Metabolic Signature Differentiated Diabetes Mellitus from Lipid Disorder in Elderly Taiwanese
Abstract
1. Introduction
2. Materials and Methods
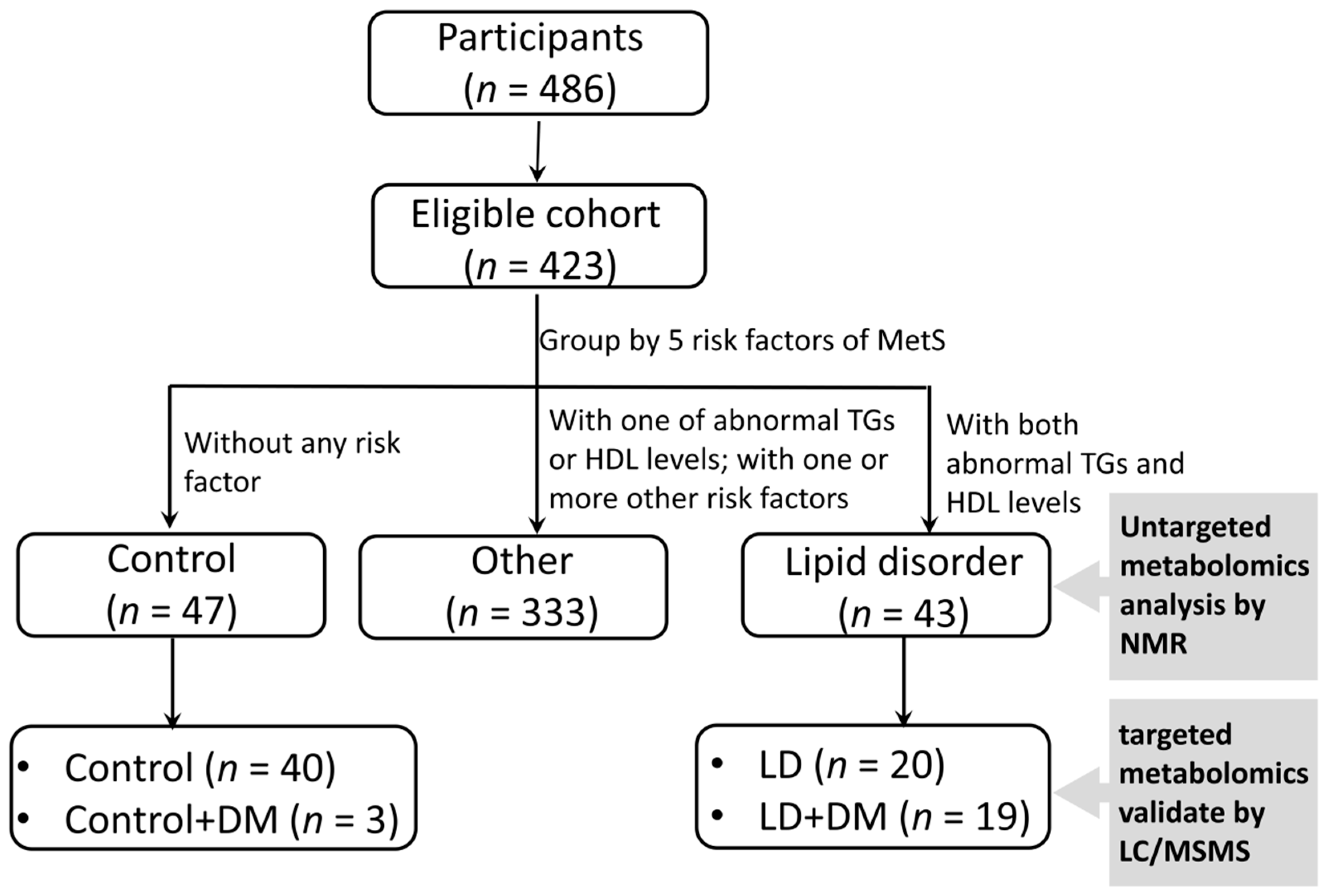
2.1. Ethics Statement and Study Populations
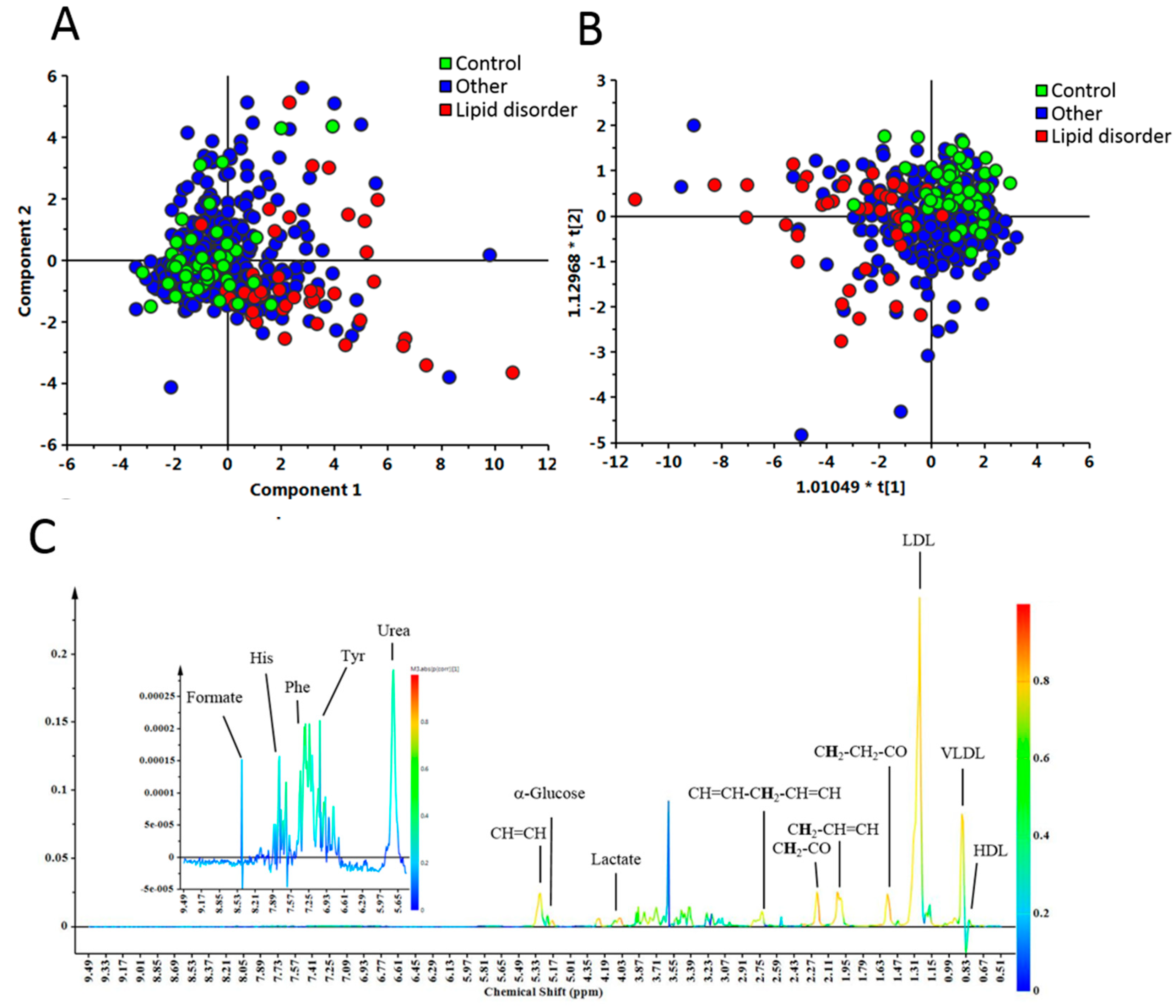
2.2. Nuclear Magnetic Resonance (NMR) Analysis of Plasma
2.3. Determining Concentrations of Plasma Metabolites with Ultra-High-Performance Liquid Chromatography-Tandem Mass Spectrometry (UPLC/MSMS)
2.4. Statistical Analysis
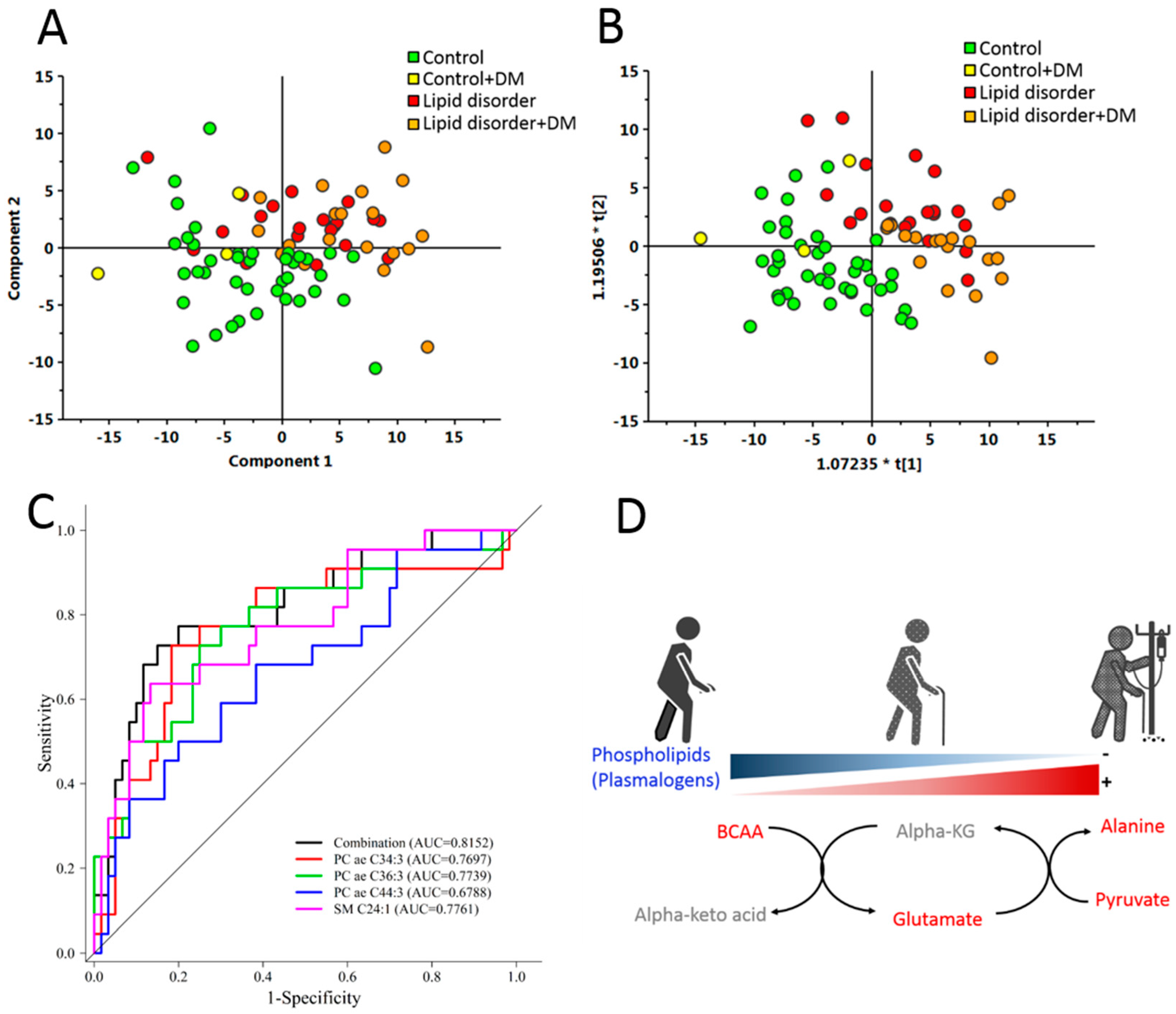
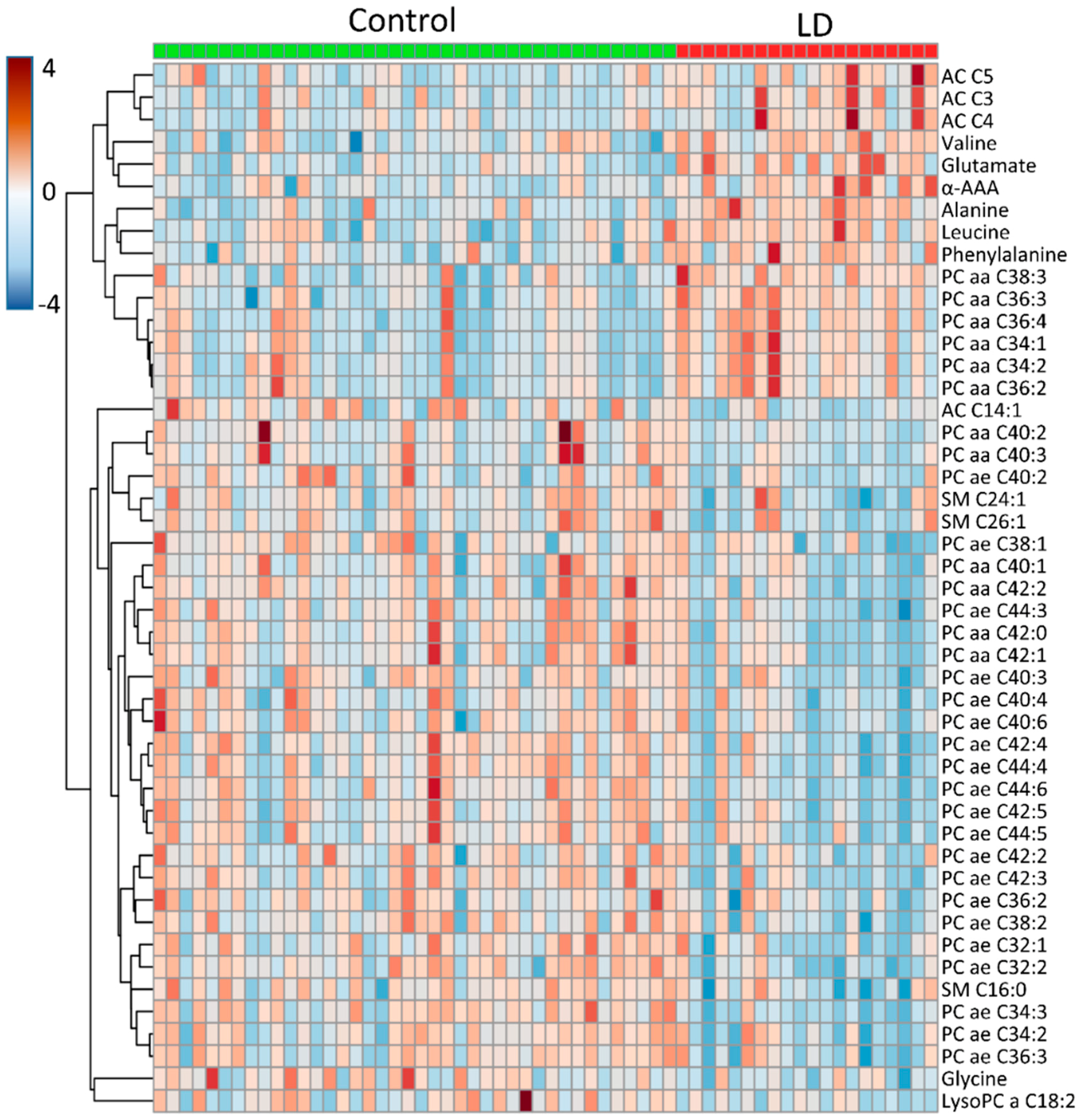
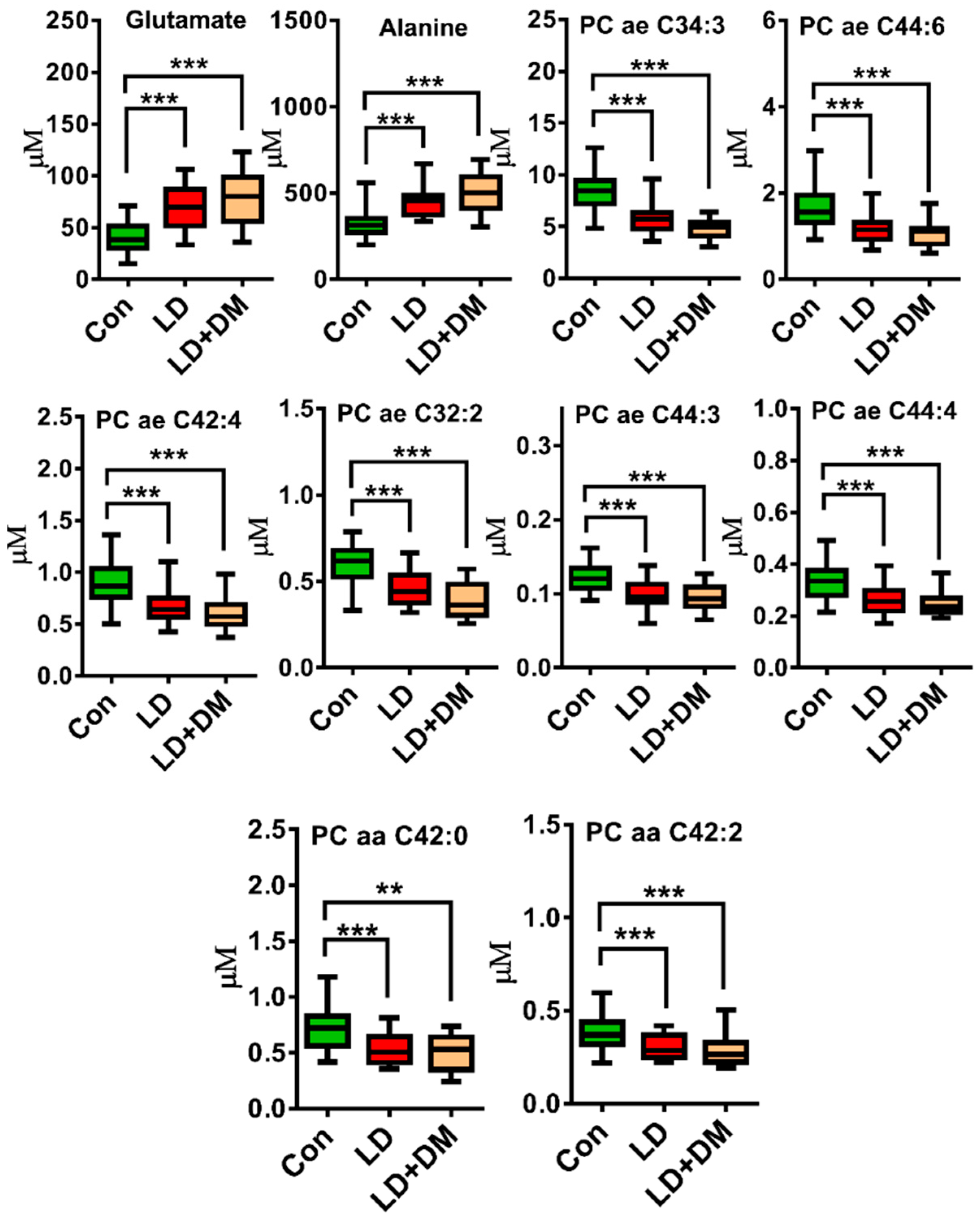
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tacutu, R.; Craig, T.; Budovsky, A.; Wuttke, D.; Lehmann, G.; Taranukha, D.; Costa, J.; Fraifeld, V.E.; de Magalhaes, J.P. Human ageing genomic resources: Integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013, 41, D1027–D1033. [Google Scholar] [CrossRef] [PubMed]
- Dato, S.; Rose, G.; Crocco, P.; Monti, D.; Garagnani, P.; Franceschi, C.; Passarino, G. The genetics of human longevity: An intricacy of genes, environment, culture and microbiome. Mech. Ageing Dev. 2017, 165, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Li, H.; Peng, X.X. Functional metabolomics: From biomarker discovery to metabolome reprogramming. Protein Cell 2015, 6, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Wang, W.; Yi, M.; Cheng, S.; Wang, D. Study of the metabolomics characteristics of patients with metabolic syndrome based on liquid chromatography quadrupole time-of-flight mass spectrometry. Ann. Endocrinol. 2018, 79, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Sevin, D.C.; Kuehne, A.; Zamboni, N.; Sauer, U. Biological insights through nontargeted metabolomics. Curr. Opin. Biotechnol. 2015, 34, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, D.; He, Y.; Chen, C.; Song, C.; Zhao, Y.; Bai, Y.; Wang, Y.; Pu, J.; Chen, J.; et al. Investigation of novel metabolites potentially involved in the pathogenesis of coronary heart disease using a uhplc-qtof/ms-based metabolomics approach. Sci. Rep. 2017, 7, 15357. [Google Scholar] [CrossRef] [PubMed]
