
3.1.3. Androgens and Gonadotrophins
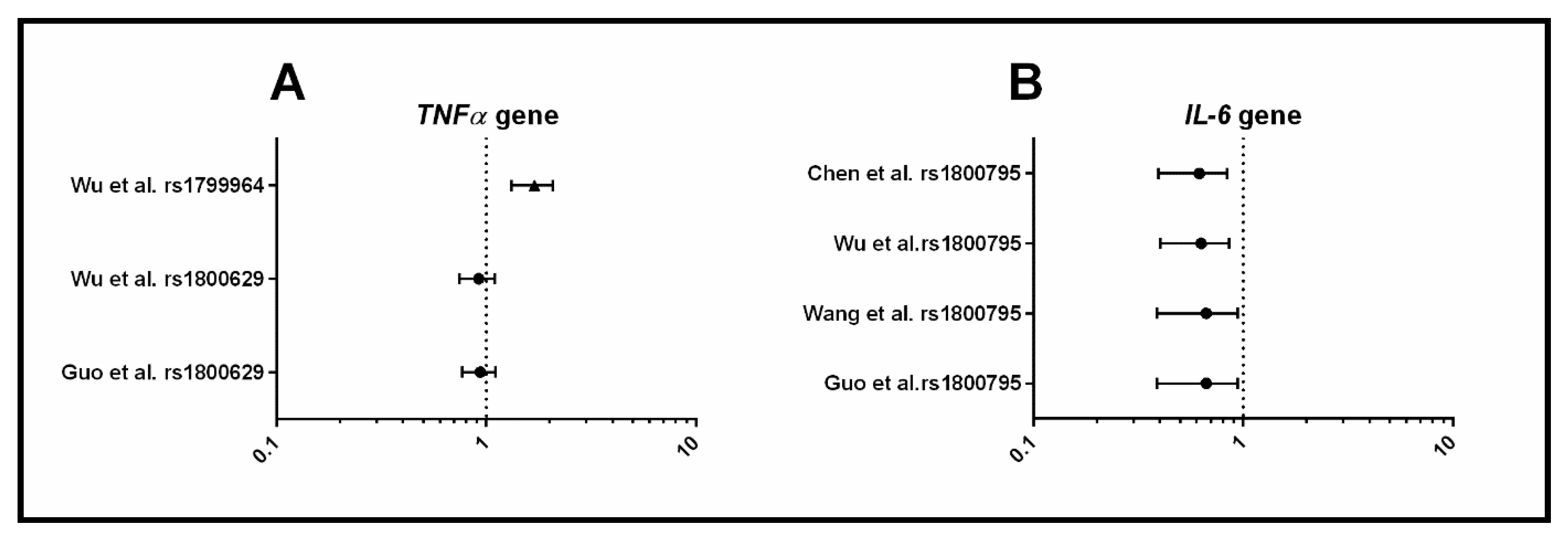
3.1.4. Inflammation
3.2. Genome-Wide Association Studies
3.2.1. Methodological Limitations of the Systematic Reviews
3.2.2. Assessment of Systematic Review Quality Using the AMSTAR Tool
4. Discussion
4.1. Metabolic Dysfunction
4.2. Dysregulation of Androgens and Gonadotrophins
4.3. Inflammation
4.4. Recommendations for Future Genetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bozdag, G.; Mumusoglu, S.; Zengin, D.; Karabulut, E.; Yildiz, B.O. The prevalence and phenotypic features of polycystic ovary syndrome: A systematic review and meta-analysis. Hum. Reprod. 2016, 31, 2841–2855. [Google Scholar] [CrossRef] [PubMed]
- Teede, H.J.; Misso, M.L.; Deeks, A.A.; Moran, L.J.; Stuckey, B.G.A.; Wong, J.L.A.; Norman, R.J.; Costello, M.F. Assessment and management of polycystic ovary syndrome: Summary of an evidence-based guideline. Med. J. Aust. 2011, 195, S65–S112. [Google Scholar] [CrossRef] [PubMed]
- Moran, L.J.; Misso, M.L.; Wild, R.A.; Norman, R.J. Impaired glucose tolerance, type 2 diabetes and metabolic syndrome in polycystic ovary syndrome: A systematic review and meta-analysis. Hum. Reprod. Update 2010, 16, 347–363. [Google Scholar] [PubMed]
- Cooney, L.G.; Lee, I.; Sammel, M.D.; Dokras, A. High prevalence of moderate and severe depressive and anxiety symptoms in polycystic ovary syndrome: A systematic review and meta-analysis. Hum. Reprod. 2017, 32, 1075–1091. [Google Scholar] [CrossRef] [PubMed]
- Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum. Reprod. 2004, 19, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Moran, L.J.; Norman, R.J.; Teede, H.J. Metabolic risk in PCOS: Phenotype and adiposity impact. Trends Endocrinol. Metab. 2015, 26, 136–143. [Google Scholar] [CrossRef]
- Cassar, S.; Misso, M.; Shaw, C.; Hopkin, W.; Teede, H.; Stepto, N. Insulin resistance in Polycystic Ovary Syndrome: A systematic review and meta-analysis of euglycaemic-hyperinsulinaemic clamp studies. Hum. Reprod. 2016, 31, 2619–2631. [Google Scholar] [CrossRef]
- Nestler, J.E.; Jakubowicz, D.J. Decreases in ovarian cytochrome P450c17 alpha activity and serum free testosterone after reduction of insulin secretion in polycystic ovary syndrome. N. Engl. J. Med. 1996, 335, 617–623. [Google Scholar] [CrossRef]
- Stepto, N.K.; Moreno-Asso, A.; McIlvenna, L.C.; Walters, K.A.; Rodgers, R.J. Molecular Mechanisms of Insulin Resistance in Polycystic Ovary Syndrome. Unraveling the Conundrum in Skeletal Muscle? J. Clin. Endocrinol. Metab. 2019, 104, 5372–5381. [Google Scholar] [CrossRef]
- Legro, R.S.; Driscoll, D.; Strauss, J.F.; Fox, J.; Dunaif, A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc. Natl. Acad. Sci. USA 1998, 95, 14956–14960. [Google Scholar] [CrossRef]
