Detection of Plasmid-Mediated β-Lactamase Genes and Emergence of a Novel AmpC (CMH-1) in Enterobacter cloacae at a Medical Center in Southern Taiwan
Abstract
:1. Introduction
2. Material and Methods
2.1. Clinical Isolates and Antimicrobial Susceptibility Testing
2.2. DNA Manipulation, PCR Amplification and Sequencing
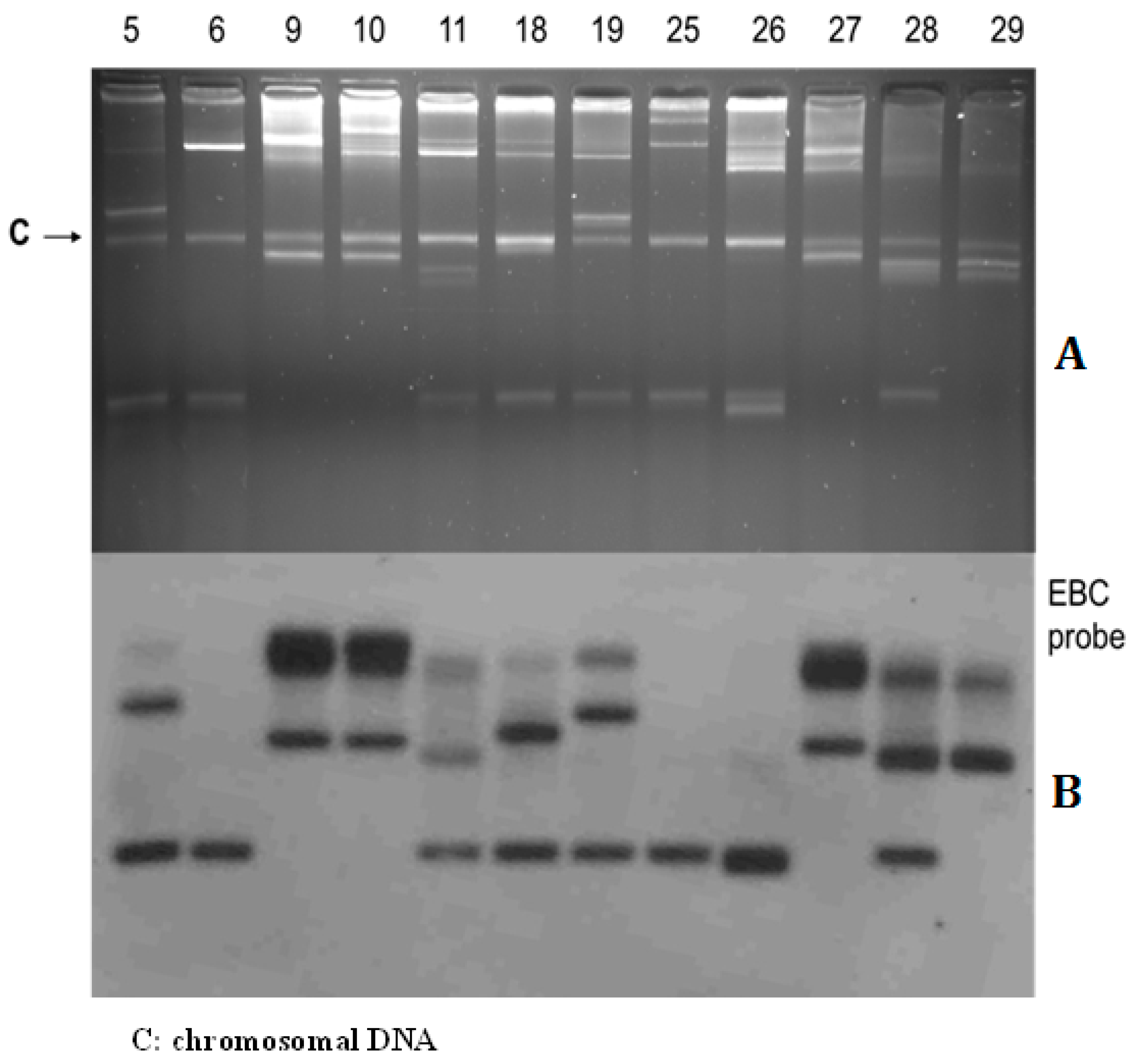
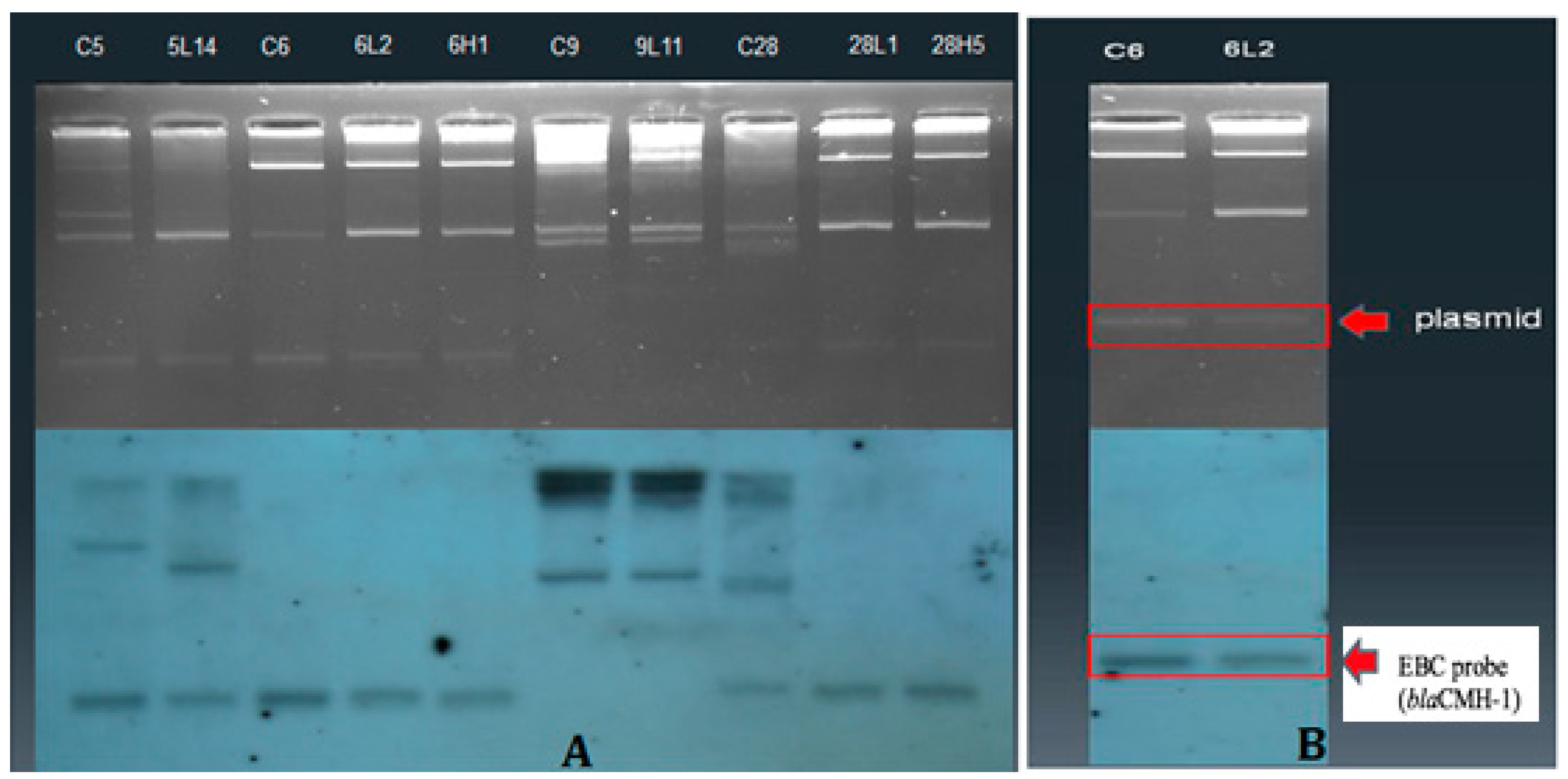
2.3. Plasmid Conjugation Experiments and Southern Hybridization
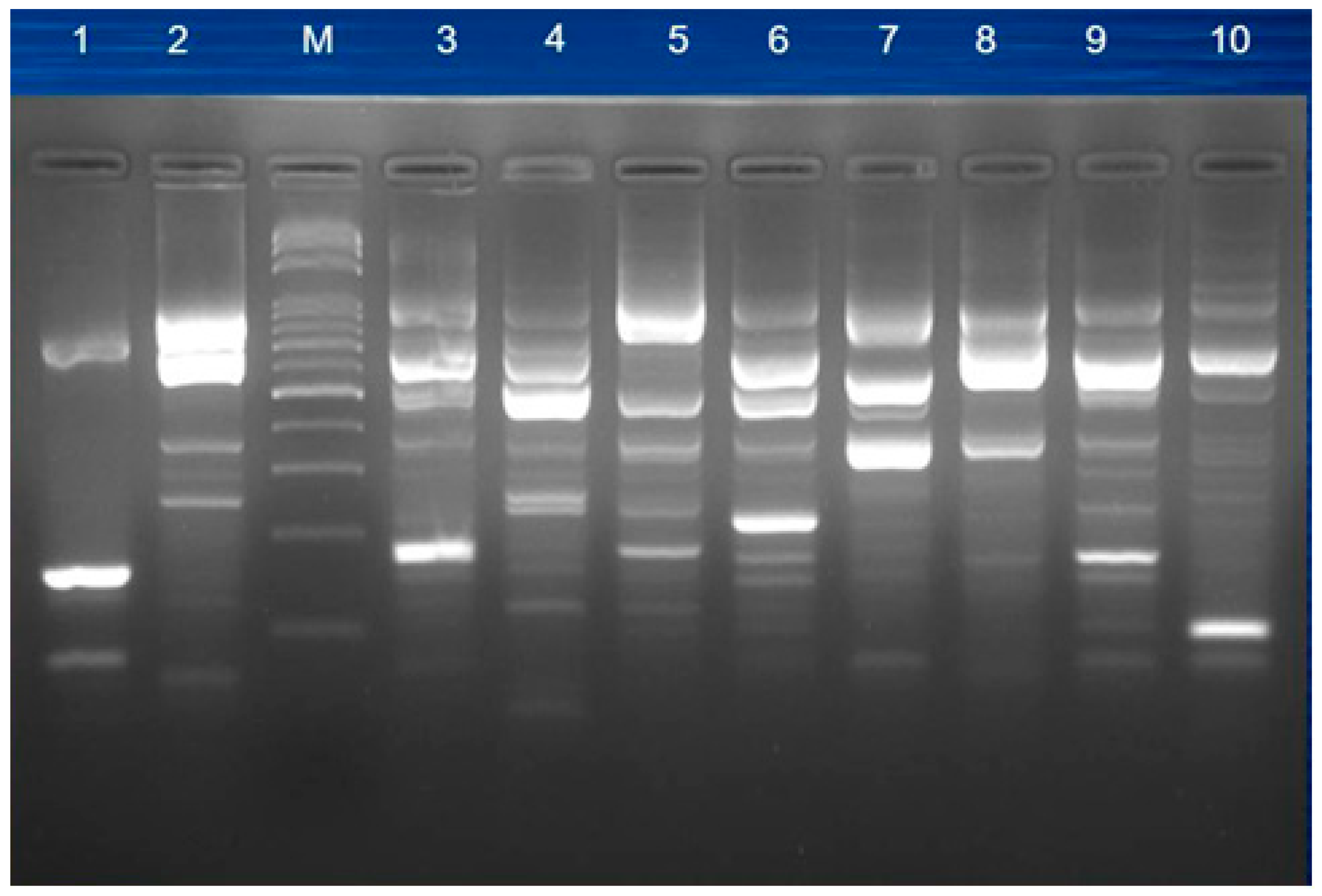
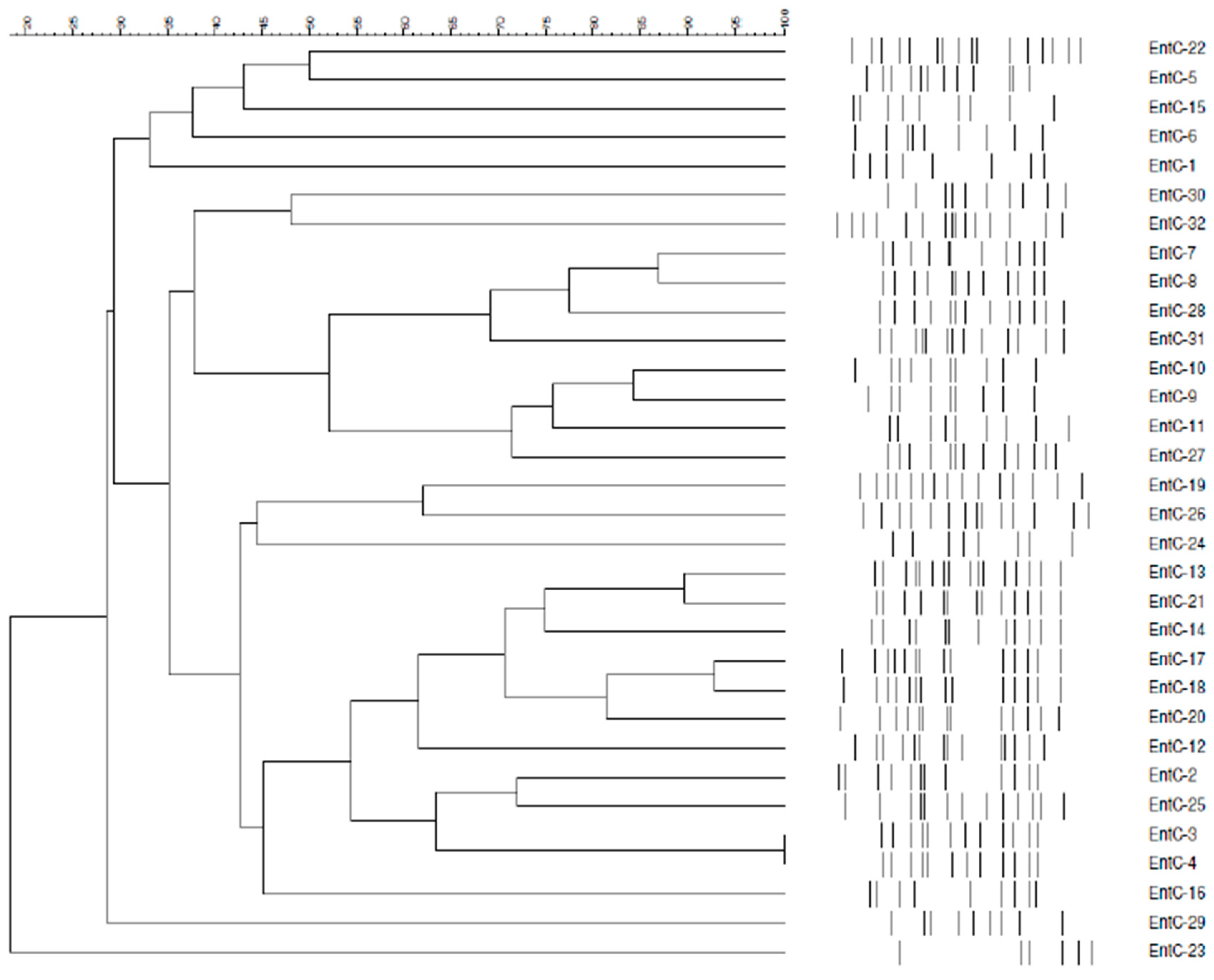
2.4. Pulsed-Field Gel Electrophoresis (PFGE)
3. Results
3.1. Bacterial Strains and Antimicrobial Susceptibility Profiles
3.2. Detection of β-Lactamase Genes on Plasmid and Antibiograms
3.3. Plasmid Profiles and Location of Resistance Gene
3.4. Molecular Typing for Genomic DNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dallenne, C.; da Costa, A.; Decre, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Manchanda, V.; Singh, N.P. Occurrence and detection of AmpC β-lactamases among Gram-negative clinical isolates using a modified three-dimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J. Antimicrob. Chemother. 2003, 51, 415–418. [Google Scholar] [CrossRef]
- Upadhyay, S.; Sen, M.R.; Bhattacharjee, A. Presence of different β-lactamase classes among clinical isolates of Pseudomonas aeruginosa expressing AmpC β-lactamase enzyme. J. Infect. Dev. Ctries. 2010, 4, 239–242. [Google Scholar]
- Paterson, D.L. Recommendation for treatment of severe infections caused by Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs). Clin. Microbiol. Infect. 2000, 6, 460–463. [Google Scholar] [CrossRef]
- Winokur, P.L.; Canton, R.; Casellas, J.M.; Legakis, N. Variations in the prevalence of strains expressing an extended-spectrum β-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific region. Clin. Infect. Dis. 2001, 32, S94–S103. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC β-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- Costa, D.; Vinue, L.; Poeta, P.; Coelho, A.C.; Matos, M.; Saenz, Y.; Somalo, S.; Zarazaga, M.; Rodrigues, J.; Torres, C. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli isolates in faecal samples of broilers. Vet. Microbiol. 2009, 138, 339–344. [Google Scholar] [CrossRef]
- Kao, C.C.; Liu, M.F.; Lin, C.F.; Huang, Y.C.; Liu, P.Y.; Chang, C.W.; Shi, Z.Y. Antimicrobial susceptibility and multiplex PCR screening of AmpC genes from isolates of Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens. J. Microbiol. Immunol. Infect. 2010, 43, 180–187. [Google Scholar] [CrossRef]
- Bush, K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 2010, 13, 558–564. [Google Scholar] [CrossRef]