- Cardellini, M.; Ballanti, M.; Davato, F.; Cardolini, I.; Guglielmi, V.; Rizza, S.; Pecchioli, C.; Casagrande, V.; Mavilio, M.; Porzio, O.; et al. 2-hydroxycaproate predicts cardiovascular mortality in patients with atherosclerotic disease. Atherosclerosis 2018, 277, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.L.; Wang, C.H.; Shiao, M.S.; Liu, M.H.; Huang, Y.Y.; Huang, C.Y.; Mao, C.T.; Lin, J.F.; Ho, H.Y.; Yang, N.I. Metabolic disturbances identified in plasma are associated with outcomes in patients with heart failure: Diagnostic and prognostic value of metabolomics. J. Am. Coll. Cardiol. 2015, 65, 1509–1520. [Google Scholar] [CrossRef]
- Wu, T.; Qiao, S.; Shi, C.; Wang, S.; Ji, G. Metabolomics window into diabetic complications. J. Diabetes Investig. 2018, 9, 244–255. [Google Scholar] [CrossRef]
- Wilkins, J.M.; Trushina, E. Application of metabolomics in alzheimer’s disease. Front. Neurol. 2017, 8, 719. [Google Scholar] [CrossRef]
- Yang, K.; Xia, B.; Wang, W.; Cheng, J.; Yin, M.; Xie, H.; Li, J.; Ma, L.; Yang, C.; Li, A.; et al. A comprehensive analysis of metabolomics and transcriptomics in cervical cancer. Sci. Rep. 2017, 7, 43353. [Google Scholar] [CrossRef]
- Kaushik, A.K.; DeBerardinis, R.J. Applications of metabolomics to study cancer metabolism. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.M.; Cooper, M.E. Mechanisms of diabetic complications. Physiol. Rev. 2013, 93, 137–188. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.J.; Liu, J.S.; Li, P.F.; Chang, H.; Hsieh, C.H.; Chang, J.B.; Hung, Y.J.; Pei, D.; Liang, Y.J.; Chen, Y.L. The relationships between hematogram and metabolic syndrome in elderly subgroups: A taiwan cohort study. Arch. Gerontol. Geriatr. 2016, 63, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Yen, Y.F.; Hu, H.Y.; Lin, I.F.; Lai, Y.J.; Su, V.Y.; Pan, S.W.; Ting, W.Y.; Su, W.J. Associations of metabolic syndrome and its components with mortality in the elderly: A cohort study of 73,547 taiwanese adults. Medicine (Baltimore) 2015, 94, e956. [Google Scholar] [CrossRef] [PubMed]