- Yilmaz, B.; Vellanki, P.; Ata, B.; Yildiz, B.O. Metabolic syndrome, hypertension, and hyperlipidemia in mothers, fathers, sisters, and brothers of women with polycystic ovary syndrome: A systematic review and meta-analysis. Fertil. Steril. 2018, 109, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Vink, J.M.; Sadrzadeh, S.; Lambalk, C.B.; Boomsma, D.I. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J. Clin. Endocrinol. Metab. 2006, 91, 2100–2104. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.G.; Urbanek, M.; Ehrmann, D.A.; Armstrong, L.L.; Lee, J.Y.; Sisk, R.; Karaderi, T.; Barber, T.M.; McCarthy, M.I.; Franks, S.; et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat. Commun. 2015, 6, 7502. [Google Scholar] [CrossRef] [PubMed]
- Day, F.R.; Hinds, D.A.; Tung, J.Y.; Stolk, L.; Styrkarsdottir, U.; Saxena, R.; Bjonnes, A.; Broer, L.; Dunger, D.B.; Halldorsson, B.V.; et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat. Commun. 2015, 6, 8464. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, H.; Shi, Y.; Cao, Y.; Yang, D.; Li, Z.; Zhang, B.; Liang, X.; Li, T.; Chen, J.; et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat. Genet. 2012, 44, 1020–1025. [Google Scholar] [CrossRef]
- Chen, Z.J.; Zhao, H.; He, L.; Shi, Y.; Qin, Y.; Shi, Y.; Li, Z.; You, L.; Zhao, J.; Liu, J.; et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat. Gene 2011, 43, 55–59. [Google Scholar] [CrossRef]
- Day, F.; Karaderi, T.; Jones, M.R.; Meun, C.; He, C.; Drong, A.; Kraft, P.; Lin, N.; Huang, H.; Broer, L.; et al. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet. 2018, 14, e1007813. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Lee, E.J.; Jin Go, M.; Sung, Y.A.; Lee, H.J.; Heon Kwak, S.; Jang, H.C.; Soo Park, K.; Lee, H.J.; Byul Jang, H.; et al. Genome-wide association study identifies GYS2 as a novel genetic factor for polycystic ovary syndrome through obesity-related condition. J. Hum. Genet. 2012, 57, 660–664. [Google Scholar] [CrossRef]
- Lee, H.; Oh, J.Y.; Sung, Y.A.; Chung, H.; Kim, H.L.; Kim, G.S.; Cho, Y.S.; Kim, J.T. Genome-wide association study identified new susceptibility loci for polycystic ovary syndrome. Hum. Reprod. 2015, 30, 723–731. [Google Scholar] [CrossRef]
- Wilkening, S.; Chen, B.; Bermejo, J.L.; Canzian, F. Is there still a need for candidate gene approaches in the era of genome-wide association studies? Genomics 2009, 93, 415–419. [Google Scholar] [CrossRef]
- Mykhalchenko, K.; Lizneva, D.; Trofimova, T.; Walker, W.; Suturina, L.; Diamond, M.P.; Azziz, R. Genetics of polycystic ovary syndrome. Expert Rev. Mol. Diagn. 2017, 17, 723–733. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Cardon, L.R. Designing candidate gene and genome-wide case-control association studies. Nat. Protoc. 2007, 2, 2492–2501. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Williams, M.G.; Eynon, N.; Ashton, K.J.; Little, J.P.; Wisloff, U.; Coombes, J.S. Genes to predict VO2max trainability: A systematic review. BMC Genom. 2017, 18, 831. [Google Scholar] [CrossRef]
- Sagoo, G.S.; Little, J.; Higgins, J.P.T. Systematic Reviews of Genetic Association Studies. PLoS Med. 2009, 6, e1000028. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M. Genetic association studies: Design, analysis and interpretation. Brief. Bioinform. 2002, 3, 146–153. [Google Scholar] [CrossRef]