- Dai, W.; Sun, S.; Yang, P.; Huang, S.; Zhang, X.; Zhang, L. Characterization of carbapenemases, extended spectrum β-lactamases and molecular epidemiology of carbapenem-non-susceptible Enterobacter cloacae in a Chinese hospital in Chongqing. Infect. Genet. Evol. 2013, 14, 1–7. [Google Scholar] [CrossRef]
- Heller, I.; Grif, K.; Orth, D. Emergence of VIM-1-carbapenemase-producing Enterobacter cloacae in Tyrol, Austria. J. Med. Microbiol. 2011, 61, 567–571. [Google Scholar] [CrossRef]
- Hamada, Y.; Watanabe, K.; Tatsuya, T.; Mezaki, K.; Takeuchi, S.; Shimizu, T.; Kirikae, T.; Ohmagari, N. Three cases of IMP-type metallo-β-lactamase-producing Enterobacter cloacae bloodstream infection in Japan. J. Infect. Chemother. 2013, 19, 956–958. [Google Scholar] [CrossRef]
- Mezzatesta, M.L.; Gona, F.; Stefani, S. Enterobacter cloacae complex: Clinical impact and emerging antibiotic resistance. Future Microbiol. 2012, 7, 887–902. [Google Scholar] [CrossRef]
- Oteo, J.; Hernandez-Almaraz, J.L.; Gil-Anton, J.; Vindel, A.; Fernandez, S.; Bautista, V.; Campos, J. Outbreak of vim-1-carbapenemase-producing Enterobacter cloacae in a pediatric intensive care unit. Pediatr. Infect. Dis. J. 2010, 29, 1144–1146. [Google Scholar] [CrossRef]
- Paterson, D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control. 2006, 34, S20–S28. [Google Scholar] [CrossRef]
- Woodford, N.; Dallow, J.W.; Hill, R.L.; Palepou, M.F.; Pike, R.; Ward, M.E.; Warner, M.; Livermore, D.M. Ertapenem resistance among Klebsiella and Enterobacter submitted in the UK to a reference laboratory. Int. J. Antimicrob. Agents 2007, 29, 456–459. [Google Scholar] [CrossRef]
- Huang, S.; Dai, W.; Sun, S.; Zhang, X.; Zhang, L. Prevalence of plasmid-mediated quinolone resistance and aminoglycoside resistance determinants among carbapeneme non-susceptible Enterobacter cloacae. PLoS ONE 2012, 7, e47636. [Google Scholar] [CrossRef]
- Yan, J.J.; Hsueh, P.R.; Ko, W.C.; Luh, K.T.; Tsai, S.H.; Wu, H.M.; Wu, J.J. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 2001, 45, 2224–2228. [Google Scholar] [CrossRef]
- Yan, J.J.; Ko, W.C.; Chuang, C.L.; Wu, J.J. Metallo-β-lactamase-producing Enterobacteriaceae isolates in a university hospital in Taiwan: Prevalence of IMP-8 in Enterobacter cloacae and first identification of VIM-2 in Citrobacter freundii. J. Antimicrob. Chemother. 2002, 50, 503–511. [Google Scholar] [CrossRef]
- Yan, J.J.; Hsueh, P.R.; Lu, J.J.; Chang, F.Y.; Ko, W.C.; Wu, J.J. Characterization of acquired β-lactamases and their genetic support in multidrug-resistant Pseudomonas aeruginosa isolates in Taiwan: The prevalence of unusual integrons. J. Antimicrob. Chemother. 2006, 58, 530–536. [Google Scholar] [CrossRef]