- Salazar, M.R.; Carbajal, H.A.; Espeche, W.G.; Leiva Sisnieguez, C.E.; Balbin, E.; Dulbecco, C.A.; Aizpurua, M.; Marillet, A.G.; Reaven, G.M. Relation among the plasma triglyceride/high-density lipoprotein cholesterol concentration ratio, insulin resistance, and associated cardio-metabolic risk factors in men and women. Am. J. Cardiol. 2012, 109, 1749–1753. [Google Scholar] [CrossRef] [PubMed]
- Quijada, Z.; Paoli, M.; Zerpa, Y.; Camacho, N.; Cichetti, R.; Villarroel, V.; Arata-Bellabarba, G.; Lanes, R. The triglyceride/HDL-cholesterol ratio as a marker of cardiovascular risk in obese children; association with traditional and emergent risk factors. Pediatr. Diabetes 2008, 9, 464–471. [Google Scholar] [CrossRef]
- Iwani, N.A.; Jalaludin, M.Y.; Zin, R.M.; Fuziah, M.Z.; Hong, J.Y.; Abqariyah, Y.; Mokhtar, A.H.; Wan Nazaimoon, W.M. Triglyceride to HDL-C ratio is associated with insulin resistance in overweight and obese children. Sci. Rep. 2017, 7, 40055. [Google Scholar] [CrossRef]
- Lenz, E.M.; Wilson, I.D. Analytical strategies in metabonomics. J. Proteome Res. 2007, 6, 443–458. [Google Scholar] [CrossRef]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Fauman, E.; Erte, I.; Perry, J.R.; Kastenmuller, G.; Shin, S.Y.; Petersen, A.K.; Hyde, C.; Psatha, M.; Ward, K.J.; et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 2013, 62, 4270–4276. [Google Scholar] [CrossRef] [PubMed]
- Wang-Sattler, R.; Yu, Z.; Herder, C.; Messias, A.C.; Floegel, A.; He, Y.; Heim, K.; Campillos, M.; Holzapfel, C.; Thorand, B.; et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 2012, 8, 615. [Google Scholar] [CrossRef] [PubMed]
- Floegel, A.; Stefan, N.; Yu, Z.; Muhlenbruch, K.; Drogan, D.; Joost, H.G.; Fritsche, A.; Haring, H.U.; Hrabe de Angelis, M.; Peters, A.; et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 2013, 62, 639–648. [Google Scholar] [CrossRef]