- Silva, V.; Grande, A.J.; Martimbianco, A.L.; Riera, R.; Carvalho, A.P. Overview of systematic reviews - a new type of study: Part I: Why and for whom? Sao Paulo Med. J. 2012, 130, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Int. J. Surg. 2010, 8, 336–341. [Google Scholar] [CrossRef]
- Tay, C.T.; Joham, A.E.; Hiam, D.S.; Gadalla, M.A.; Pundir, J.; Thangaratinam, S.; Teede, H.J.; Moran, L.J. Pharmacological and surgical treatment of nonreproductive outcomes in polycystic ovary syndrome: An overview of systematic reviews. Clin. Endocrinol. 2018, 89, 535–553. [Google Scholar] [CrossRef]
- Gilbert, E.W.; Tay, C.T.; Hiam, D.S.; Teede, H.J.; Moran, L.J. Comorbidities and complications of polycystic ovary syndrome: An overview of systematic reviews. Clin. Endocrinol. 2018, 89, 683–699. [Google Scholar] [CrossRef]
- Hardy, G.H. Mendelian Proportions in a Mixed Population. Science 1908, 28, 49–50. [Google Scholar] [CrossRef]
- Zintzaras, E.; Lau, J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J. Clin. Epidemiol. 2008, 61, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Shea, B.J.; Reeves, B.C.; Wells, G.; Thuku, M.; Hamel, C.; Moran, J.; Moher, D.; Tugwell, P.; Welch, V.; Kristjansson, E.; et al. AMSTAR 2: A critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ 2017, 358, j4008. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Lv, P.P.; Yu, T.T.; Jin, M.; Shen, J.M.; Wang, X.; Zhou, F.; Jiang, S.W. The association between polymorphism of INSR and polycystic ovary syndrome: A meta-analysis. Int. J. Mol. Sci. 2015, 16, 2403–2425. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Zheng, Y.; Yang, J.; Zheng, N. Association of TNF-alpha, IL-6 and IL-1beta gene polymorphisms with polycystic ovary syndrome: A meta-analysis. BMC Genet. 2015, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Yu, L.; Guo, X.; Gao, W.; Jiang, Z. Associations of adiponectin gene polymorphisms with polycystic ovary syndrome: A meta-analysis. Endocrine 2012, 42, 299–306. [Google Scholar] [CrossRef]
- Li, Y.; Liu, F.; Luo, S.; Hu, H.; Li, X.H.; Li, S.W. Polymorphism T->C of gene CYP17 promoter and polycystic ovary syndrome risk: A meta-analysis. Gene 2012, 495, 16–22. [Google Scholar] [CrossRef]
- Qiu, L.; Liu, J.; Hei, Q.M. Association between two polymorphisms of follicle stimulating hormone receptor gene and susceptibility to polycystic ovary syndrome: A meta-analysis. Chin. Med. Sci. J. 2015, 30, 44–50. [Google Scholar] [CrossRef]
- Ramos, R.B.; Fabris, V.C.; Brondani Lde, A.; Spritzer, P.M. Association between rs7903146 and rs12255372 polymorphisms of transcription factor 7-like 2 gene and polycystic ovary syndrome: A systematic review and meta-analysis. Endocrine 2015, 49, 635–642. [Google Scholar] [CrossRef]
- Ruan, Y.; Ma, J.; Xie, X. Association of IRS-1 and IRS-2 genes polymorphisms with polycystic ovary syndrome: A meta-analysis. Endocr. J. 2012, 59, 601–609. [Google Scholar] [CrossRef]
- Shen, W.; Li, T.; Hu, Y.; Liu, H.; Song, M. Calpain-10 genetic polymorphisms and polycystic ovary syndrome risk: A meta-analysis and meta-regression. Gene 2013, 531, 426–434. [Google Scholar] [CrossRef]