- Tseng, S.P.; Tsai, J.C.; Teng, L.J.; Hsueh, P.R. Dissemination of transposon Tn6001 in carbapenem-non-susceptible and extensively drug-resistant Pseudomonas aeruginosa in Taiwan. J. Antimicrob. Chemother. 2009, 64, 1170–1174. [Google Scholar] [CrossRef]
- Lee, C.H.; Chia, J.H.; Chu, C.; Wu, T.L.; Liu, J.W.; Su, L.H. In vivo selection of OmpK35-deficient mutant after cefuroxime therapy for primary liver abscess caused by Klebsiella pneumoniae. J. Antimicrob. Chemother. 2006, 58, 857–860. [Google Scholar] [CrossRef]
- Lee, C.H.; Chu, C.; Liu, J.W.; Chen, Y.S.; Chiu, C.J.; Su, L.H. Collateral damage of flomoxef therapy: In vivo development of porin deficiency and acquisition of blaDHA-1 leading to ertapenem resistance in a clinical isolate of Klebsiella pneumoniae producing CTX-M-3 and SHV-5 β-lactamases. J. Antimicrob. Chemother. 2007, 60, 410–413. [Google Scholar] [CrossRef]
- Liu, Y.F.; Yan, J.J.; Ko, W.C.; Tsai, S.H.; Wu, J.J. Characterization of carbapenem-non-susceptible Escherichia coli isolates from a university hospital in Taiwan. J. Antimicrob. Chemother. 2008, 61, 1020–1023. [Google Scholar] [CrossRef]
- Yang, F.C.; Yan, J.J.; Hung, K.H.; Wu, J.J. Characterization of ertapenem-resistant Enterobacter cloacae in a Taiwanese university hospital. J. Clin. Microbiol. 2011, 50, 223–226. [Google Scholar] [CrossRef]
- Yan, J.J.; Ko, W.C.; Tsai, S.H.; Wu, H.M.; Jin, Y.T.; Wu, J.J. Dissemination of CTX-M-3 and CMY-2 β-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J. Clin. Microbiol. 2000, 38, 4320–4325. [Google Scholar]
- Yan, J.J.; Hsueh, P.R.; Lu, J.J.; Chang, F.Y.; Shyr, J.M.; Wan, J.H.; Liu, Y.C.; Chuang, Y.C.; Yang, Y.C.; Tsao, S.M.; et al. Extended-spectrum β-lactamases and plasmid-mediated AmpC enzymes among clinical isolates of Escherichia coli and Klebsiella pneumoniae from seven medical centers in Taiwan. Antimicrob. Agents Chemother. 2006, 50, 1861–1864. [Google Scholar] [CrossRef]
- Su, P.A.; Wu, L.T.; Cheng, K.C.; Ko, W.C.; Chuang, Y.C.; Yu, W.L. Screening extended-spectrum β-lactamase production in Enterobacter cloacae and Serratia marcescens using antibiogram-based methods. J. Microbiol. Immunol. Infect. 2010, 43, 26–34. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; 16th Informational Supplement; CLSI Document M100-S16; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2006. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; CLSI M100–S24; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2014. [Google Scholar]
- Kado, C.I.; Liu, S.T. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 1981, 145, 1365–1373. [Google Scholar]
- Rasmussen, B.A.; Bush, K. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 1997, 41, 223–232. [Google Scholar] [CrossRef]