- Liu, J.; Semiz, S.; van der Lee, S.J.; van der Spek, A.; Verhoeven, A.; van Klinken, J.B.; Sijbrands, E.; Harms, A.C.; Hankemeier, T.; van Dijk, K.W.; et al. Metabolomics based markers predict type 2 diabetes in a 14-year follow-up study. Metabolomics 2017, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Cheng, M.L.; Fan, C.M.; Hong, C.Y.; Shiao, M.S. Nicotinuric acid: A potential marker of metabolic syndrome through a metabolomics-based approach. Diabetes Care 2013, 36, 1729–1731. [Google Scholar] [CrossRef]
- Suhre, K.; Meisinger, C.; Doring, A.; Altmaier, E.; Belcredi, P.; Gieger, C.; Chang, D.; Milburn, M.V.; Gall, W.E.; Weinberger, K.M.; et al. Metabolic footprint of diabetes: A multiplatform metabolomics study in an epidemiological setting. PLoS ONE 2010, 5, e13953. [Google Scholar] [CrossRef] [PubMed]
- Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive summary of the third report of the national cholesterol education program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III). JAMA 2001, 285, 2486–2497. [Google Scholar] [CrossRef]
- American Diabetes Association. Standards of medical care in diabetes. Diabetes Care 2004, 27 (Suppl. 1), S15–S35. [Google Scholar] [CrossRef]
- Tan, C.E.; Ma, S.; Wai, D.; Chew, S.K.; Tai, E.S. Can we apply the national cholesterol education program adult treatment panel definition of the metabolic syndrome to asians? Diabetes Care 2004, 27, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Anthony, C.; Dona, B.J.; Schäfer, H.; Humpfer, E.; Spraul, M.; Lewis, M.R.; Pearce, J.T.M.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Precision high-throughput proton nmr spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Anal. Chem. 2014, 86, 9887–9894. [Google Scholar]
- Tai, E.S.; Tan, M.L.; Stevens, R.D.; Low, Y.L.; Muehlbauer, M.J.; Goh, D.L.; Ilkayeva, O.R.; Wenner, B.R.; Bain, J.R.; Lee, J.J.M.; et al. Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men. Diabetologia 2010, 53, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhu, L.; Cui, X.; Feng, L.; Zhao, X.; He, S.; Ping, F.; Li, W.; Li, Y. The triglyceride to high-density lipoprotein cholesterol (TG/HDL-C) ratio as a predictor of insulin resistance but not of beta cell function in a chinese population with different glucose tolerance status. Lipids Health Dis. 2016, 15, 104. [Google Scholar] [CrossRef]