- Shen, W.; Li, T.; Hu, Y.; Liu, H.; Song, M. CYP1A1 gene polymorphisms and polycystic ovary syndrome risk: A meta-analysis and meta-regression. Genet Test Mol. Biomark. 2013, 17, 727–735. [Google Scholar] [CrossRef]
- Shen, W.; Li, T.; Hu, Y.; Liu, H.; Song, M. Common polymorphisms in the CYP1A1 and CYP11A1 genes and polycystic ovary syndrome risk: A meta-analysis and meta-regression. Arch. Gynecol. Obstet. 2014, 289, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.J.; Li, T.R.; Hu, Y.J.; Liu, H.B.; Song, M. Relationships between TCF7L2 genetic polymorphisms and polycystic ovary syndrome risk: A meta-analysis. Metab. Syndr. Relat. Disord. 2014, 12, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Xie, X.; Jia, Y.; Li, S. Associations of insulin receptor and insulin receptor substrates genetic polymorphisms with polycystic ovary syndrome: A systematic review and meta-analysis. J. Obstet. Gynaecol. Res. 2016, 42, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tong, X.; Ji, Y.; Li, H.; Lu, W.; Song, Z. Meta-analysis of the correlation between IL-6 -174 G/C polymorphism and polycystic ovarian syndrome. J. Obstet. Gynaecol. Res. 2015, 41, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yu, K.; Yang, Z. Associations between TNF-alpha and interleukin gene polymorphisms with polycystic ovary syndrome risk: A systematic review and meta-analysis. J. Assist. Reprod. Genet. 2015, 32, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, Z.; Huang, J.; Jin, M. Association between rs1800795 polymorphism in the interleukin-6 gene and the risk of polycystic ovary syndrome: A meta-analysis. Medicine (Baltimore) 2018, 97, e11558. [Google Scholar] [CrossRef]
- Gao, J.; Xue, J.D.; Li, Z.C.; Zhou, L.; Chen, C. The association of DENND1A gene polymorphisms and polycystic ovary syndrome risk: A systematic review and meta-analysis. Arch. Gynecol. Obstet. 2016, 294, 1073–1080. [Google Scholar] [CrossRef]
- Liao, D.; Yu, H.; Han, L.; Zhong, C.; Ran, X.; Wang, D.; Mo, L. Association of PON1 gene polymorphisms with polycystic ovarian syndrome risk: A meta-analysis of case-control studies. J. Endocrinol. Investig. 2018, 41, 1289–1300. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Z.; Hao, C.; Tian, Y.; Fu, J. Effects of ADIPOQ polymorphisms on PCOS risk: A meta-analysis. Reprod. Biol. Endocrinol. 2018, 16, 120. [Google Scholar] [CrossRef]
- Pabalan, N.; Montagna, E.; Singian, E.; Tabangay, L.; Jarjanazi, H.; Barbosa, C.P.; Bianco, B. Associations of Polymorphisms in Anti-Mullerian Hormone (AMH Ile49Ser) and its Type II Receptor (AMHRII -482 A>G) on Reproductive Outcomes and Polycystic Ovary Syndrome: A Systematic Review and Meta-Analysis. Cell Physiol. Biochem. 2016, 39, 2249–2261. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.S.; Liang, G.Y.; Xia, B.R.; Liu, D.Y.; Kong, D.; Jin, X.M. Association of insulin gene variable number of tandem repeats regulatory polymorphism with polycystic ovary syndrome. Hum. Immunol. 