- Bellais, S.; Girlich, D.; Karim, A.; Nordmann, P. EBR-1, a novel Ambler subclass B1 β-lactamase from Empedobacter brevis. Antimicrob. Agents Chemother. 2002, 46, 3223–3227. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P. Acquired carbapenem-hydrolyzing β-lactamases and their genetic support. Curr. Pharm. Biotechnol. 2002, 3, 117–127. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatileβ-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Perez-Perez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Poirel, L.; Heritier, C.; Tolun, V.; Nordmann, P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef]
- Eckert, C.; Gautier, V.; Saladin-Allard, M.; Hidri, N.; Verdet, C.; Ould-Hocine, Z.; Barnaud, G.; Delisle, F.; Rossier, A.; Lambert, T.; et al. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 2004, 48, 1249–1255. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.T.; Hächler, H.; Kayser, F.H. Detection of genes coding for extended-spectrum SHV β-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur. J. Clin. Microbiol. Infect. Dis. 1996, 15, 398–402. [Google Scholar] [CrossRef]
- Song, W.; Kim, J.S.; Kim, H.S.; Jeong, S.H.; Yong, D.; Lee, K.M. Emergence of Escherichia coli isolates producing conjugative plasmid-mediated DHA-1 β-lactamase in a Korean university hospital. J. Hosp. Infect. 2006, 63, 459–464. [Google Scholar] [CrossRef]
- Bellais, S.; Aubert, D.; Naas, T.; Nordmann, P. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing β-lactamases in Chryseobacterium meningosepticum. Antimicrob. Agents Chemother. 2000, 44, 1878–1886. [Google Scholar] [CrossRef]
- Stumpf, A.N.; Roggenkamp, A.; Hoffmann, H. Specificity of enterobacterial repetitive intergenic consensus and repetitive extragenic palindromic polymerase chain reaction for the detection of clonality within the Enterobacter cloacae complex. Diagn. Microbiol. Infect. Dis. 2005, 53, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Dice, L.R. Measures of the amount of ecological association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Sneath, P.H.; Sokal, R.R. Numerical Taxonomy. The Principles and Practice of Numerical Classification; Taylor & Francis, Ltd.: San Francisco, CA, USA, 1973. [Google Scholar]
- Juan, C.; Macia, M.D.; Gutierrez, O.; Vidal, C.; Perez, J.L.; Oliver, A. Molecular mechanisms of β-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 2005, 49, 4733–4738. [Google Scholar] [CrossRef] [PubMed]
- Mammeri, H.; Poirel, L.; Nordmann, P. In vivo selection of a chromosomally encoded β-lactamase variant conferring ceftazidime resistance in Klebsiella oxytoca. Antimicrob. Agents Chemother. 2003, 47, 3739–3742. [Google Scholar] [CrossRef] [PubMed]