- Ren, X.; Chen, Z.A.; Zheng, S.; Han, T.; Li, Y.; Liu, W.; Hu, Y. Association between triglyceride to hdl-c ratio (TG/HDL-C) and insulin resistance in chinese patients with newly diagnosed type 2 diabetes mellitus. PLoS ONE 2016, 11, e0154345. [Google Scholar] [CrossRef] [PubMed]
- Adeva, M.M.; Calvino, J.; Souto, G.; Donapetry, C. Insulin resistance and the metabolism of branched-chain amino acids in humans. Amino Acids 2012, 43, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Drabkova, P.; Sanderova, J.; Kovarik, J.; kandar, R. An assay of selected serum amino acids in patients with type 2 diabetes mellitus. Adv. Clin. Exp. Med. 2015, 24, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Felig, P.; Wahren, J.; Sherwin, R.; Palaiologos, G. Amino acid and protein metabolism in diabetes mellitus. Arch. Intern. Med. 1977, 137, 507–513. [Google Scholar] [CrossRef]
- Oberlinner, C.; Zober, A.; Nawroth, P.P.; Humpert, P.M.; Morcos, M. Alanine-aminotransferase levels predict impaired glucose tolerance in a worksite population. Acta Diabetol. 2010, 47, 161–165. [Google Scholar] [CrossRef]
- Otter, S.; Lammert, E. Exciting times for pancreatic islets: Glutamate signaling in endocrine cells. Trends Endocrinol. Metab. TEM 2016, 27, 177–188. [Google Scholar] [CrossRef]
- Meikle, P.J.; Summers, S.A. Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat. Rev. Endocrinol. 2017, 13, 79–91. [Google Scholar] [CrossRef]
- Meikle, P.J.; Wong, G.; Barlow, C.K.; Weir, J.M.; Greeve, M.A.; MacIntosh, G.L.; Almasy, L.; Comuzzie, A.G.; Mahaney, M.C.; Kowalczyk, A.; et al. Plasma lipid profiling shows similar associations with prediabetes and type 2 diabetes. PLoS ONE 2013, 8, e74341. [Google Scholar] [CrossRef] [PubMed]
- Lessig, J.; Fuchs, B. Plasmalogens in biological systems: Their role in oxidative processes in biological membranes, their contribution to pathological processes and aging and plasmalogen analysis. Curr. Med. Chem. 2009, 16, 2021–2041. [Google Scholar] [CrossRef] [PubMed]