2014, 75, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liang, W.; Fang, M.; Yu, J.; Ni, Y.; Li, Z. Association of the CAG repeat polymorphisms in androgen receptor gene with polycystic ovary syndrome: A systemic review and meta-analysis. Gene 2013, 524, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.X.; Li, Y.; Hu, R.; Wang, F.M.; Zhang, X.M.; Guan, B. Anti-Mullerian hormone gene polymorphism is associated with androgen levels in Chinese polycystic ovary syndrome patients with insulin resistance. J. Assist. Reprod. Genet. 2016, 33, 199–205. [Google Scholar] [CrossRef]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular signals of epigenetic states. Science 2010, 330, 612–616. [Google Scholar] [CrossRef]
- Huang, T.; Shu, Y.; Cai, Y.D. Genetic differences among ethnic groups. BMC Genom. 2015, 16, 1093. [Google Scholar] [CrossRef]
- Valkenburg, O.; Lao, O.; Schipper, I.; Louwers, Y.; Uitterlinden, A.G.; Kayser, M.; Laven, J.S. Genetic ancestry affects the phenotype of normogonadotropic anovulatory (WHOII) subfertility. J. Clin. Endocrinol. Metab. 2011, 96, E1181–E1187. [Google Scholar] [CrossRef]
- Mersha, T.B.; Abebe, T. Self-reported race/ethnicity in the age of genomic research: Its potential impact on understanding health disparities. Hum. Genom. 2015, 9, 1. [Google Scholar] [CrossRef]
- Shorakae, S.; Boyle, J.; Teede, H. Polycystic ovary syndrome: A common hormonal condition with major metabolic sequelae that physicians should know about. Intern. Med. J. 2014, 44, 720–726. [Google Scholar] [CrossRef]
- Cassar, S.; Teede, H.J.; Moran, L.J.; Joham, A.E.; Harrison, C.L.; Strauss, B.J.; Stepto, N.K. Polycystic ovary syndrome and anti-Mullerian hormone: Role of insulin resistance, androgens, obesity and gonadotrophins. Clin. Endocrinol. 2014, 81, 899–906. [Google Scholar] [CrossRef]
- Dumesic, D.A.; Oberfield, S.E.; Stener-Victorin, E.; Marshall, J.C.; Laven, J.S.; Legro, R.S. Scientific Statement on the Diagnostic Criteria, Epidemiology, Pathophysiology, and Molecular Genetics of Polycystic Ovary Syndrome. Endocr. Rev. 2015, 36, 487–525. [Google Scholar] [CrossRef] [PubMed]
- Pau, C.T.; Mosbruger, T.; Saxena, R.; Welt, C.K. Phenotype and Tissue Expression as a Function of Genetic Risk in Polycystic Ovary Syndrome. PLoS ONE 2017, 12, e0168870. [Google Scholar] [CrossRef] [PubMed]
- Laven, J.S.E. Follicle Stimulating Hormone Receptor (FSHR) Polymorphisms and Polycystic Ovary Syndrome (PCOS). Front. Endocrinol. (Lausanne) 2019, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Zeggini, E.; Scott, L.J.; Saxena, R.; Voight, B.F.; Marchini, J.L.; Hu, T.; de Bakker, P.I.; Abecasis, G.R.; Almgren, P.; Andersen, G.; et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008, 40, 638–645. [Google Scholar] [CrossRef]
- Cui, L.; Zhao, H.; Zhang, B.; Qu, Z.; Liu, J.; Liang, X.; Zhao, X.; Zhao, J.; Sun, Y.; Wang, P.; et al. Genotype–phenotype correlations of PCOS susceptibility SNPs identified by GWAS in a large cohort of Han Chinese women. Hum. Reprod. 2012, 28, 538–544. [Google Scholar] [CrossRef]
- Gonzalez, F. Inflammation in Polycystic Ovary Syndrome: Underpinning of insulin resistance and ovarian dysfunction. Steroids 2012, 77, 300–305. [Google Scholar] [CrossRef]