- Mohd Khari, F.I.; Karunakaran, R.; Rosli, R.; Tee Tay, S. Genotypic and phenotypic detection of AmpC β-lactamases in Enterobacter spp. isolated from a teaching hospital in Malaysia. PLoS ONE 2016, 11, e0150643. [Google Scholar] [CrossRef] [PubMed]
- Papanicolaou, G.A.; Medeiros, A.A.; Jacoby, G.A. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 1990, 34, 2200–2209. [Google Scholar] [CrossRef] [PubMed]
- Bradford, P.A.; Urban, C.; Mariano, N.; Projan, S.J.; Rahal, J.J.; Bush, K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 1997, 41, 563–569. [Google Scholar] [CrossRef]
- Wang, S.; Xiao, S.Z.; Gu, F.F.; Tang, J.; Guo, X.K.; Ni, Y.X.; Qu, J.M.; Han, L.Z. Antimicrobial susceptibility and molecular epidemiology of clinical Enterobacter cloacae bloodstream isolates in Shanghai, China. PLoS ONE 2017, 12, e0189713. [Google Scholar] [CrossRef]
- Ingti, B.; Laskar, M.A.; Choudhury, S.; Maurya, A.P.; Paul, D.; Talukdar, A.D.; Choudhury, M.D.; Dhar, D.; Chakravarty, A.; Bhattacharjee, A. Molecular and in silico analysis of a new plasmid-mediated AmpC β-lactamase (CMH-2) in clinical isolates of Klebsiella pneumoniae. Infect. Genet. Evol. 2017, 48, 34–39. [Google Scholar] [CrossRef]
- Yu, W.L.; Winokur, P.L.; von Stein, D.; Pfaller, M.A.; Wang, J.H.; Jones, R.N. First description of Klebsiella pneumoniae harboring CTX-M β-lactamases (CTX-M-14 and CTX-M-3) in Taiwan. Antimicrob. Agents Chemother. 2002, 46, 1098–1100. [Google Scholar] [CrossRef]
- Wu, L.T.; Wu, H.J.; Chung, J.G.; Chuang, Y.C.; Cheng, K.C.; Yu, W.L. Dissemination of Proteus mirabilis isolates harboring CTX-M-14 and CTX-M-3 β-lactamases at 2 hospitals in Taiwan. Diagn. Microbiol. Infect. Dis. 2006, 54, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.T.; Tsou, M.F.; Wu, H.J.; Chen, H.E.; Chuang, Y.C.; Yu, W.L. Survey of CTX-M-3 extended-spectrum β-lactamase (ESBL) among cefotaxime-resistant Serratia marcescens at a medical center in middle Taiwan. Diagn. Microbiol. Infect. Dis. 2004, 49, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.L.; Cheng, K.C.; Chi, C.J.; Chen, H.E.; Chuang, Y.C.; Wu, L.T. Characterisation and molecular epidemiology of extended-spectrum β-lactamase-producing Enterobacter cloacae isolated from a district teaching hospital in Taiwan. Clin. Microbiol. Infect. 2006, 12, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.M.; Yu, W.L.; Huang, M.; Liu, J.J.; Chen, I.C.; Chen, H.F.; Wu, L.T. Characterization of IS26-composite transposons and multidrug resistance in conjugative plasmids from Enterobacter cloacae. Microbiol. Immunol. 2015, 59, 516–525. [Google Scholar] [CrossRef] [PubMed]
| Genes | Primers | Oligonucleotide Sequence (5′–3′) | Reference |