- Jurgens, G.; Fell, A.; Ledinski, G.; Chen, Q.; Paltauf, F. Delay of copper-catalyzed oxidation of low density lipoprotein by in vitro enrichment with choline or ethanolamine plasmalogens. Chem. Phys. Lipids 1995, 77, 25–31. [Google Scholar] [CrossRef]
- Yan-Do, R.; Duong, E.; Manning Fox, J.E.; Dai, X.; Suzuki, K.; Khan, S.; Bautista, A.; Ferdaoussi, M.; Lyon, J.; Wu, X.; et al. A glycine-insulin autocrine feedback loop enhances insulin secretion from human beta-cells and is impaired in type 2 diabetes. Diabetes 2016, 65, 2311–2321. [Google Scholar] [CrossRef] [PubMed]
- Meikle, P.J.; Wong, G.; Tsorotes, D.; Barlow, C.K.; Weir, J.M.; Christopher, M.J.; MacIntosh, G.L.; Goudey, B.; Stern, L.; Kowalczyk, A.; et al. Plasma lipidomic analysis of stable and unstable coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2723–2732. [Google Scholar] [CrossRef] [PubMed]
- Maeba, R.; Maeda, T.; Kinoshita, M.; Takao, K.; Takenaka, H.; Kusano, J.; Yoshimura, N.; Takeoka, Y.; Yasuda, D.; Okazaki, T.; et al. Plasmalogens in human serum positively correlate with high- density lipoprotein and decrease with aging. J. Atheroscler. Thromb. 2007, 14, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Nishimukai, M.; Maeba, R.; Yamazaki, Y.; Nezu, T.; Sakurai, T.; Takahashi, Y.; Hui, S.P.; Chiba, H.; Okazaki, T.; Hara, H. Serum choline plasmalogens, particularly those with oleic acid in sn-2, are associated with proatherogenic state. J. Lipid Res. 2014, 55, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Maeba, R.; Kojima, K.I.; Nagura, M.; Komori, A.; Nishimukai, M.; Okazaki, T.; Uchida, S. Association of cholesterol efflux capacity with plasmalogen levels of high-density lipoprotein: A cross-sectional study in chronic kidney disease patients. Atherosclerosis 2018, 270, 102–109. [Google Scholar] [CrossRef]
- Xu, F.; Tavintharan, S.; Sum, C.F.; Woon, K.; Lim, S.C.; Ong, C.N. Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry-based metabolomics. J. Clin. Endocrinol. Metab. 2013, 98, E1060–E1065. [Google Scholar] [CrossRef]