- Shorakae, S.; Teede, H.; de Courten, B.; Lambert, G.; Boyle, J.; Moran, L.J. The Emerging Role of Chronic Low-Grade Inflammation in the Pathophysiology of Polycystic Ovary Syndrome. Semin. Reprod. Med. 2015, 33, 257–269. [Google Scholar] [CrossRef]
- Duleba, A.J.; Dokras, A. Is PCOS an inflammatory process? Fertil. Steril. 2012, 97, 7–12. [Google Scholar] [CrossRef]
- Samy, N.; Hashim, M.; Sayed, M.; Said, M. Clinical significance of inflammatory markers in polycystic ovary syndrome: Their relationship to insulin resistance and body mass index. Dis. Markers 2009, 26, 163–170. [Google Scholar] [CrossRef]
- Teede, H.J.; Misso, M.L.; Costello, M.F.; Dokras, A.; Laven, J.; Moran, L.; Piltonen, T.; Norman, R.J.; Andersen, M.; Azziz, R.; et al. Recommendations from the international evidence-based guideline for the assessment and management of polycystic ovary syndrome. Fertil. Steril. 2018, 110, 364–379. [Google Scholar] [CrossRef]
- Hiam, D.; Simar, D.; Laker, R.; Altintas, A.; Gibson-Helm, M.; Fletcher, E.; Moreno-Asso, A.; Trewin, A.J.; Barres, R.; Stepto, N.K. Epigenetic reprogramming of immune cells in women with PCOS impact genes controlling reproductive function. J. Clin. Endocrinol. Metab. 2019. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jia, P.; Wolfinger, R.D.; Chen, X.; Zhao, Z. Gene set analysis of genome-wide association studies: Methodological issues and perspectives. Genomics 2011, 98, 1–8. [Google Scholar] [CrossRef] [PubMed]
- De, R.; Bush, W.S.; Moore, J.H. Bioinformatics challenges in genome-wide association studies (GWAS). Methods Mol. Biol. 2014, 1168, 63–81. [Google Scholar] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.I.; Abecasis, G.R.; Cardon, L.R.; Goldstein, D.B.; Little, J.; Ioannidis, J.P.; Hirschhorn, J.N. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef]
- Dapas, M.; Sisk, R.; Legro, R.S.; Urbanek, M.; Dunaif, A.; Hayes, M.G. Family-based quantitative trait meta-analysis implicates rare noncoding variants in DENND1A in polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2019. [Google Scholar] [CrossRef]
- Crespo, R.P.; Bachega, T.; Mendonca, B.B.; Gomes, L.G. An update of genetic basis of PCOS pathogenesis. Arch. Endocrinol. Metab. 2018, 62, 352–361. [Google Scholar] [CrossRef]
- Hebbring, S. Genomic and Phenomic Research in the 21st Century. Trends Genet. 2019, 35, 29–41. [Google Scholar] [CrossRef]
- Rotimi, C.N.; Jorde, L.B. Ancestry and disease in the age of genomic medicine. N. Engl. J. Med. 2010, 363, 1551–1558. [Google Scholar] [CrossRef]
- Bloom, M.S.; Schisterman, E.F.; Hediger, M.L. The use and misuse of matching in case-control studies: The example of polycystic ovary syndrome. Fertil. Steril. 2007, 88, 707–710. [Google Scholar] [CrossRef]
- Thakkinstian, A.; McElduff, P.; D’Este, C.; Duffy, D.; Attia, J. A method for meta-analysis of molecular association studies. Stat. Med. 2005, 24, 1291–1306. [Google Scholar] [CrossRef] [PubMed]

| Study | Diagnostic Criteria | Gene Locus | SNPs | Nearest Gene |
|---|---|---|---|---|
| Chen et al., 2011 [11] | Rotterdam | 2p16.3 | rs13405728 | LHCGR, STON1-GTF2A1L |
| 2p21 | rs12468394 | THADA | ||
| rs13429458 | ||||
| rs12478601 | ||||
| 9q33.3 | rs10818854 | DENND1A | ||