|---|---|---|---|
| Class A Carbapenemases | |||
| SME | SME-F | AGATAGTAAATTTTATAG | [36] |
| SME-R | CTCTAACGCTAATAG | ||
| IMI | IMI-F | ATAGCCATCCTTGTTTAGCTC | [36] |
| IMI-R | TCTGCGATTACTTATCCTC | ||
| KPC | KPC-F | ATGTCACTGTATCGCCGTCT | [36] |
| KPC-R | TTTTCAGAGCCTTACTGCCC | ||
| GES | GES-F | GTTTTGCAATGTGCTCAACG | [36] |
| GES-R | TGCCATAGCAATAGGCGTAG | ||
| Class B metallo-β-lactamases | |||
| IMP-1 | IMP-1-F | TGAGCAAGTTATCTGTATTC | [36] |
| IMP-1-R | TTAGTTGCTTGGTTTTGATG | ||
| IMP-2 | IMP-2-F | GGCAGTCGCCCTAAAACAAA | [36] |
| IMP-2-R | TAGTTACTTGGCTGTGATGG | ||
| VIM-1 | VIM-1-F | TTATGGAGCAGCAACCGATGT | [36] |
| VIM-1-R | CAAAAGTCCCGCTCCAACGA | ||
| VIM-2 | VIM-2-F | AAAGTTATGCCGCACTCACC | [36] |
| VIM-2-R | TGCAACTTCATGTTATGCCG | ||
| NDM | NDM-F | TCTCGACAATGCCGGGTTT | In this study |
| NDM-R | GAGATTGCCGAGCGACTT | ||
| AmpC β-lactamases | |||
| CMY-2 | AmpC-1B | TTTTCAAGAATGCGCCAGGC | In this study |
| AmpC-1C | CTGCTGCTGACAGCCTCTTT | ||
| DHA-1 | DHA-1A | CTGATGAAAAAATCGTTATC | In this study |
| DHA-1B | ATTCCAGTGCACTCAAAATA | ||
| MOX | MOXMF | GCTGCTCAAGGAGCACAGGAT | [37] |
| MOXMR | CACATTGACATAGGTGTGGTGC | ||
| CIT | CITMF | TGGCCAGAACTGACAGGCAAA | [37] |
| CITMR | TTTCTCCTGAACGTGGCTGGC | ||
| DHA | DHAMF | AACTTTCACAGGTGTGCTGGGT | [37] |
| DHAMR | CCGTACGCATACTGGCTTTGC | ||
| ACC | ACCMF | AACAGCCTCAGCAGCCGGTTA | [37] |
| ACCMR | TTCGCCGCAATCATCCCTAGC | ||
| FOX | FOXMF | AACATGGGGTATCAGGGAGATG | [37] |
| FOXMR | CAAAGCGCGTAACCGGATTGG | ||
| EBC | EBCMF | TCGGTAAAGCCGATGTTGCGG | [37] |
| EBCMR | CTTCCACTGCGGCTGCCAGTT | ||
| CMH-1 | CMH-1F | ATGATGACAAAATCCCTAAGCTG | In this study a |
| CMH-1R | TTACTGTAGCGCGTCGAGGATA | ||
| MIR-6 | MIR-6F | ATGATGACAAAATCCCTAAGCTG | In this study a |
| MIR-6R | TTACTGCAGCGCGTCGACG | ||
| Class D Oxacillinases | |||
| OXA-48 | OXA-48-F | GATGTGTCATAGTATTCGTCG | [38] |
| OXA-48-R | TCACAACAACTAAAAGCACTG | ||
| OXA-1 | OXA-1A | TCAACTTTCAAGATCGCA | [38] |
| OXA-1B | GTGTGTTTAGAATGGTGA | ||
| OXA-9 | OXA-9A | TTCGTTTCCGCCACTCTCCC | [38] |
| OXA-9B | ACGAGAATATCCTCTCGTGC | ||
| ESBL genes | |||
| SHV | SHV specific F | GATCCACTATCGCCAGCAGG | In this study |
| SHV specific R | ACCACAATGCGCTCTGCTTTG | ||
| CTX-M-1 group | CTX-M-1F | GGTTAAAAAATCACTGCGTC | [39] |
| CTX-M-1R | TTGGTGAGATTTTAGCCGC | ||
| CTX-M-2 group | CTX-M-2F | TGGGTTACGATTTTCGCCGC | [39] |
| CTX-M-2R | TGGGTTACGATTTTCGCCGC | ||
| CTX-M-9 group | CTX-M-9F | ATGGTGACAAAGAGAGTGCA | [39] |
| CTX-M-9R | CCCTTCGGCGATGATTCTC | ||
| TEM | TEM-F | ATGAGTATTCAACATTTCCG | [39] |
| TEM-R | CCAATGCTTAATCAGTGAGG | ||
| MIC (mg/L) | |||||
|---|---|---|---|---|---|
| R Criteria | Range | MIC50 | MIC90 | R (%) | |
| Amikacin | ≥64 | ≤4–32 | ≤4 | 8 | 0 |