- Wu, T.W.; Chan, H.L.; Hung, C.L.; Lu, I.J.; Wang, S.D.; Wang, S.W.; Wu, Y.J.; Wang, L.Y.; Yeh, H.I.; Wei, Y.H.; et al. Differential patterns of effects of age and sex on metabolic syndrome in taiwan: Implication for the inadequate internal consistency of the current criteria. Diabetes Res. Clin. Pract. 2014, 105, 239–244. [Google Scholar] [CrossRef]
| ID | All (n = 486) | Control (n = 57) | LD (n = 49) | Other (n = 380) | p-Value |
|---|---|---|---|---|---|
| Age (years) | 81.2 ± 7.1 | 80.1 ± 7.3 | 79.8 ± 7.7 | 81.6 ± 7.0 | 0.8291 |
| Height (cm) | 157.6 ± 8.7 | 157.1 ± 8.0 | 154.6 ± 7.7 | 158.1 ± 8.8 | 0.1064 |
| Weight (kg) | 57.9 ± 10.8 | 49.8 ± 8.0 | 62.0 ± 11.8 | 58.6 ± 10.4 | <0.001 |
| BMI (kg/m2) | 23.2 ± 3.5 | 20.1 ± 2.4 | 25.8 ± 3.9 | 23.3 ± 3.2 | <0.001 |
| Waist circumference (cm) | 86.8 ± 9.9 | 75.7 ± 5.1 | 91.9 ± 11.7 | 87.8 ± 9.1 | <0.001 |
| SBP (mmHg) | 134.5 ± 20.8 | 115.6 ± 8.6 | 139.4 ± 21.0 | 136.7 ± 20.6 | <0.001 |
| DBP (mmHg) | 72.0 ± 10.0 | 66.1 ± 8.0 | 72.8 ± 13.7 | 72.8 ± 9.4 | 0.0036 |
| Glucose (mg/dL) | 99.0 ± 18.1 | 87.6 ± 6.6 | 110.2 ± 27.6 | 99.2 ± 16.8 | <0.001 |
| Hb-A1c (%) | 6.0 ± 0.7 | 5.6 ± 0.3 | 6.4 ± 0.8 | 6.0 ± 0.7 | <0.001 |
| T-cholesterol (mg/dL) | 184.7 ± 35.4 | 195.4 ± 33.2 | 193.9 ± 40.5 | 181.9 ± 34.5 | 0.8275 |
| Triglyceride (mg/dL) | 102.9 ± 53.5 | 80.3 ± 27.9 | 212.2 ± 57.7 | 92.2 ± 37.3 | <0.001 |
| HDL-C (mg/dL) | 54.1 ± 13.7 | 62.2 ± 12.2 | 40.4 ± 5.4 | 54.7 ± 13.4 | <0.001 |
| LDL-C (mg/dL) | 107.4 ± 29.4 | 112.8 ± 28.6 | 109.8 ± 35.8 | 106.2 ± 28.6 | 0.6296 |
| Albumin (g/dL) | 4.3 ± 0.3 | 4.3 ± 0.2 | 4.5 ± 0.2 | 4.3 ± 0.3 | 0.0001 |
| Total protein (g/dL) | 7.0 ± 0.4 | 6.9 ± 0.4 | 7.2 ± 0.4 | 7.0 ± 0.4 | 0.0005 |
| AST/GOT (U/L) | 27.0 ± 11.4 | 29.1 ± 18.3 | 27.2 ± 10.4 | 26.6 ± 10.2 | 0.5050 |
| ALT/GPT (U/L) | 19.3 ± 13.6 | 18.1 ± 17.0 | 21.3 ± 10.2 | 19.2 ± 13.5 | 0.2447 |
| ALK-P (U/L) | 66.2 ± 21.1 | 63.2 ± 19.0 | 67.3 ± 22.2 | 66.5 ± 21.3 | 0.3043 |
| Total bilirubin (mg/dL) | 0.7 ± 0.3 | 0.7 ± 0.3 | 0.6 ± 0.2 | 0.7 ± 0.3 | 0.0036 |
| BUN (mg/dL) | 17.9 ± 7.4 | 16.2 ± 4.4 | 20.9 ± 10.6 | 17.8 ± 7.2 | 0.0051 |
| Creatinine (mg/dL) | 0.9 ± 0.5 | 0.8 ± 0.4 | 1.1 ± 0.7 | 0.9 ± 0.5 | 0.0473 |