| rs2479106 | ||||
| rs10986105 | ||||
| Shi et al., 2012 [15] | Rotterdam | 2p16.3 | rs13405728 | LHCGR, STON1-GTF2A1L |
| 2p16.3 | rs2268361 | FSHR | ||
| rs2349415 | ||||
| 2p21 | rs12468394 | THADA | ||
| rs13429458 | ||||
| rs12478601 | ||||
| 9q33.3 | rs10818854 | DENND1A | ||
| rs2479106 | ||||
| rs10986105 | ||||
| 9q22.32 | rs4385527 | C9orf3 | ||
| rs3802457 | ||||
| 11q22.1 | rs18974116 | YAP1 | ||
| 12q13.2 | rs705702 | RAB5B, SUOX | ||
| 12q14.3 | rs2272046 | HMGA2 | ||
| 16q12.1 | rs4784165 | TOX3 | ||
| 19p13.3 | rs2059807 | INSR | ||
| 20q13.2 | rs6022786 | SUMO1P1 | ||
| Lee et al., 2015 [19] | Rotterdam | 8q24.2 | rs10505648 | KHDRBS3, LINC02055 |
| Hwang et al., 2012 [18] | Rotterdam | 12p12.2 | rs10841843 | GYS2 |
| rs6487237 | ||||
| rs7485509 | ||||
| Hayes et al., 2015 [13] | NIH | 8p32.1 | rs804279 | GATA4, NEIL2 |
| 9q22.32 | rs10993397 | C9orf3 | ||
| 11p14.1 | rs11031006 | ARL14EP, FSHB | ||
| Day et al., 2015 [14] | NIH | 2q.34 | rs1351592 | ERBB4 |
| 11q22.1 | rs11225154 | YAP1 | ||
| 2q21 | rs7563201 | THADA | ||
| 11p14.1 | rs11031006 | FSHB | ||
| 5q31.1 | rs13164856 | RAD50 | ||
| 12q21.2 | rs1275468 | KRR1 |
| Study | Diagnostic Criteria | Gene Locus | SNPs | Nearest Gene |
|---|---|---|---|---|
| Day et al. [17] | Rotterdam NIH | 2q21 | rs7563201 | THADA * |
| 2q.34 | rs2178575 | ERBB4 | ||
| 5q31.1 | rs13164856 | IRF1/RAD50 | ||
| 8p32.1 | rs804279 | GATA4/NEIL2 | ||
| 9p24.1 | rs10739076 | PLGRKT | ||
| 9q22.32 | rs7864171 | C9orf3 * | ||
| 9q33.3 | rs9696009 | DENND1A * | ||
| 11p14.1 | rs11031005 | ARL14EP/FSHB | ||
| 11q22.1 | rs11225154 | YAP1 * | ||
| 11q23.2 | rs1784692 | ZBTB16 | ||
| 12q13.2 | rs2271194 | ERBB3/RAB5B | ||
| 12q21.2 | rs1795379 | KRR1 | ||
| 16q12.1 | rs8043701 | TOX3 * | ||
| 20q11.21 | rs853854 | MAPRE1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiam, D.; Moreno-Asso, A.; Teede, H.J.; Laven, J.S.E.; Stepto, N.K.; Moran, L.J.; Gibson-Helm, M. The Genetics of Polycystic Ovary Syndrome: An Overview of Candidate Gene Systematic Reviews and Genome-Wide Association Studies. J. Clin. Med. 2019, 8, 1606. https://doi.org/10.3390/jcm8101606
Hiam D, Moreno-Asso A, Teede HJ, Laven JSE, Stepto NK, Moran LJ, Gibson-Helm M. The Genetics of Polycystic Ovary Syndrome: An Overview of Candidate Gene Systematic Reviews and Genome-Wide Association Studies. Journal of Clinical Medicine. 2019; 8(10):1606. https://doi.org/10.3390/jcm8101606
Chicago/Turabian StyleHiam, Danielle, Alba Moreno-Asso, Helena J. Teede, Joop S.E. Laven, Nigel K. Stepto, Lisa J. Moran, and Melanie Gibson-Helm. 2019. "The Genetics of Polycystic Ovary Syndrome: An Overview of Candidate Gene Systematic Reviews and Genome-Wide Association Studies" Journal of Clinical Medicine 8, no. 10: 1606. https://doi.org/10.3390/jcm8101606
APA StyleHiam, D., Moreno-Asso, A., Teede, H. J., Laven, J. S. E., Stepto, N. K., Moran, L. J., & Gibson-Helm, M. (2019). The Genetics of Polycystic Ovary Syndrome: An Overview of Candidate Gene Systematic Reviews and Genome-Wide Association Studies. Journal of Clinical Medicine, 8(10), 1606. https://doi.org/10.3390/jcm8101606