| Amoxicillin/clavulanic acid | ≥32 | ≤8–32 | >32 | >32 | 93 |
| Ampicillin | ≥32 | ≤2–16 | >16 | >16 | 95 |
| Aztreonam | ≥16 | ≤1–16 | 4 | >16 | 54 |
| Cefazolin | ≥8 | ≤2–32 | >32 | >32 | 95 |
| Cefepime | ≥16 | ≤1–16 | ≤1 | 16 | 17 |
| Cefotaxime | ≥4 | ≤1–32 | 16 | >32 | 54 |
| Cefoxitin | ≥32 | ≤4–16 | >16 | >16 | 95 |
| Ceftazidime | ≥16 | ≤1–16 | 4 | >16 | 54 |
| Cefuroxime | ≥32 | 4–16 | >16 | >16 | 95 |
| Ciprofloxacin | ≥4 | ≤0.06–2 | ≤0.06 | >2 | 20 |
| Colistin | >2 a | ≤0.5–2 | 1 | >2 | 12 |
| Doripenem | ≥4 | ≤0.25–2 | 0.25 | 0.25 | 0 |
| Ertapenem | ≥2 | ≤0.25–2 | 0.5 | 1 | 7 |
| Gentamicin | ≥16 | ≤1–8 | 1 | >8 | 24 |
| Imipenem | ≥4 | ≤0.25–2 | 0.5 | 1 | 0 |
| Meropenem | ≥4 | ≤0.25–0.5 | 0.25 | 0.25 | 0 |
| Piperacillin/tazobactam | ≥128 | ≤4–64 | 8 | >64 | 29 |
| Tigecycline | >2 b | ≤0.25–2 | 1 | >2 | 24 |
| Strain No. | Plasmid-Mediated β-Lactamase Gene(s) | Resistance Profiles (I) CAZ/CTX/CRO/FEP | Resistance Profiles (II) IPM/CIP/GM/AN | Resistance Code |
|---|---|---|---|---|
| EntC-1 | ACT-like | R/R/R/S (A) | S/S/S/S (a) | Aa (I) |
| EntC-2 | ACT-like | R/R/R/S (A) | S/S/R/S (b) | Ab (II) |
| EntC-3 | ACT-like/TEM-1/SHV-12 a | R/R/R/S (A) | S/S/S/S (a) | Aa (I) |
| EntC-4 | ACT-like | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-5 | ACT-like | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-6 | TEM-1/CMH-1 | R/R/R/S (A) | S/S/R/S (b) | Ab (II) |
| EntC-7 | ACT-like | R/R/R/S (A) | S/S/S/S (a) | Aa (I) |
| EntC-8 | ACT-like | R/R/R/S (A) | S/S/S/S (a) | Aa (I) |
| EntC-9 | ACT-like/TEM-1/SHV-12 | R/R/R/S (A) | S/R/R/S (c) | Ac (IV) |
| EntC-10 | TEM/ACT-like | R/R/R/R (C) | S/R/R/S (c) | Cc (V) |
| EntC-11 | ACT-like/TEM-1/SHV-12 a | R/R/R/R (C) | S/R/S/S (d) | Cd (VI) |
| EntC-12 | ACT-like/TEM-1/SHV-12 a | R/R/R/S (A) | S/S/R/S (b) | Ab (II) |
| EntC-13 | ACT-like/TEM-1/SHV-12 a | R/R/R/S (A) | S/S/S/S (a) | Aa (I) |
| EntC-14 | ACT-like | R/R/R/S (A) | S/S/S/S (a) | Aa (I) |
| EntC-15 | ACT-like | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-16 | ACT-like/TEM-1 | R/R/R/S (A) | S/S/R/S (b) | Ab (II) |
| EntC-17 | ACT-like/SHV-12 a | R/R/R/R (C) | S/R/R/S (c) | Cc (V) |
| EntC-18 | ACT-like/SHV-12 a | R/R/R/R (C) | S/R/S/S (d) | Cd (VI) |
| EntC-19 | ACT-like | R/R/R/S (A) | S/S/S/S (a) | Aa (I) |
| EntC-20 | ACT-like/SHV-12 a | R/R/R/R (C) | S/R/R/S (c) | Cc (V) |
| EntC-21 | ACT-like | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-22 | ACT-like | R/R/R/S (A) | S/S/S/S (a) | Aa (I) |
| EntC-23 | Not identified | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-24 | Not identified | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-25 | ACT-like/DHA-1/SHV-12 | R/R/R/R (C) | S/S/R/S (b) | Cb (VII) |