| Uric acid (mg/dL) | 5.7 ± 1.5 | 5.0 ± 1.3 | 6.7 ± 2.0 | 5.7 ± 1.5 | <0.001 |
| Hypertension (%) | 62.6 (304) | 38.6 (22) | 77.6 (38) | 64.2 (244) | <0.001 |
| Hyperlipidemia (%) | 29.8 (145) | 15.8 (9) | 44.9 (22) | 30.0 (114) | 0.0012 |
| Metabolic syndrome (%) | 28.6 (139) | 0 (0) | 95.9 (47) | 24.2 (92) | <0.001 |
| DM (%) | 24.3 (118) | 5.3 (3) | 49.0 (24) | 23.9 (91) | <0.001 |
| CAD (%) | 7.6 (37) | 0 (0) | 10.2 (5) | 8.4 (32) | 0.0237 |
| Stroke (%) | 7.6 (37) | 5.3 (3) | 10.2 (5) | 7.6 (29) | 0.3530 |
| CKD (%) | 9.9 (48) | 3.5 (2) | 22.4 (11) | 9.2 (35) | 0.0049 |
| Variable | Odds Ratio (95% CI) | p-Value |
|---|---|---|
| Age | 1.016 (0.924, 1.118) | 0.7421 |
| Lipid disorder | 29.096 (2.635, 321.309) | 0.0059 |
| PC ae C34:3 | 0.488 (0.228, 1.046) | 0.0651 |
| PC ae C44:3 | <0.001 (<0.001, 0.359) | 0.0453 |
| SM C24:1 | 1.024 (1.001, 1.048) | 0.0439 |
| PC ae C36:3 | 3.112 (1.104, 8.771) | 0.0318 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo, C.-J.; Tang, H.-Y.; Huang, C.-Y.; Lin, C.-M.; Ho, H.-Y.; Shiao, M.-S.; Cheng, M.-L. Metabolic Signature Differentiated Diabetes Mellitus from Lipid Disorder in Elderly Taiwanese. J. Clin. Med. 2019, 8, 13. https://doi.org/10.3390/jcm8010013
Lo C-J, Tang H-Y, Huang C-Y, Lin C-M, Ho H-Y, Shiao M-S, Cheng M-L. Metabolic Signature Differentiated Diabetes Mellitus from Lipid Disorder in Elderly Taiwanese. Journal of Clinical Medicine. 2019; 8(1):13. https://doi.org/10.3390/jcm8010013
Chicago/Turabian StyleLo, Chi-Jen, Hsiang-Yu Tang, Cheng-Yu Huang, Chih-Ming Lin, Hung-Yao Ho, Ming-Shi Shiao, and Mei-Ling Cheng. 2019. "Metabolic Signature Differentiated Diabetes Mellitus from Lipid Disorder in Elderly Taiwanese" Journal of Clinical Medicine 8, no. 1: 13. https://doi.org/10.3390/jcm8010013
APA StyleLo, C.-J., Tang, H.-Y., Huang, C.-Y., Lin, C.-M., Ho, H.-Y., Shiao, M.-S., & Cheng, M.-L. (2019). Metabolic Signature Differentiated Diabetes Mellitus from Lipid Disorder in Elderly Taiwanese. Journal of Clinical Medicine, 8(1), 13. https://doi.org/10.3390/jcm8010013