| EntC-26 | ACT-like/CTX-M-3 a | R/R/R/R (C) | S/R/R/S (c) | Cc (V) |
| EntC-27 | ACT-like | R/R/R/S (A) | S/R/S/S (d) | Ad (VIII) |
| EntC-28 | ACT-like | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-29 | MIR-6 | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-30 | ACT-like/SHV-12 | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-31 | ACT-like | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| EntC-32 | CMH-1 | R/R/R/S (A) | S/S/S/S (a) | Aa (I) |
| EntC-33 to EntC-41 | Not identified | S/S/S/S (B) | S/S/S/S (a) | Ba (III) |
| Strain | Source | AmpC | CAZ | CTX | CRO | FEP | IPM | CIP | GM | TGC | CL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli J53 | 2 | 0.25 | 0.25 | 0.06 | 2 | 2 | 1 | 1 | 0.5 | ||
| EntC-6 | Blood | CMH-1 | 128 | 64 | 128 | 2 | 0.5 | 1 | >64 | 1 | >16 |
| 6L2 | CMH-1 | 256 | 32 | 16 | 8 | 2 | <0.03 | >64 | 1 | 1 | |
| EntC-5 | Blood | ACT-like | 4 | 2 | 4 | 1 | 0.5 | <0.03 | 1 | 2 | 1 |
| 5L14 | ACT-like | >256 | 128 | 256 | 8 | 0.5 | 0.5 | >64 | 8 | 1 | |
| 5H15 | ACT-like | 256 | 32 | 16 | 8 | 4 | 4 | >64 | 1 | 1 | |
| EntC-28 | Blood | ACT-like | 1 | 0.5 | 1 | 0.13 | 1 | <0.3 | 1 | 1 | 2 |
| 28L1 | ACT-like | 256 | 16 | 16 | 8 | 2 | 2 | >64 | 1 | 1 | |
| 28H5 | ACT-like | 256 | 16 | 16 | 8 | 4 | 4 | >64 | 2 | 1 | |
| EntC-32 | Blood | CMH-1 | 128 | 128 | 128 | 0.5 | 1 | 0.25 | 1 | 1 | >16 |
| EntC-29 | Blood | MIR-6 | 0.13 | 0.13 | 1 | 0.03 | 1 | <0.31 | 1 | 1 | 1 |
| ATCC E. coli 25922 | 1 | 0.13 | 0.13 | 0.63 | 0.25 | <0.03 | 2 | 0.25 | 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ku, Y.-H.; Lee, M.-F.; Chuang, Y.-C.; Yu, W.-L. Detection of Plasmid-Mediated β-Lactamase Genes and Emergence of a Novel AmpC (CMH-1) in Enterobacter cloacae at a Medical Center in Southern Taiwan. J. Clin. Med. 2019, 8, 8. https://doi.org/10.3390/jcm8010008
Ku Y-H, Lee M-F, Chuang Y-C, Yu W-L. Detection of Plasmid-Mediated β-Lactamase Genes and Emergence of a Novel AmpC (CMH-1) in Enterobacter cloacae at a Medical Center in Southern Taiwan. Journal of Clinical Medicine. 2019; 8(1):8. https://doi.org/10.3390/jcm8010008
Chicago/Turabian StyleKu, Yee-Huang, Mei-Feng Lee, Yin-Ching Chuang, and Wen-Liang Yu. 2019. "Detection of Plasmid-Mediated β-Lactamase Genes and Emergence of a Novel AmpC (CMH-1) in Enterobacter cloacae at a Medical Center in Southern Taiwan" Journal of Clinical Medicine 8, no. 1: 8. https://doi.org/10.3390/jcm8010008
APA StyleKu, Y.-H., Lee, M.-F., Chuang, Y.-C., & Yu, W.-L. (2019). Detection of Plasmid-Mediated β-Lactamase Genes and Emergence of a Novel AmpC (CMH-1) in Enterobacter cloacae at a Medical Center in Southern Taiwan. Journal of Clinical Medicine, 8(1), 8. https://doi.org/10.3390/jcm8010008





